Abstract
Signal-regulatory proteins (SIRPs) comprise a novel transmembrane glycoprotein family involved in the negative regulation of receptor tyrosine kinase-coupled signaling pathways. To analyze the expression and function of SIRPs, we prepared soluble recombinant fusion proteins of the extracellular regions of SIRP1 and SIRP2, as well as a variety of monoclonal antibodies (MoAbs) against these domains. The antibodies reacted predominantly with monocytes, granulocytes, dendritic cells, and their precursors, as well as with bone marrow CD34+, AC133+, CD90+hematopoietic stem/progenitor cells. In contrast, SIRP expression was absent or significantly reduced on the majority of myeloid blasts from patients with acute myeloid leukemia (AML) or chronic myeloid leukemia (CML). Functional studies showed that the extracellular domains of SIRP1 and SIRP2 support adhesion of a number of primary hematopoietic cells and cell lines. This interaction could be blocked by 4 of 7 SIRP1-reactive MoAbs. In addition, SIRP1 and SIRP2 competed for the same cell binding site, suggesting a common widely expressed SIRP ligand. In an approach to identify this molecule, MoAbs were generated against the SIRP-binding cell line CCRF-CEM, and MoAb CC2C6 was selected because of its capacity to inhibit cell binding to SIRP1. Further analysis showed that this antibody recognized CD47, a ubiquitously expressed plasma membrane protein previously implicated in integrin function, host defense action, and neutrophil migration. In this study, we identify CD47 as the extracellular ligand for human SIRP and show that these two counterreceptors are involved in cellular adhesion.
PROLIFERATION and differentiation of hematopoietic cells are regulated by growth factors and their transmembrane receptors, many of which are tyrosine kinases (RTKs).1,2 Recently, a new family of signal-regulatory proteins (SIRPs) has been identified as negative regulators for several RTK-coupled signaling pathways.3,4 These molecules are also termed SHPS-1 (src homology 2 domain-containing phosphatase substrate-1), BIT (brain immunoglobulin-like molecule with a tyrosine-based activation motif), P84, and MFR (macrophage fusion receptor). SIRPs are transmembrane glycoproteins consisting of a large extracellular region with 3 immunoglobulin-like domains, a single hydrophobic transmembrane region, and a cytoplasmic tail containing 2 immunoreceptor tyrosine-based inhibitory motifs (ITIMs).3In humans, at least 15 SIRP members have been identified, which can be divided into 2 subgroups, according to the presence (SIRPα) or the absence (SIRPβ) of the ITIM-containing cytoplasmic domain.3 These ITIM regions, which are also present on other inhibitory receptor molecules, including CD22, the newly described killer inhibitory receptors (KIRs), FcγRIIB, and CTLA4,5 can interact with the src homology 2 domain-containing phosphatases SHP-1 and SHP-2, and therefore influence signal transduction cascades.3,4,6 The best characterized member of the human SIRP family, SIRPα1, is a substrate of activated RTKs. Its overexpression leads to a reduced responsiveness to the RTK ligands epidermal growth factor (EGF), insulin, and platelet-derived growth factor (PDGF).3
Although several molecules interacting with the cytoplasmic domain of the SIRPα subfamily members have been identified,3,4,7,9little is known about the cellular distribution, extracellular ligand, and physiologic function of SIRP in human tissues. In rat, mouse, and cattle, SIRP expression was observed in neurons and myeloid cells (macrophages, monocytes, granulocytes, dendritic cells).7-10 Sano et al11-13 could show that BIT, the rat homologue of SIRPα, is a major substrate of tyrosine kinases in vitro, and that cerebral cortical neurons could extend their neurites on BIT-coated substrate. Therefore, they suggested that BIT functions as a membrane signaling molecule in the brain and participates in cell-cell interactions.13 A similar expression and function was also described for the neural brain adhesion molecule P84, the mouse homologue of human SIRPα1.14,15 Further studies in the rat showed a selective SIRP expression in myeloid and neuronal cells and an induction of nitric oxide production by functional SIRP-reactive antibodies in macrophages.10 Moreover, Saginario et al16 showed that rat SIRP is involved in macrophage adhesion and fusion. Finally, Brooke et al8 demonstrated that cattle SIRP promoted the attachment of CD4+ T lymphocytes to myeloid antigen presenting cells and is involved in T-cell activation and proliferation.
In this study, we describe the differential expression and adhesive capacity of human SIRP and its extracellular ligand on hematopoietic cells. For this purpose, recombinant proteins representing the extracellular domains of SIRPα1 and SIRPα2 and SIRP-reactive monoclonal antibodies (MoAbs) were produced. Using these reagents, we show that SIRP is predominantly expressed on myeloid cells and hematopoietic stem/progenitor cells and provide evidence that hematopoietic cells strongly adhere to SIRPα1 and SIRPα2 in a competitive way. Finally, we generated an antibody against the extracellular counterreceptor for SIRPs on these cells and identified CD47, an integrin-associated protein (IAP) with multiple functions in immunological and neuronal processes,17-24 as the SIRP binding partner.
MATERIALS AND METHODS
Cells
Bone marrow (BM) or peripheral blood (PB) cells from healthy donors and patients with acute or chronic myeloid leukemia (AML or CML) were obtained after informed consent according to the guidelines of the local ethics committee in Tübingen. Buffy coat PB cells from normal volunteers were obtained from the Transfusion Department, Tübingen, according to institutional guidelines. Mononuclear cells were isolated on a Ficoll-Hypaque density gradient (1.077 g/mL) by collecting the interphase cells. For immunofluorescence labeling of PB lymphocytes, monocytes, and granulocytes, lysing reagent without fixative from Immunotech (Marseille, France) was used to remove erythrocytes.
Selection of CD34+ cells from 5 to 10 × 107 Ficoll-Hypaque isolated BM cells was performed on a magnetic activated cell sorting (MACS) column according to the instructions of the manufacturer (Miltenyi Biotec, Bergisch Gladbach, Germany). Purity of selected CD34+ cells was 96% to 99% and the recovery was greater than 65%.
The human leukemic cell lines HL60, KG1a, K562, M07e, 207, Daudi, CCRF-CEM, Jurkat, and Molt-4 were obtained from ATCC (American Type Culture Collection, Rockville, MD). The murine myeloma cell line, SP2/0, and the human leukemic cell lines, EM-2, U937, and LAMA-84, were obtained from the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany). The megakaryoblastic cell line, MEG-01, was a gift from Dr H. Saito (Nagoya, Japan),25 the erythroblastic cell line, TF-1, was kindly provided by Dr T. Kitamura (Tokyo, Japan),26 and the B-lymphoblastic cell line, OCI-Ly8, was a gift from Dr I.G.H. Schmidt-Wolf (Berlin, Germany).27
NIH-3T3 cells were transfected with the complete coding sequence of the human SIRPα1 cDNA (NIH-3T3/huSIRPα1) as described previously.3 The ovarian carcinoma cell line, OV10, was transfected with human CD47 (OV10/huCD47) as described.28
All cell lines were grown in RPMI 1640 culture medium (GIBCO-BRL, Eggenstein, Germany) supplemented with 10% fetal calf serum (FCS) and antibiotics. The culture medium for the erythroblastic cell line, TF-1, was supplemented with 10 ng/mL interleukin-3 (IL-3; Genzyme, Cambridge, MA). All cells were cultured at 37°C and 5% CO2.
Dendritic cells were generated in vitro as described.29Briefly, monocytes were isolated from mononuclear PB cells by adhesion to culture dishes for 2 hours at 37°C. After removing nonadherent cells, RPMI 1640 medium supplemented with 10% FCS, antibiotics, IL-4 (1,000 IU/mL) (Genzyme) and granulocyte-macrophage colony-stimulating factor (GM-CSF) (Leukomax, 10 ng/mL) (Novartis, Basel, Switzerland) was added, and dendritic cells were collected after 7 days of culture.
Recombinant SIRPα1 and SIRPα2 Proteins
The construction of the pSj26(mod) cloning vector was designed for the eukaryotic expression and secretion of recombinant SIRPα1 and SIRPα2 fusion proteins and was derived from the pCDNA3 cloning vector (Invitrogen, Groningen, The Netherlands) by the following procedure: the complete DNA sequence coding for Schistosoma japonicum glutathione-S-transferase (GST) was amplified from the vector pGEX 2TK (Pharmacia Biotech, Freiburg, Germany) by polymerase chain reaction (PCR) using the following primers: sense: 5′-CCG CTC GAG CGT CGT GCA TCT GTT GAT GAT GAT GAT AAG ATG TCC CCT ATA CTA-3′; antisense: 5′-GCG GGG CCC TTA TTT TGG AGG ATG GTC G-3′. The PCR product was digested with Xho1 andApa1 and cloned into pCDNA3 at these sites. This vector is now termed pSj26(mod).
The extracellular domain of SIRPα1 or SIRPα2 was amplified from clones previously obtained3 using the following primers: sense: 5′-GCG GAA TTC GCC ACC ATG GAG CCC GCC GGC-3′; antisense: 5′-GCG CTC GAG CCG TTC ATT AGA TCC-3′. The PCR product was digested with EcoR1 and Xho1 and cloned into pSj26(mod) at these sites.
The resultant pSj26(mod)-SIRPα1/2ex expression plasmids were transfected into 293 cells by the calcium phosphate method.30 Cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FCS. After selection with 1 mg/mL G418 (Sigma, Deisenhofen, Germany) for 2 weeks, 10 surviving clones were tested for expression and secretion of fusion protein by Western blot with antibody against the extracellular domain of SIRPα1.3 High-expressing clones were used to produce SIRPα1ex and SIRPα2ex. Medium was collected from confluent cultures daily. One liter of collected medium was sterile filtered and incubated with 1 mL glutathione Sepharose (Pharmacia Biotech) overnight at 4°C. The Sepharose was separated and washed with phosphate-buffered saline (PBS). Elution was performed with 5 mL 10 mmol/L glutathione at 20°C. The eluents were dialyzed 1:106 (vol/vol) in PBS/10% glycerol. Protein concentration was determined using MicroBCA protein determination kit (Pierce, Rockford, IL).
Recombinant SIRPα1ex and SIRPα2ex fusion proteins were biotinylated by adding biotin-7-NHS (Boehringer Mannheim, Mannheim, Germany) in dimethyl sulfoxide (DMSO) to the protein solution in PBS at a molar ratio of 20:1. After 2 hours of incubation, unbound biotin-7-NHS was separated from the biotinylated protein by gel-filtration on a Sephadex G25 column (Pharmacia Biotech).
Immunization and Hybridoma Production
MoAbs SE5A5, SE7C2, SE8A3, SE11A6, SE12B6, and SE12C3 were raised by immunization of 4- to 8-week-old female Balb/c mice with recombinant GST fusion protein containing the whole extracellular domain of SIRPα1. The mice were injected intramuscularly 3 times in 14-day intervals with 50 μg protein diluted 1:2 in ABM-2 adjuvans (PanSystems, Aidenbach, Germany). The spleens were removed 4 days after the last injection for fusion with the SP2/0 myeloma cell line. The resulting hybridomas were grown in RPMI 1640 culture medium containing 10% FCS, antibiotics, and hypoxanthine, aminopterin, and thymidine (HAT) (Sigma). Culture supernatants were screened by flow cytometric analysis on NIH-3T3/huSIRPα1 cells, and positive hybridomas secreting antibodies selectively recognizing the transfectant cell line, but not the parental NIH-3T3 cells, were cloned by limiting dilution. The SIRPα1-reactive clones were cultured in serum-free medium supplemented with 1% Nutridoma (Boehringer Mannheim), and antibodies were purified from supernatants using Protein G Sepharose columns (Pharmacia Biotech). Using the same immunization protocol, MoAb P3C4 was raised by immunization with recombinant GST fusion protein containing only the N-terminal immunoglobulin-like domain of SIRPα1.
MoAb CC2C6 was generated by immunizing a Balb/c mouse with the T-lymphoblastic cell line, CCRF-CEM, 4 times every 2 weeks, using 107 cells for each intraperitoneal injection. The MoAb was selected because it inhibited the binding of soluble SIRPα1ex to CCRF-CEM cells. The specificity of MoAb CC2C6 for CD47 was confirmed by its selective reaction with OV10/huCD47, but not with OV10 cells. The isotypes of the MoAbs were determined by flow cytometry analysis using phycoerythrin (PE)-conjugated isotype-specific secondary antisera for staining (Southern Biotechnology, Birmingham, AL). All isotypes were IgG1 except in the case of MoAb P3C4 (IgG2a).
Immunoprecipitation and Western Blot Analysis
Crude protein extracts of NIH-3T3/huSIRPα1 cells were obtained by solubilizing cellular proteins with lysis buffer (50 mmol/L sodium borate, pH 8.0, 150 mmol/L NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1 mg/mL phenylmethyl sulfonyl fluoride [PMSF], 1 μg/mL aprotinin, 1 μg/mL leupeptin) for 60 minutes on ice. After preclearing the protein extracts with 50 μL Protein A Sepharose (Sigma), 5 μg of SIRPα1-reactive MoAbs were incubated with the supernatant for 1 hour at 4°C. As a negative control, a nonreacting MoAb of the same isotype was used. Immunoprecipitation was performed overnight at 4°C using 100 μL Protein A Sepharose solution for each MoAb. The antibody-Sepharose complexes were washed 6 times with Tris-buffered saline (TBS: 10 mmol/L TrisHCl pH 7.5, 100 mmol/L NaCl), and bound proteins were eluted with reducing Laemmli sample buffer.31After boiling the proteins for 5 minutes, they were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Electrophoresed proteins were transferred to nitrocellulose, and the membranes were blocked with 3% bovine serum albumin (BSA) in TTBS (TBS + 0.1% Tween 20). A polyclonal antiserum against human SIRPα13 was used as primary antibody (1:5,000 in TTBS + 0.1% BSA). After incubation for 1 hour at room temperature (RT), the membranes were washed with TTBS and then incubated with alkaline phosphatase-conjugated goat anti-rabbit antiserum (1:1,000 in TTBS + 0.1% BSA) for 1 hour at RT. After washing the membranes, bound antibodies were detected using BCIB/NBT (5-bromo-4-chloro-3-indolylphosphate p-toluidine salt/nitro blue tetrazolium chloride) Sigma Fast tablets (Sigma). Immunoprecipitations with recombinant SIRPα2ex fusion protein were performed as described above, using 1 μg of SIRPα2ex solution instead of protein extracts.
Immunofluorescence Labeling and Flow Cytometry Analysis
Indirect staining of cells.
Cells from growing cell lines or primary mononuclear cells from BM and PB were washed in fluorescence-activated cell sorting (FACS) buffer (PBS supplemented with 0.1% BSA and 0.1% sodium azide) before incubation with 20% human AB serum for 10 minutes at 4°C to prevent unspecific binding of mouse antibodies. Cells were then incubated with 10 μg/mL of the primary antibody for 30 minutes on ice. After washing 2 times with FACS buffer, cells were stained with PE-conjugated goat anti-mouse IgG1 or IgG2a antiserum (Southern Biotechnology) for 30 minutes at 4°C. After washing twice, cells were suspended in FACS buffer and analyzed on a FACSCalibur flow cytometer (Becton Dickinson, Heidelberg, Germany). To analyze the expression of the SIRP ligand, hematopoietic cells were incubated with 100 μg/mL biotinylated recombinant SIRPα1ex or SIRPα2ex fusion protein for 30 minutes on ice. After 2 washing steps, the cells were labeled with streptavidin (SA)-PE (DAKO, Hamburg, Germany).
Two-color staining of BM and PB cells.
Mononuclear BM cells were labeled with MoAb P3C4 (IgG2a) and fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG2a-specific antiserum (Caltag, San Francisco, CA) as described above. In addition, PE-conjugated antibodies against CD3 (SK7), CD19 (4G7), CD33 (P67.6), CD34 (8G12), CD56 (My31) (Becton Dickinson), CD117 (104D2),32 and glycophorin A (11E4B7.6) (Immunotech), as well as the MoAb AC133-PE (Miltenyi Biotec) were used for fluorescence labeling of the cells. Dendritic cells were double-stained with MoAb P3C4 plus PE-conjugated goat anti-mouse IgG2a-specific antiserum, and CD83-FITC (HB15A) (Immunotech). The stained cells were analyzed on a FACSCalibur flow cytometer using the Cellquest software (Becton Dickinson).
Three-color staining of CD34+ BM cells.
CD34+ cells were selected by MACS and labeled with anti-CD34–PerCP (8G12) (Becton Dickinson), the SIRP-reactive MoAb P3C4 (IgG2a), and the PE-conjugated antibodies against CD19 (4G7) (Becton Dickinson), CD71 (RVS-10) (Cymbus Biotechnology, Hants, UK), CD90 (5E10) (Pharmingen, San Diego, CA), CD117 (104D2),32 or the nonconjugated CD164-specific antibody 103B2 (IgG3).33 34After washing, the cells were incubated with the F(ab)′2 fragments of an FITC-conjugated IgG2a-specific goat anti-mouse antiserum (Caltag) and a PE-conjugated IgG3-specific goat anti-mouse antiserum (Medac, Hamburg, Germany) to stain MoAb P3C4 and MoAb 103B2. After washing, 25,000 cells of each probe were analyzed on a FACSCalibur flow cytometer using the Cellquest software.
Competitive binding experiments.
In a first step, leukemic cell lines or primary PB cells were preincubated either with 100 μg/mL recombinant SIRPα1ex or SIRPα2ex protein or with 10 μg/mL of anti-CD47 antibodies BRIC126,35 1/1A4,35 2D3,36 or CC2C6 (described in this report) for 30 minutes on ice. The cells were then stained with CD47-FITC (B6H12) (Pharmingen) or indirectly with anti-CD47 antibodies and PE-conjugated goat anti-mouse IgG1 or IgG2b antisera as described above (Southern Biotechnology). Alternatively, cells were labeled with 100 μg/mL biotinylated SIRPα1ex or SIRPα2ex protein and stained with SA-PE. After washing twice, cells were analyzed on a flow cytometer.
Cell Adhesion Assay
Adhesion of leukemic cell lines and primary hematopoietic cells to SIRPα1ex and SIRPα2ex was performed as described previously.37 Briefly, various dilutions of the SIRPα1ex and SIRPα2ex fusion protein or of GST protein alone were immobilized onto nitrocellulose-coated plastic dishes (35-mm diameter) by air-drying at RT. Nonspecific binding of cells to nitrocellulose was prevented by blocking with 1% BSA solution in PBS. A total of 3 × 106 hematopoietic cells in serum-free medium was allowed to adhere to the immobilized protein for 1 hour at 37°C. Nonadherent cells were removed by gently rinsing the dishes with warm PBS. Specific cell binding was evaluated under a Zeiss Axiovert microscope (Carl Zeiss, Göttingen, Germany). Photographs of representative fields were taken.
To inhibit cell adhesion, immobilized SIRPα1ex protein was preincubated with the different SIRPα1-reactive MoAbs (described in this report) for 30 minutes at 37°C before cell adherence in the presence of the antibodies. Alternatively, leukemic cells were preincubated with CD47-specific MoAbs BRIC126,351/1A4,35 2D3,36 or CC2C6 for 30 minutes at RT before incubation with immobilized SIRPα1ex and SIRPα2ex protein.
RESULTS
Generation of Recombinant SIRP Proteins and SIRP-Reactive MoAbs
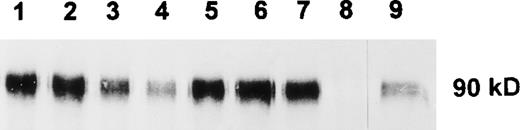
To analyze the function and cell surface expression of human SIRP molecules, recombinant GST fusion proteins containing the extracellular domains of SIRPα1 (SIRPα1ex) and SIRPα2 (SIRPα2ex) were prepared. A panel of MoAbs was raised by immunization of Balb/c mice either with the recombinant construct containing the complete extracellular or the N-terminal immunoglobulin-like domain of SIRPα1. FACS analysis showed that 6 MoAbs against the entire extracellular domain of SIRPα1 (SE5A5, SE7C2, SE8A3, SE11A6, SE12B6, and SE12C3) and 1 MoAb (P3C4) against the first immunoglobulin-like domain specifically reacted with the NIH-3T3/huSIRPα1 transfectant cells,3 but not with parental NIH-3T3 cells (data not shown). Immunoprecipitation of NIH-3T3/huSIRPα1 proteins with the 7 MoAbs followed by Western blot analysis with a polyclonal antibody against the N-terminal immunoglobulin-like domain of SIRPα13 showed that all MoAbs were able to precipitate SIRPα1 as a 90-kD band (Fig 1), which corresponds well with published data.3
Immunoprecipitation of SIRP1 and Western blotting. SIRP1 was immunoprecipitated from cellular extracts of NIH-3T3/huSIRP1 cells with MoAbs SE5A5 (lane 1), SE7C2 (lane 2), SE8A3 (lane 3), SE11A6 (lane 4), SE12B6 (lane 5), SE12C3 (lane 6), and P3C4 (lane 7). A nonbinding MoAb was used as a negative control (lane 8). Precipitated protein was separated by 12% SDS-PAGE and immunoblotted with a polyclonal antibody against SIRP1.3 NIH-3T3/ huSIRP1 extract was used as a positive control for the Western blot (lane 9).
Immunoprecipitation of SIRP1 and Western blotting. SIRP1 was immunoprecipitated from cellular extracts of NIH-3T3/huSIRP1 cells with MoAbs SE5A5 (lane 1), SE7C2 (lane 2), SE8A3 (lane 3), SE11A6 (lane 4), SE12B6 (lane 5), SE12C3 (lane 6), and P3C4 (lane 7). A nonbinding MoAb was used as a negative control (lane 8). Precipitated protein was separated by 12% SDS-PAGE and immunoblotted with a polyclonal antibody against SIRP1.3 NIH-3T3/ huSIRP1 extract was used as a positive control for the Western blot (lane 9).
The SIRPα1-reactive MoAbs were also tested for potential cross-reactivity with SIRPα2. Immunoprecipitation of recombinant SIRPα2ex protein with the MoAbs showed that MoAbs SE5A5, SE8A3, SE12B6, SE12C3, and P3C4 cross-react with SIRPα2, whereas MoAbs SE7C2 and SE11A6 do not detect SIRPα2, indicating a superior selectivity of these reagents (data not shown). Because other members of the SIRP family have very similar extracellular domains, it is likely that SIRPα1-reactive MoAbs also cross-react with other SIRP members. Therefore, our MoAbs raised against SIRPα1 are referred to as specific for SIRP throughout this report.
SIRP Is Differentially Expressed on PB and BM Mononuclear Cells
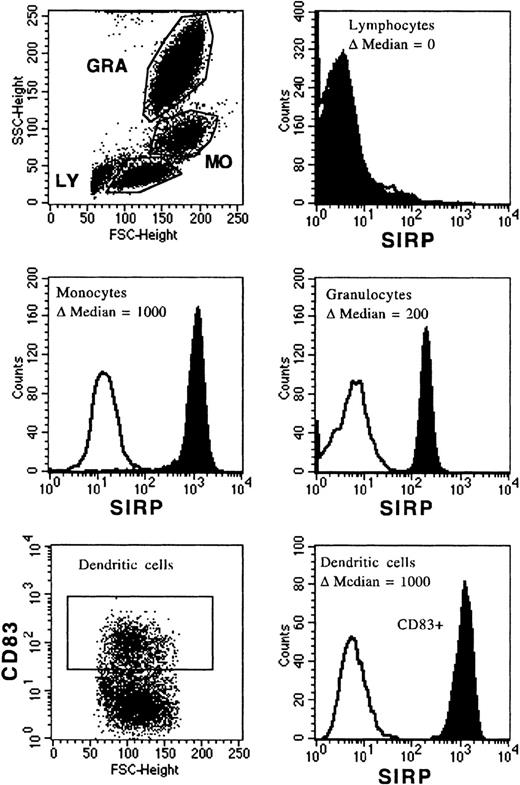
The MoAb, P3C4, was used to analyze SIRP surface expression on PB and BM mononuclear cells. Figure 2 shows a broad distribution of SIRP on mature PB cells, with the strongest expression on monocytes (Δmedian fluorescence of 1,000 ± 50), an intermediate expression on granulocytes (Δmedian fluorescence of 200 ± 50), and almost no expression on lymphocytes (Δmedian fluorescence of 0 to 3) as assessed by flow cytometry. CD83+ dendritic cells, derived from monocytes cultured with IL-4 and GM-CSF, were also strongly positive for SIRP (Δmedian fluorescence of 1,000 ± 50).
Expression of SIRP on PB and dendritic cells. Mononuclear PB cells were immunolabeled with MoAb P3C4 (IgG2a) and PE-conjugated goat anti-mouse IgG2a-specific antiserum (filled histogram). Nonbinding IgG2a antibody was used as negative control (black line). Cells were analyzed on a FACSCalibur flow cytometer and gated on lymphocytes (LY), monocytes (MO), or granulocytes (GRA); respectively (n = 6). In vitro generated dendritic cells derived from monocytes were labeled with MoAb P3C4 and stained with PE-conjugated goat anti-mouse IgG2a-specific antiserum and anti-CD83–FITC (IgG1). The histogram shows SIRP expression gated on CD83+ dendritic cells (n = 3).
Expression of SIRP on PB and dendritic cells. Mononuclear PB cells were immunolabeled with MoAb P3C4 (IgG2a) and PE-conjugated goat anti-mouse IgG2a-specific antiserum (filled histogram). Nonbinding IgG2a antibody was used as negative control (black line). Cells were analyzed on a FACSCalibur flow cytometer and gated on lymphocytes (LY), monocytes (MO), or granulocytes (GRA); respectively (n = 6). In vitro generated dendritic cells derived from monocytes were labeled with MoAb P3C4 and stained with PE-conjugated goat anti-mouse IgG2a-specific antiserum and anti-CD83–FITC (IgG1). The histogram shows SIRP expression gated on CD83+ dendritic cells (n = 3).
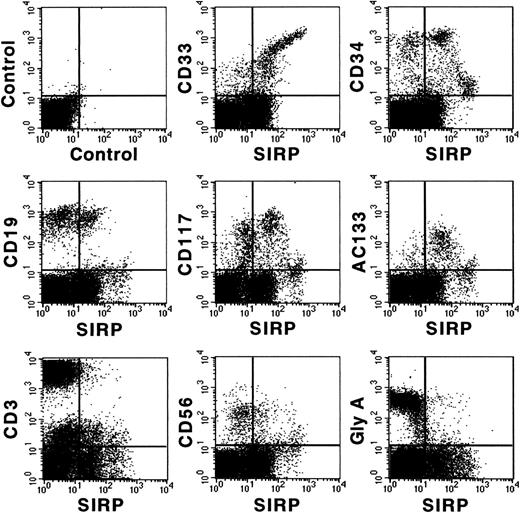
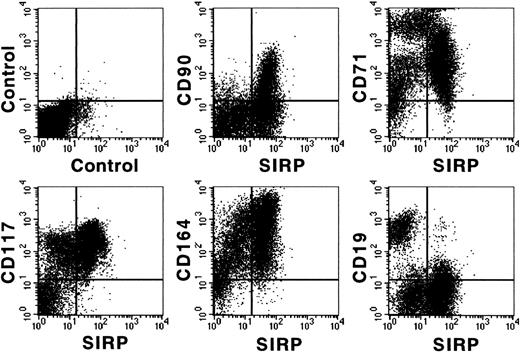
Figure 3 shows coexpression of SIRP and selected CD molecules on mononuclear BM subsets. The strongest expression of SIRP was detected on CD33+ and CD34low myelo/monocytic precursors. In addition, SIRP is also expressed on CD19+ B lymphoid, as well as on immature CD117+ and AC133+ progenitor subsets. In contrast, no detectable SIRP expression was found on CD3+ T cells, CD56+ natural killer cells, or glycophorin A+ erythroid cells. Hence, SIRP expression in BM is mainly found on myeloid and CD34+ stem and progenitor cells. To further analyze SIRP+ subsets within the stem/progenitor compartment, CD34+ BM cells were purified by MACS and stained with antibodies against CD34, SIRP, and selected CD markers. Figure 4 shows that SIRP is expressed on CD34+CD90+, CD34+CD117+, and CD34+CD164bright stem cell and myeloid subsets, but not on CD34+CD19+ B-cell subsets or CD34+CD71bright erythroid progenitor cells, suggesting that SIRP may also play a role in the regulation of stem cell growth and differentiation.
Coexpression of CD antigens and SIRP on BM subsets. Mononuclear BM cells were analyzed by 2-color immunofluorescence. Cells were labeled with MoAb P3C4 (IgG2a) and stained with FITC-conjugated goat anti-mouse IgG2a-specific antiserum, as well as with PE-conjugated MoAbs against CD33, CD34, CD19, CD117, CD3, CD56, glycophorin A (Gly A), and AC133 antigen. Gated mononuclear cells were analyzed on a FACSCalibur flow cytometer (n = 6).
Coexpression of CD antigens and SIRP on BM subsets. Mononuclear BM cells were analyzed by 2-color immunofluorescence. Cells were labeled with MoAb P3C4 (IgG2a) and stained with FITC-conjugated goat anti-mouse IgG2a-specific antiserum, as well as with PE-conjugated MoAbs against CD33, CD34, CD19, CD117, CD3, CD56, glycophorin A (Gly A), and AC133 antigen. Gated mononuclear cells were analyzed on a FACSCalibur flow cytometer (n = 6).
Coexpression of CD antigens and SIRP on CD34+ BM cells. CD34+ cells were isolated from BM aspirates of healthy donors using MACS separation columns and analyzed by 3-color immunofluorescence. Cells were labeled with CD34-PerCP and MoAb P3C4 (IgG2a), as well as with PE-conjugated MoAbs against CD19, CD71, CD90, CD117, or the nonconjugated CD164-specific MoAb 103B2 (IgG3). After washing, the cells were stained with FITC-conjugated goat anti-mouse IgG2a-specific antiserum and with PE-conjugated IgG3-specific goat anti-mouse antiserum. Immunolabeled cells were analyzed on a FACSCalibur flow cytometer (n = 3).
Coexpression of CD antigens and SIRP on CD34+ BM cells. CD34+ cells were isolated from BM aspirates of healthy donors using MACS separation columns and analyzed by 3-color immunofluorescence. Cells were labeled with CD34-PerCP and MoAb P3C4 (IgG2a), as well as with PE-conjugated MoAbs against CD19, CD71, CD90, CD117, or the nonconjugated CD164-specific MoAb 103B2 (IgG3). After washing, the cells were stained with FITC-conjugated goat anti-mouse IgG2a-specific antiserum and with PE-conjugated IgG3-specific goat anti-mouse antiserum. Immunolabeled cells were analyzed on a FACSCalibur flow cytometer (n = 3).
SIRP Expression Is Reduced on Primary Leukemic Blasts
In the next step, we analyzed SIRP expression on AML and CML blasts. In contrast to normal myeloid cells, all of the 4 analyzed myeloid CML blast samples and 26 of 59 of the tested AML blasts (44%) were SIRP−, whereas 15 of 59 of the AML blasts (25%) expressed low levels of SIRP and only 18 of 59 blasts (31%) expressed SIRP to a similar level as normal myeloid or monocytic cells in the BM. Notably, 17 of these 18 samples were of the M4/M5 monocytic subtype according to the French-American-British (FAB) classification. All immature leukemic blasts of M0 or M1 FAB types are SIRP− or express SIRP at reduced levels. Representative examples of these groups are shown in Fig 5. These data suggest an aberrant regulation of SIRP expression on many leukemic blasts.
Reduced SIRP expression on leukemic blasts. Mononuclear BM or PB cells from AML and CML patients were immunolabeled with CD45-FITC and MoAb P3C4 (IgG2a), followed by PE-conjugated goat anti-mouse IgG2a-specific antiserum (filled histogram). Nonbinding IgG2a antibody was used as negative control (black line). The histograms show representative examples of SIRP expression gated on CD45low leukemic blasts. AML samples were divided into 3 groups: no expression (▵median of 0 to 5), reduced expression (▵median of 10 to 150), and normal expression of SIRP (▵median over 150). The number of analyzed samples is given in brackets.
Reduced SIRP expression on leukemic blasts. Mononuclear BM or PB cells from AML and CML patients were immunolabeled with CD45-FITC and MoAb P3C4 (IgG2a), followed by PE-conjugated goat anti-mouse IgG2a-specific antiserum (filled histogram). Nonbinding IgG2a antibody was used as negative control (black line). The histograms show representative examples of SIRP expression gated on CD45low leukemic blasts. AML samples were divided into 3 groups: no expression (▵median of 0 to 5), reduced expression (▵median of 10 to 150), and normal expression of SIRP (▵median over 150). The number of analyzed samples is given in brackets.
Hematopoietic Cells Adhere to Recombinant SIRPα1ex and SIRPα2ex
Because SIRP molecules consist of a large extracellular region with 3 immunoglobulin-like domains, it was likely that SIRPs interact with an extracellular ligand. To analyze this potential interaction of SIRP with cellular components on normal and malignant hematopoietic cells, attachment assays with immobilized SIRPα1ex and SIRPα2ex fusion proteins were performed. All tested myelo/monoblastic, erythroblastic, megakaryoblastic, B- and T-lymphoblastic cell lines, as well as primary PB cells, strongly adhered to SIRPα1ex and SIRPα2ex at a concentration of 1 mg/mL, but not to GST alone (data not shown), as evaluated by microscopy. Serial dilutions of the fusion proteins resulted in detectable adhesion of leukemic cells down to 20 μg/mL SIRPex protein, compared with background binding of cells. Interestingly, B- and T-lymphoblastic cell lines adhered at SIRPα1ex and SIRPα2ex dilutions down to 2 μg/mL, indicating that these cells have a higher number of binding sites for SIRP.
Cell Adhesion to SIRPα1ex and SIRPα2ex Is Inhibited by SIRP-Reactive MoAbs
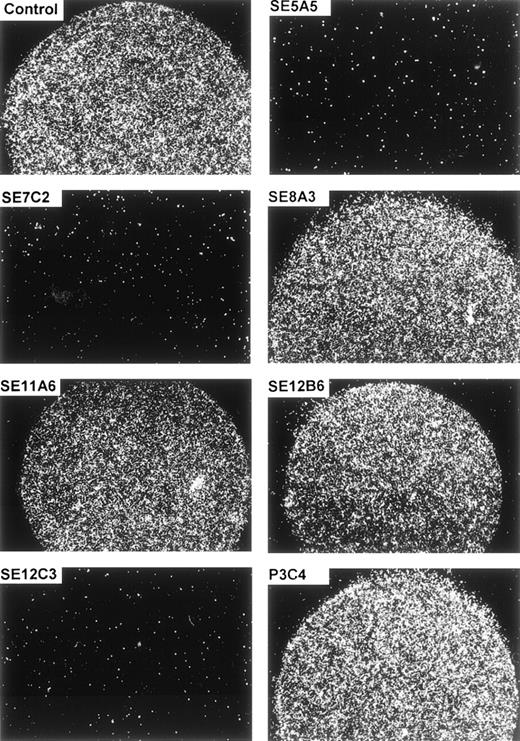
The specificity of the cellular adhesion to SIRPα1ex and SIRPα2ex could be further demonstrated by inhibition studies. Figure 6 and Table 1 show that preincubation of the immobilized protein with MoAbs SE5A5, SE7C2, and SE12C3 completely inhibited the cell binding to SIRPα1ex, whereas MoAbs SE8A3, SE11A6, SE12B6, and P3C4, as well as a nonbinding control antibody, did not interfere with the cell adherence to SIRPα1ex. The adhesion-blocking MoAbs, SE5A5 and SE12C3, also prevented cell binding to immobilized SIRPα2ex, whereas MoAb, SE7C2, with superior specificity for SIRPα1 did not inhibit cell attachment to SIRPα2ex (Table 1). Interestingly, MoAb SE12B6, which did not interfere with cell adhesion to SIRPα1ex, caused a complete inhibition of cell binding to SIRPα2ex. Together, these data show that normal and malignant hematopoietic cells strongly adhere to SIRPα1 and SIRPα2, and that this adhesion can be blocked by some of the SIRP-reactive MoAbs.
Inhibition of cell adhesion to SIRP1ex by SIRP-reactive MoAbs. Immobilized SIRP1ex (2 μL of 20 μg/mL solution) was preincubated for 30 minutes at 37°C with SIRP1-reactive MoAbs SE5A5, SE7C2, SE8A3, SE11A6, SE12B6, SE12C3, and P3C4 or a nonbinding control antibody. Adhesion of Jurkat cells to SIRP1ex was analyzed by incubating the cells for 1 hour in the presence of the antibodies. Specific cell binding is shown in the round protein drop and was evaluated and photographed using a Zeiss Axiovert microscope (original magnification × 40) (n = 3).
Inhibition of cell adhesion to SIRP1ex by SIRP-reactive MoAbs. Immobilized SIRP1ex (2 μL of 20 μg/mL solution) was preincubated for 30 minutes at 37°C with SIRP1-reactive MoAbs SE5A5, SE7C2, SE8A3, SE11A6, SE12B6, SE12C3, and P3C4 or a nonbinding control antibody. Adhesion of Jurkat cells to SIRP1ex was analyzed by incubating the cells for 1 hour in the presence of the antibodies. Specific cell binding is shown in the round protein drop and was evaluated and photographed using a Zeiss Axiovert microscope (original magnification × 40) (n = 3).
Inhibition of Cell Adhesion to Immobilized SIRP 1ex and SIRP2ex by SIRP-Reactive MoAbs
| SIRP-Reactive MoAb . | Inhibition of Cell Adhesion to . | |
|---|---|---|
| SIRPα1ex . | SIRPα2ex . | |
| SE5A5 | + | + |
| SE7C2 | + | − |
| SE8A3 | − | − |
| SE11A6 | − | − |
| SE12B6 | − | + |
| SE12C3 | + | + |
| P3C4 | − | − |
| SIRP-Reactive MoAb . | Inhibition of Cell Adhesion to . | |
|---|---|---|
| SIRPα1ex . | SIRPα2ex . | |
| SE5A5 | + | + |
| SE7C2 | + | − |
| SE8A3 | − | − |
| SE11A6 | − | − |
| SE12B6 | − | + |
| SE12C3 | + | + |
| P3C4 | − | − |
Cell adhesion assays on immobilized SIRPα1ex and SIRPα2ex were performed as described in Fig 6.
Differential Expression of Extracellular SIRPα1 and SIRPα2 Ligand(s)
To further study the expression of putative SIRP ligand(s), mononuclear PB and BM cells were labeled with biotinylated SIRPα1ex and SIRPα2ex proteins, stained with SA-PE and analyzed by flow cytometry. As a control, biotinylated GST protein was used. FACS histograms of PB cells gated on lymphocytes (low side scatter, SSC), monocytes (intermediate SSC), or granulocytes (high SSC), respectively, show that all PB subsets are stained with biotinylated SIRPα1ex (Fig 7). The strongest expression of the recognized molecule was found on lymphocytes, whereas monocytes expressed the SIRP ligand at intermediate levels, and granulocytes were only weakly positive. Similar results were obtained when PB cells were labeled with SIRPα2ex plus SA-PE (data not shown), suggesting an equal distribution or identity of SIRPα1 and SIRPα2 ligands. Labeling of BM cells with biotinylated SIRPα1ex and SIRPα2ex showed that all cell subsets, including early hematopoietic stem and progenitor cells (CD34+, CD117+, and AC133+) express an extracellular ligand for SIRP (data not shown).
Binding of biotinylated SIRP1ex to PB cells. Mononuclear PB cells were incubated for 30 minutes with biotinylated SIRP1ex protein and labeled with SA-PE. Histograms show binding of biotinylated SIRP1ex to gated lymphocytes (LY), monocytes (MO), or granulocytes (GRA), analyzed on a FACSCalibur flow cytometer. Results are presented as the ▵ median fluorescence intensity of the biotinylated SIRP1ex signal (filled histogram) versus the control signal (black line), where biotinylated GST was used (n = 6).
Binding of biotinylated SIRP1ex to PB cells. Mononuclear PB cells were incubated for 30 minutes with biotinylated SIRP1ex protein and labeled with SA-PE. Histograms show binding of biotinylated SIRP1ex to gated lymphocytes (LY), monocytes (MO), or granulocytes (GRA), analyzed on a FACSCalibur flow cytometer. Results are presented as the ▵ median fluorescence intensity of the biotinylated SIRP1ex signal (filled histogram) versus the control signal (black line), where biotinylated GST was used (n = 6).
To analyze whether SIRPα1ex and SIRPα2ex bind to the same ligand, the T-lymphoblastic cell line, CCRF-CEM, was preincubated with nonbiotinylated SIRPα1ex (SIRPα2ex) before labeling with biotinylated SIRPα2ex (SIRPα1ex) and staining with SA-PE. SIRPα1ex and SIRPα2ex completely inhibited the binding of SIRPα2ex-biotin and SIRPα1ex-biotin, respectively, indicating the presence of a common ligand for these 2 SIRP family members (data not shown).
MoAb CC2C6 Recognizes an Extracellular SIRP Ligand and Inhibits Cell Adhesion to SIRPα1ex and SIRPα2ex
To identify the extracellular ligand for SIRPα1 and SIRPα2 on hematopoietic cells, the SIRP-negative, but strongly SIRPα1ex- and SIRPα2ex-binding CCRF-CEM cells were used to raise MoAbs that inhibit the binding of SIRPα1ex and SIRPα2ex to these cells. Figure 8A shows that 1 MoAb, CC2C6, completely blocked this binding. Figure 8B shows that this antibody also inhibited CCRF-CEM cell adhesion to immobilized SIRPα1ex and SIRPα2ex protein, as assessed by attachment assays. These results suggest that MoAb CC2C6 binds to an extracellular SIRP ligand and show that MoAb CC2C6 inhibits the interaction of SIRPα1ex and SIRPα2ex with this ligand.
Inhibition of SIRP1ex binding by CD47-specific MoAbs CC2C6 and BRIC126. (A) CCRF-CEM cells were preincubated with CD47-specific MoAbs CC2C6, and BRIC126 (filled histogram), or a nonbinding control antibody (black line) and then labeled with biotinylated SIRP1ex and stained with SA-PE. Inhibition of SIRP1ex binding to CCRF-CEM was analyzed by flow cytometry (n = 3). (B) CCRF-CEM cells were preincubated with CD47-specific MoAbs CC2C6, BRIC126, 2D3, or a nonbinding control antibody. Adhesion of pretreated CCRF-CEM cells to immobilized SIRP1ex (2 μL of 20 μg/mL solution) was analyzed by microscopy (original magnification × 40) (n = 3).
Inhibition of SIRP1ex binding by CD47-specific MoAbs CC2C6 and BRIC126. (A) CCRF-CEM cells were preincubated with CD47-specific MoAbs CC2C6, and BRIC126 (filled histogram), or a nonbinding control antibody (black line) and then labeled with biotinylated SIRP1ex and stained with SA-PE. Inhibition of SIRP1ex binding to CCRF-CEM was analyzed by flow cytometry (n = 3). (B) CCRF-CEM cells were preincubated with CD47-specific MoAbs CC2C6, BRIC126, 2D3, or a nonbinding control antibody. Adhesion of pretreated CCRF-CEM cells to immobilized SIRP1ex (2 μL of 20 μg/mL solution) was analyzed by microscopy (original magnification × 40) (n = 3).
MoAb CC2C6 Identifies CD47 as the Extracellular SIRP Ligand
To exclude or confirm a potential specificity of MoAb CC2C6 for CD molecules, the cellular reactivities of this antibody and more than 400 antibodies against 166 different CD molecules (obtained from the Sixth International Human Leucocyte Differentiation Antigen workshop in Kobe, Japan) were compared. Interestingly, the reactivity patterns of MoAb CC2C6 and the CD47-specific MoAbs BRIC126 and 1/1A4 showed a striking similarity on all tested cell lines (not shown). Moreover, the CD47-specific workshop MoAbs and the commercially available CD47-specific MoAb B6H12 blocked the binding of MoAb CC2C6, as well as biotinylated SIRPα1ex and SIRPα2ex to CCRF-CEM cells, and vice versa. In cell attachment assays, the CD47-specific MoAbs BRIC126 and 1/1A4 inhibited the adhesion of CCRF-CEM cells to SIRPα1ex and SIRPα2ex to a similar extent as MoAb CC2C6 (Fig 8B). In contrast, a nonblocking CD47-specific MoAb, 2D3, had no effect on binding of CCRF-CEM cells to SIRPα1ex (Fig 8B). These data prompted us to analyze the reactivity of MoAb CC2C6 with the ovarian carcinoma cell line, OV10, transfected with the complete coding sequence for CD47 (OV10/huCD47). As expected, MoAb CC2C6 selectively binds to OV10/huCD47, but not to OV10 cells (Fig9A). Moreover, Fig 9B shows the same differential reactivity of MoAb CC2C6 with PB leukocytes as observed with biotinylated SIRPα1ex and SIRPα2ex (Fig 7), although staining with CC2C6 was stronger than with soluble recombinant SIRPs, indicating that nonsaturating concentrations of these proteins were used for labeling. Hence, MoAb CC2C6 recognizes CD47, the extracellular ligand for SIRPα1 and SIRPα2.
Reactivity of MoAb CC2C6 with OV10/huCD47 and PB cells. OV10/huCD47 and wild-type OV10 cells (A), as well as mononuclear PB cells (B) were immunolabeled with MoAb CC2C6 or with a nonbinding control antibody, stained with PE-conjugated goat anti-mouse IgG1-specific antiserum, and analyzed by flow cytometry. The reactivity of MoAb CC2C6 on PB cells was analyzed on gated lymphocytes (LY), monocytes (MO), or granulocytes (GRA). The results are presented as the ▵ median fluorescence intensity of the CC2C6 signal (filled histogram) versus the control signal (black line) (n = 3).
Reactivity of MoAb CC2C6 with OV10/huCD47 and PB cells. OV10/huCD47 and wild-type OV10 cells (A), as well as mononuclear PB cells (B) were immunolabeled with MoAb CC2C6 or with a nonbinding control antibody, stained with PE-conjugated goat anti-mouse IgG1-specific antiserum, and analyzed by flow cytometry. The reactivity of MoAb CC2C6 on PB cells was analyzed on gated lymphocytes (LY), monocytes (MO), or granulocytes (GRA). The results are presented as the ▵ median fluorescence intensity of the CC2C6 signal (filled histogram) versus the control signal (black line) (n = 3).
DISCUSSION
To explore the function and tissue distribution of human SIRP and its extracellular ligand, recombinant proteins containing the extracellular domains of SIRPα1 (SIRPα1ex) and SIRPα2 (SIRPα2ex), as well as SIRP-reactive MoAbs, were generated. The analyses with these probes showed a predominant expression of SIRP on normal mature myeloid cells and CD34+ hematopoietic stem/progenitor cells and suggested the presence of a ubiquitously expressed counterreceptor for SIRP on hematopoietic cells. The interaction of SIRP with its extracellular ligand mediates adhesion of these cells, which can be blocked by some SIRP-reactive MoAbs. To further identify the cognate ligand, we raised cell surface-specific MoAbs and selected 1 antibody, CC2C6, which inhibits the interaction of soluble SIRPα1ex and SIRPα2ex with the SIRP-negative cell line CCRF-CEM. By comparing the reactivity profiles of MoAbs submitted to the Sixth Human Leukocyte Differentiation Antigen Workshop with the profile of the ligand-specific MoAb CC2C6, we tentatively assigned this MoAb as specific for CD47 and confirmed this specificity by its selective recognition of OV10/huCD47 cells.28
SIRP family members, including the cytoplasmic domain-deficient SIRPβ isoforms, are highly homologous in their extracellular domains. SIRPα1 and SIRPα2, for example, exhibit sequence divergence only in the first immunoglobulin-like domain.3 Therefore, it was likely that some of our SIRPα1-reactive MoAbs also cross-react with other SIRP isoforms. In fact, immunoprecipitation experiments showed that 5 of the 7 SIRPα1-reactive MoAbs cross-reacted with SIRPα2, whereas the remaining 2 antibodies showed a superior specificity for SIRPα1. Interestingly, some cell types, including primary lymphocytes, showed differential immunostaining patterns with the 7 SIRPα1-reactive MoAbs from negative to weakly positive. This indicates that SIRP family members are differentially expressed on cells and that most of the SIRPα1-reactive MoAbs cross-react with other SIRPs. Because the cross-reactivity profiles of our MoAbs with all SIRP family members is not yet known, our antibodies were assigned as specific for SIRP.
Previous studies with rat, mouse, and cattle cells demonstrated expression of SIRP molecules mainly on neuronal and myeloid cells.7-10 This corresponds well with the strong SIRP expression we observed on the surface of human monocytes, granulocytes, and dendritic cells. Further analyses showed that SIRP is also expressed on the surface of BM CD33+ and CD34+myeloid progenitor cells, a subset of CD19+ B lymphocytes, and on early CD34+, CD117+, and CD90+ hematopoietic stem/progenitor cells. This suggests that SIRP is not only involved in the function of myeloid cells, but may also play an important role in the regulation of stem cell differentiation.
Interestingly, SIRP expression is significantly reduced on myeloid cells from different leukemias. All tested myeloid CML blasts and almost half of the analyzed AML samples were negative for SIRP. In addition, the blasts of 15 of 59 AML patients expressed reduced SIRP levels, and only 18 of 59 AML samples consisting of myeloid/monocytic blasts expressed SIRP at apparently normal levels. In this context, it is of interest that overexpression of SIRPα1 in NIH-3T3 cells leads to a reduced proliferation and responsiveness to growth factors like epidermal growth factor (EGF), insulin, and platelet-derived growth factor (PDGF).3Therefore, it is possible that the reduced SIRP expression in most leukemic blasts is either the cause or a consequence of aberrant proliferation of these cells. Further studies are in progress to determine whether defective SIRP genes are a reason for deregulated proliferation or whether overexpression of RTKs results in reduced SIRP functions, which ultimately may lead to malignant cell growth.
The presence of 3 immunoglobulin-like loops within the extracellular domain of SIRP3 suggested that this molecule interacts either with a soluble ligand or with a membrane molecule on other cells, and recent studies have shown that SIRP is indeed involved in cell-cell interactions. Brooke et al8 demonstrated that the interaction of cattle SIRP on monocytes with an unknown molecule on CD4+ T cells is important for T-cell activation and proliferation, and Sano et al13 showed that rat SIRP is involved in neuronal contacts, as well as in the outgrowth of rat neurons. Therefore, the existence of an extracellular ligand for human SIRP was very likely. In support of this hypothesis, we found that at least 2 of the 15 known human SIRP members, SIRPα1 and SIRPα2, interact with an extracellular ligand on leukemic cell lines and normal hematopoietic cells and mediate adhesion of these cells. The specificity of this interaction could be confirmed by the complete inhibition of cell adhesion to immobilized SIRPα1ex by 3 of 7 SIRPα1-reactive MoAbs (SE5A5, SE7C2, and SE12C3). Two of the cross-reacting MoAbs (SE5A5 and SE12C3) also blocked the adhesion of cells to SIRPα2ex, whereas MoAb SE7C2 with superior specificity for SIRPα1 inhibited cell binding only to SIRPα1ex. Surprisingly, the cross-reacting MoAb SE12B6 inhibited cell adhesion to SIRPα2ex, but not to SIRPα1ex. An explanation might be that different epitopes among SIRP family members are responsible for ligand interaction, or that different ligand epitopes are involved in the selective binding of SIRPα1 and SIRPα2.
In the search for a SIRP ligand-specific antibody, we generated MoAbs against the SIRP-negative, but highly SIRPα1ex-binding T-lymphoblastic cell line CCRF-CEM. MoAb CC2C6 was a candidate antibody because of its capacity to inhibit the adhesion of CCRF-CEM cells to SIRPα1ex. Comparison of expression patterns of known CD antibodies and subsequent screening on the OV10/huCD47 transfectant cell line showed that MoAb CC2C6 recognizes CD47, also known as integrin-associated protein (IAP).19 In line with our findings, Jiang et al38 very recently identified CD47 as a predominant binding partner of rat neuronal adhesion protein P84, the homologue of human SIRPα proteins.
Inhibition studies with CD47-specific MoAbs BRIC126, 1/1A4, and B6H12 showed that these antibodies blocked the binding of MoAb CC2C6, as well as that of biotinylated SIRPα1ex and SIRPα2ex to CCRF-CEM cells, whereas another CD47-specific MoAb, 2D3, did not interfere with SIRPα1ex and SIRPα2ex binding. The inhibitory activities of these antibodies correspond well with previous observations that MoAbs BRIC126 and B6H12 have a comitogenic effect on CD3-activated T cells and induce aggregation of Jurkat cells and primary thymocytes, whereas MoAb 2D3 failed to induce such phenomena.36 This suggests that CD47-specific MoAbs BRIC126, B6H12, 1/1A4, and CC2C6 recognize the same or a related functionally important epitope on this molecule, and that this epitope (but not the 2D3 epitope) is directly involved in the binding of SIRPα1 and SIRPα2.
CD47 is a pentaspan membrane glycoprotein broadly expressed in all tissues.17,39,40 So far, only the OV10 subclone of the human ovarian carcinoma cell line OVM1, is CD47-negative.22In many tissues, CD47 is associated with αvβ3 integrins and involved in the regulation of integrin function.17,41,42However, in the hematopoietic system, coexpression of αvβ3 integrins is restricted to platelets and megakaryocytes.43This suggests also integrin-independent functions of CD47.39,40 Several reports have shown that CD47 (1) acts as a receptor for thrombospondin 1,22,44 (2) is involved in adhesion-dependent processes like neutrophil phagocytosis,17 (3) transendothelial and transepithelial migration of neutrophils,20,21 (4) adhesion to vitronectin,28 (5) platelet aggregation,45 and (6) costimulation of T-cell activation.23 The identification of the extracellular counterreceptor for CD47 and its involvement in cellular adhesion might provide further insight into some of these functional aspects.
Preliminary data from transendothelial migration assays indicate that not only CD47-specific MoAbs B6H12 and CC2C6, but also SIRPα1-reactive MoAbs, reduce the migration of neutrophils across endothelial monolayers. These observations are in line with the results described by Cooper et al20 who showed inhibition of neutrophil migration across human umbilical vein endothelial cell monolayers by the CD47-specific MoAb B6H12. Because neutrophils, as well as endothelial cells, express CD47 and SIRP (unpublished observation, February 1999), an interaction of these 2 counterreceptors might be important for this migration. The fact that only the functionally active CD47-specific MoAbs B6H12 and CC2C6 can completely block the interaction between CD47 and SIRP, whereas the nonfunctionally MoAb 2D3 had no effect on this interaction suggests that binding of SIRP to CD47 might be essential for several of the functions described for CD47. So far, a costimulatory effect on CD4+ T-cell activation could be independently described for CD4723 and for cattle SIRP.8 Preliminary results from our experiments indicate that this might be true also for human SIRP. Our SIRP-reactive MoAbs, which are able to block the interaction with CD47, as well as the nonblocking antibodies, are valuable tools to study the involvement of SIRP during various immunological processes in vitro.
At present, the relevance of CD47-SIRP interactions in cellular signaling is not known. So far, 4 alternatively spliced CD47 isoforms19,46 and 15 SIRP family members with heterogeneous extracellular domains have been identified.3 Therefore, defined CD47 isoforms with specific SIRP molecules might interact in a similar way as the structurally related killer inhibitory receptors (KIRs), which are counterreceptors for the polymorphic major histocompatibility complex (MHC) class I molecules.47 These specific interactions might then trigger intracellular signal transduction cascades and lead to a variety of physiological functions. The significance of 1 of these functions, namely the migration of neutrophils, has been demonstrated with CD47−/− knock-out mice.48 These animals have an early defect in neutrophil accumulation at the site of infection. In vitro, CD47−/− neutrophils are deficient in β3 integrin-dependent ligand binding, oxidative burst activation, and Fc receptor-mediated phagocytosis, suggesting a key role of CD47 in host defense. As SIRP is the counterreceptor for CD47, it is likely that SIRP−/− knock-out mice show similar defects as CD47−/− animals. Future studies with SIRP-deficient mice or embryonic stem cells might address this question and provide insight into the role of SIRP in the regulation of immunological processes and hematopoiesis.
ACKNOWLEDGMENT
The authors thank H. Letzkus for excellent assistance in the generation of SIRP- and CD47-specific MoAbs. We would also like to thank Dr P. Brossart for kindly providing in vitro generated dendritic cells and Dr C. Faul for the well-organized supply of bone marrow cells.
Supported by the Deutsche Forschungsgemeinschaft (SFB510, project A1), by a grant from the research program of the University Clinic of Tübingen (fortüne, project 433), and by a grant from the José Carreras Foundation (Carreras/Bü-1).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Hans-Jörg Bühring, PhD, Medizinische Klinik II, Otfried-Müller-Str 10, 72076 Tübingen, Germany; e-mail: hjbuehri@med.uni-tuebingen.de.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal