Abstract
We genetically analyzed a case of hypofibrinogenemia that showed no bleeding or thrombotic tendency. Direct sequencing of a polymerase chain reaction-amplified γ-chain gene segment showed a novel nucleotide substitution. This heterozygous mutation encodes both Cys (TGT) and Arg (CGT) at residue 153. To examine the basis for the fibrinogen deficiency, we prepared expression vectors containing mutant γ-chain DNAs encoding γ153R and γ153A for in vitro expression in Chinese hamster ovary (CHO) cells. Enzyme-linked immunosorbent assay and immunoblot analysis of the culture media and cell lysates showed that CHO cells transfected with γ153R or γ153A synthesized the variant γ-chain, but did not secrete variant fibrinogen into the culture medium. Metabolic pulse-chase experiments showed that fibrinogen assembly was impaired when either variant γ-chain was expressed. In cells expressing normal fibrinogen, assem- bly intermediates and intact fibrinogen were seen in cell lysates prepared after short (3 minutes) or long (1 hour) incubation with 35S-methionine. Neither intermediates nor intact fibrinogen was seen with the variant γ-chains. These data suggest that γ-chains have an important early role in fibrinogen assembly. Thus, our results support the model for fibrinogen assembly proposed by Huang et al (J Biol Chem 268:8919, 1993), in which the first step in assembly is the formation of γ or βγ dimers, or both. This model implies that γCys153 has a critical role in the formation of these early assembly intermediates. We concluded that the γ153Cys→Arg substitution does not allow fibrinogen assembly and secretion, and this is manifest in vivo as a fibrinogen deficiency. We designated this variant as fibrinogen Matsumoto IV.
FIBRINOGEN IS A PLASMA glycoprotein consisting of 2 copies of 3 polypeptide chains, Aα, Bβ, and γ, linked by an extensive network of 29 intrachain or interchain disulfide bonds.1,2 Fibrinogen is the coagulation factor with the highest concentration in human plasma (1.5 to 3.0 g/L). Congenital dysfibrinogenemia, first reported in 1958,3 is defined as the presence of abnormal fibrinogen molecules in plasma. Approximately 300 families with presumed dysfibrinogenemias have been reported,4,5 most of whom show the functional abnormalities either in release of fibrinopeptide A or in fibrin monomer polymerization.4,5 Congenital hypofibrinogenemia and afibrinogenemia are defined by reduced or immeasurable levels of plasma fibrinogen. The first case of afibrinogenemia was reported in 1920,6 and since then approximately 150 cases7have been reported. The first case of hypofibrinogenemia was reported in 1935,8 and 30 to 40 cases9 have been reported subsequently. In this report, we describe analysis of the hypofibrinogenemia Matsumoto IV.
Quantitative deficiencies have been identified in other proteins of the coagulation and fibrinolytic systems, including protein C,10 protein S,11 antithrombin III,12 and factor VII.13 In these cases, genetic analysis of patients' DNA or RNA has identified several abnormalities that cause reduced levels of protein in plasma. Missense mutations, nonsense mutations, frameshift mutations, splice-site abnormalities, or others have all been associated with protein deficiency. Protein-deficient cases of Cys substituted for Arg, the substitution described here, have been reported for protein C,14 protein S,11 and antithrombin III.15 Furthermore, Sugahara et al14 have reported a transient expression experiment in which secretion of protein C 331Cys→Arg in COS-7 cells was dramatically impaired.
Genetic analyses of quantitative deficiencies in fibrinogen have been reported for several hypodysfibrinogenemias in which vaiant proteins were present in the plasma, although at reduced concentrations.16-19 In all of these cases, the variant fibrinogens were carboxy-terminal truncations of the Aα-chain. Recently, an extraordinary afibrinogenemic family was described in which 4 affected individuals had homozygous deletions of approximately 11 kb in the Aα-chain gene.20 These deletions apparently arose on 3 distinct ancestral chromosomes. In these cases, fibrinogen was not detected in the plasma, because no Aα-chains were synthesized. We report here genetic analysis of the hypofibrinogenemia Matsumoto IV, in which no variant fibrinogen was detected in the plasma. We found a novel missense mutation that resulted in impaired secretion of the altered fibrinogen when expressed in cultured Chinese hamster ovary (CHO) cells. Our discovery of impaired secretion in a γ-chain missense variant may be analogous to that of fibrinogen Brescia, which has been described in a published abstract.21
MATERIALS AND METHODS
Patient identification.
The propositus was a 21-year-old healthy woman who had no history of bleeding or thrombosis. Routine medical examination showed a remarkably low concentration of plasma fibrinogen. After acquiring informed consent from the patient, we collected blood from the propositus for biochemical and genetic analyses. None of her family members had a history of a bleeding or thrombotic tendency. Unfortunately, we were not able to analyze other family members.
Coagulation screening tests.
Nine volumes of blood were collected into plastic tubes containing 1 vol of 3.2% trisodium citrate. Plasma was separated by centrifugation at 1,500g for 10 minutes at 4°C. The buffy coat was collected and extracted to prepare genomic DNA. Prothrombin time (PT), activated partial thromboplastin time (APTT), and the fibrinogen concentration, which was determined by the thrombin time method, were measured with an automated analyzer CA-5000 (TOA Medical Electronics Co Ltd, Kobe, Japan). The immunologic fibrinogen concentration was determined by a latex photometric immunoassay using antifibrinogen antibody-coated latex particles (DIAYATORON, Tokyo, Japan).22
Immunoblot analysis of plasma fibrinogen.
Polymerase chain reaction (PCR) amplification and DNA sequence analysis.
To amplify 5 exons (exon I through V) of the Aα-chain gene, 8 exons (exon I through VIII) of the Bβ-chain, and 10 exons (exon I through X) of the γ-chain gene, we designed pairs of primers corresponding to the appropriate introns; 2 additional primers were designed for sequencing for Aα exon V. The primers are shown in Table 1. Genomic DNA was isolated from peripheral blood leukocytes as described.24 Aliquots of approximately 1 μg of the DNA were amplified by PCR in 25 μL of 10 mmol/L Tris-HCl, pH 8.4, containing 1.5 mmol/L MgCl2, 0.2 μmol/L of each forward and reverse primer, 200 μmol/L of each of dATP, dTTP, dCTP, and dGTP (Idaho Technology, Idaho Falls, ID), and 0.6 U Taq DNA polymerase (Perkin Elmer Cetus, Norwalk, CT). Thirty cycles of amplification were performed on a PC-800 DNA thermal cycler (Astec, Fukuoka, Japan) under the following conditions: denaturation at 94°C for 1 minute, annealing at various temperatures from 50°C to 59°C for 1 minute (Table 1), and extension at 72°C for 1 minute. PCR products were electrophoresed on a 2% agarose gel and stained with ethidium bromide. DNA fragments were extracted from the gels and directly sequenced as described22 using a Taq Dye Deoxy Terminator Sequencing Kit (Applied Biosystems, Foster City, CA) and a 373A DNA sequencer (Applied Biosystems).
Primers Used for PCR Amplification and Sequencing of 3 Fibrinogen Polypeptide Genes
| Peptide and Exon(s) . | Forward (5′-3′) . | Reverse (5′-3′) . | Annealing Temperature (°C) . | PCR Product (bp) . |
|---|---|---|---|---|
| Aα | ||||
| I | GTC TAG GAG CCA GCC CCA CC | GTG TCA GGA CAT AGA GCA GG | 59 | 180 |
| II and III | GCC CTG TGT CTG CTC TCC TTA ATC | ACG AAA ACA AAA GCT CCC TC | 56 | 1,070 |
| IV and V(1) | TTT GCT GGC AAT TAC AGA CA | CCT GGG GCT TTC CGT CTC TG* | 56 | 1,144 |
| V(2) | AGA CCT GGT GGA AAT GAG ATT AC* | GTT AAG AAG GAA ATG CAA GGG | 53 | 1,206 |
| V(3) | TGG CAC TCT GAA TCT GGA AG*,† | CCC AGG GTG ATG AGA ACT GG*,† | ||
| Bβ | ||||
| I | GGA TGG TTT CTT GGA GC | GCT AAG CCA TCC TCA TCT | 50 | 335 |
| II and III | GAT TCA CTA TCA CCA ACC AGC CAG | CAT GCA TTC TCG TGG CAG TC | 50 | 1,088 |
| IV and V | CTG CTT GGT GAT AGC TCA GT | CTG GCC TTG TTT CCT GGC AT | 50 | 1,078 |
| VI and VII | GAA TGG ACA GGG GAT TCA GA | AAG TGC CCA GGA AGT GGT AG | 50 | 808 |
| VIII | GTG CAC ACG AGT GTA GCA GT | GAA CGC TTC TCC TTC CTT AC | 50 | 842 |
| γ | ||||
| I and II | GGA GCC TGA GAG GTG ACA GTG C | CCA GTT CAC ACA CAA AGG GAG AAA C | 50 | 321 |
| III and IV | TTT CTC TTT TAG TAT GTT GC | TCA ACA TAA TCA GGC ATA AT | 50 | 779 |
| V and VI | CTT ATT TTT GTC TTC TTA TT | TGT GCC TCA GTT TCC TTT TC | 50 | 784 |
| VII | ATT TTC TCC TTT TGC TCT TG | CCC AAG AAC CAA ACA GAC TC | 50 | 372 |
| VIII | AGA TCC CTG AGG AGG GTC AG | CTA AGA AAG GAA AAC ATA CC | 50 | 347 |
| IX and X | AGA CTT GCA GAG GTA AAA AG | GCT TTG CAA GTC CAT TGT CC | 55 | 880 |
| Peptide and Exon(s) . | Forward (5′-3′) . | Reverse (5′-3′) . | Annealing Temperature (°C) . | PCR Product (bp) . |
|---|---|---|---|---|
| Aα | ||||
| I | GTC TAG GAG CCA GCC CCA CC | GTG TCA GGA CAT AGA GCA GG | 59 | 180 |
| II and III | GCC CTG TGT CTG CTC TCC TTA ATC | ACG AAA ACA AAA GCT CCC TC | 56 | 1,070 |
| IV and V(1) | TTT GCT GGC AAT TAC AGA CA | CCT GGG GCT TTC CGT CTC TG* | 56 | 1,144 |
| V(2) | AGA CCT GGT GGA AAT GAG ATT AC* | GTT AAG AAG GAA ATG CAA GGG | 53 | 1,206 |
| V(3) | TGG CAC TCT GAA TCT GGA AG*,† | CCC AGG GTG ATG AGA ACT GG*,† | ||
| Bβ | ||||
| I | GGA TGG TTT CTT GGA GC | GCT AAG CCA TCC TCA TCT | 50 | 335 |
| II and III | GAT TCA CTA TCA CCA ACC AGC CAG | CAT GCA TTC TCG TGG CAG TC | 50 | 1,088 |
| IV and V | CTG CTT GGT GAT AGC TCA GT | CTG GCC TTG TTT CCT GGC AT | 50 | 1,078 |
| VI and VII | GAA TGG ACA GGG GAT TCA GA | AAG TGC CCA GGA AGT GGT AG | 50 | 808 |
| VIII | GTG CAC ACG AGT GTA GCA GT | GAA CGC TTC TCC TTC CTT AC | 50 | 842 |
| γ | ||||
| I and II | GGA GCC TGA GAG GTG ACA GTG C | CCA GTT CAC ACA CAA AGG GAG AAA C | 50 | 321 |
| III and IV | TTT CTC TTT TAG TAT GTT GC | TCA ACA TAA TCA GGC ATA AT | 50 | 779 |
| V and VI | CTT ATT TTT GTC TTC TTA TT | TGT GCC TCA GTT TCC TTT TC | 50 | 784 |
| VII | ATT TTC TCC TTT TGC TCT TG | CCC AAG AAC CAA ACA GAC TC | 50 | 372 |
| VIII | AGA TCC CTG AGG AGG GTC AG | CTA AGA AAG GAA AAC ATA CC | 50 | 347 |
| IX and X | AGA CTT GCA GAG GTA AAA AG | GCT TTG CAA GTC CAT TGT CC | 55 | 880 |
Primer in exon.
Primer only for sequencing.
Endonuclease restriction digestion of PCR-amplified DNA.
Twenty microliters of PCR-amplified fibrinogen γ-chain exon VI products (784 bp) was precipitated by 1.1 μL of 3 mol/L sodium acetate and 44 μL of absolute ethanol and then left for overnight at −20°C. The precipitates were washed with frozen 80% ethanol, dissolved in 10 μL water, and completely digested with 1.25 UMbo I (New England Biolaboratory, Beverly, MA) at 37°C. The samples were electrophoresed on 2% agarose gel, and DNA fragments were visualized by ethidium bromide.
Construction of mutant expression vectors.
The fibrinogen γ-chain expression vector, pMLP-γ,25 was altered by oligonucleotide-directed mutagenesis using the Transformer Site-Directed Mutagenesis kit from Clontech Laboratories (Palo Alto, CA).26 Briefly, single-stranded pMLP-γ was annealed with a 5′-phosphorylated mutagenesis primer (see below) and a 5′-phosphorylated selection primer (5′-TCTAGGGCCCAGGCTTGTTTGC) that deleted a unique HindIII site in the vector. The second strand was synthesized using T4 DNA polymerase and closed with T4 DNA ligase. The mixture was treated withHindIII, and the products were transfected into competent BMH 71-18 mut S cells. Using the QIAGEN Plasmid Kit (QIAGEN, Inc, Chatsworth, CA), plasmid DNA was purified from transfected bacteria grown in broth overnight. Isolated DNA was again linearized withHindIII, and the products were transfected into competent DH5α cells. Transfectants were grown overnight on LB-ampicillin plates. DNA was prepared from ampicillin-resistant colonies, and plasmids lacking HindIII sites were sequenced using a 373A DNA sequencer. The complete γ-chain cDNA was sequenced using 2 forward and 2 reverse primers. One forward primer, which lies upstream from the cDNA, and one reverse primer, which lies downstream from the cDNA, were as described.26 The second forward and reverse primers were, respectively, 5′-GACGCTGCTACTTTGAAGTCC, which encodes amino acids residues 80-86, and 5′-TTGTCACTAGGATCATCGCC, which encodes amino acids residues 296-302. Two mutant vectors were constructed using the following mutagenesis primers (the altered bases are underlined) to change 153Cys to Arg (5′-TGGGAAAGATCGTCAAGACAT) and to change 153Cys to Ala (5′-TGGGAAAGATGCTCAAGACATT). The expression plasmids were called pMLP-γ153R and pMLP-γ153A.
Recombinant protein expression.
CHO cells were cultured in Dulbecco's modified Eagle's medium (DMEM)-Ham's nutrient mixture F12 (F12) supplemented with 5% bovine calf serum (Hyclone Laboratories, Logan, UT)/5% Nu-serum (Becton Dickinson Labware, Bedford, MA)/10 IU/mL penicillin/10 μg/mL streptomycin. CHO cell lines that expressed normal human fibrinogen Aα- and Bβ-chains, CHO-AαBβ cells,25were obtained by cotransfecting as described the plasmids pMLP-Aα, pMLP-Bβ, and pRSVneo into CHO cells.27 Plasmid DNAs for CHO cell transfection were prepared from large scale cultures with the QIAGEN kit. Transfected cells were selected with G418 (GIBCO BRL, Rockville, MD), and fibrinogen polypeptide expression was examined by immunoblot analysis of cell lysates. One cell line that synthesized high and approximately equivalent levels of both Aα- and Bβ-chains was selected for further transfection. Each of the variant pMLP-γ vectors and original pMLP-γ vector25 was cotransfected with the histidinol selection plasmid (pMSVhis) into the CHO-AαBβ cell line, using the standard calcium-phosphate coprecipitation method.27,28 Colonies were selected on both G418 and histidinol (Aldrich Chemical Co, Milwaukee, WI). Individual colonies were expanded and examined for fibrinogen synthesis as described.27 Three lines were analyzed for the work described here: γ153C-14, γ153R-24, and γ153A-10, which synthesize normal human fibrinogen, variant fibrinogen with Arg at position 153, and variant fibrinogen with Ala at position 153.
Immunoblot analysis and enzyme-linked immunosorbent assay (ELISA).
The SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblot analysis of fibrinogen or individual polypeptides were performed using a modified method.27 Immunoblots were developed with a rabbit antihuman fibrinogen antibody (DAKO, Carpinteria, CA) or a rabbit antihuman fibrinogen γ-chain antibody (Chemicon International Inc, Temecula, CA),29 and cross-reacting species were visualized with alkaline-phosphatase conjugated-goat antirabbit IgG antibody (EY Laboratories Inc, San Mateo, CA) with the substrate 5-bromo-4-chloro-3-indolyl-phosphate p-toluidine salt (Sigma Chemical Co, St Louis, MO) and nitroblue tetrazolium (Wako Pure Chemical, Osaka, Japan). Fibrinogen concentrations in cell lysates or in the cell culture media were determined by ELISA, as described.27
Culture medium for immunological analysis was prepared as follows. The cells were grown to confluence in 60-mm dishes, medium was removed, the cells were washed twice with phosphate-buffered saline (PBS), 3 mL of serum-free medium was added, and the cells were cultured for 7 days. The conditioned medium was harvested for immunoblot analysis or ELISA. Cell lysates for immunological analysis were prepared as follows. Cells were grown to confluence in a 60-mm culture dish, the medium was removed, and the cells were harvested in trypsin-EDTA solution (Sigma). Harvested cells were washed twice with PBS and lysed in either 60 μL of Laemmli sample buffer for immunoblot analysis or in 120 μL of 0.1% IGEPAL CA-630 (nonionic detergent; Sigma) and 10 mmol/L phenylmethylsulfonyl fluoride (PMSF; Sigma) for ELISA.
RNA isolation and reverse transcription (RT)-PCR.
CHO cells were harvested and washed as described for the immunological analyses and resuspended in 100 μL of PBS. Total RNA was isolated using IsogenLS (Nippon Gene, Tokyo, Japan) according to the manufacturer's protocol. RT of known amounts of RNA from CHO cells was performed with oligo-dT and Moloney's murine leukemia virus (M-MLV) reverse transcriptase (GIBCO BRL), and PCR of γ-chain c-DNA was performed with primers (sense, 5′-ATGAGTTGGTCCTTGCACCC-3′; antisense, 5′-AAGGTTCCTGGCACTGTGCTT-3′) designed to cover amino acid residues from −26 to 138. The control primers for human glyceraldehyde phosphate dehydrogenase (GAPDH) were as follows: sense, 5′-ACCACAGTCCATGCCATCAC-3′; antisense, 5′-TCCACCACCCTGTTGCTGTA-3′. PCR was performed for 25 cycles (94°C for 1 minute, 54°C for 1 minute, and 72°C for 1 minute) for γ-chain c-DNA and for 25 cycles (94°C for 1 minute, 62°C for 1 minute, and 72°C for 1 minute) for GAPDH c-DNA using 10, 25, 50, 100, 200, or 500 ng of input RNA. The PCR products were analyzed by 2% agarose gel electrophoresis and visualized by ethidium bromide staining.
Pulse-chase analysis of protein synthesis using [35S]-methionine.
Pulse-chase studies were performed by the procedure described by Yamamoto et al.30 In brief, for metabolic radiolabeling of methionine residues, γ153C-14, γ153R-24, and γ153A-10 cell lines were grown to confluence in 60-mm dishes. The cells were cultured for 16 hours without G418 and histidinol, washed twice with PBS, and incubated with 3 mL methionine-free DMEM (GIBCO-BRL) for 30 minutes at 37°C. The medium was replaced with 1 mL of methionine-free DMEM supplemented with 1.5 MBq (40 μCi) or 2.2 MBq (60 μCi) L-[35S]-methionine (ICN Radiochemicals, Irvine, CA), and the cells were incubated in 5% CO2 for 60 or 3 minutes at 37°C. After the pulse, the cells were rinsed twice with PBS, 1 mL of fresh DMEM containing 20 mmol/L unlabeled L-methionine (Wako) was added, and the cells were incubated for various time periods. The 60-minute pulse was followed by a 0-, 1-, 2-, 3-, 4-, 6-, 8-, or 24-hour chase, and the 3-minute pulse was followed by a 0-, 5-, 10-, 20-, or 60-minute chase. The media were harvested, and cell lysates were prepared in 120 μL lysis buffer containing 1% IGEPAL CA-630, 150 mmol/L NaCl, 5 mmol/L EDTA, and 10 mmol/L PMSF in 50 mmol/L Tris-HCl buffer, pH 8.0. Each 100 μL of medium or cell lysate was added to an equivalent amount of 1:1,000 diluted rabbit antifibrinogen polyclonal antibody (DAKO) and incubated overnight at 4°C. The specificity of the immunoprecipitation was tested by adding 90 μg of fibrinogen to the medium and cell lysate from the γ153C-14 60-minute pulse and 6-hour chase samples. The immune complexes were incubated with protein A-Sepharose (HiTrap Protein A; Pharmacia Biotech, Uppsala, Sweden) for 30 minutes at 37°C and were collected by centrifugation. The precipitates were washed with PBS containing 0.05% IGEPAL CA-630, 30 μL of Laemmli's sample buffer was added, and the samples were heated at 100°C for 5 minutes. The proteins were resolved on SDS-polyacrylamide gradient gels (4% to 12%) under nonreducing or reducing conditions. The gel was dried and radioactive bands were detected with the Fujix Bio-Imaging Analyzer BAS1500 System (Fuji Photo Film Co, Tokyo, Japan).
RESULTS
Characterization of the patient.
Routine screening assays showed impaired coagulation, with a prolonged PT and normal APTT. The PT was 14.1 seconds, whereas the normal range was 10.0 to 12.0 seconds. The APTT was 36.0 seconds, whereas the normal range was 24.0 to 37.0 seconds. The plasma fibrinogen concentration determined by both the thrombin time method (0.81 g/L) and the immunologic method (0.87 g/L) was obviously lower than normal (1.50 to 3.00 g/L). SDS-PAGE and immunoblot analysis of plasma fibrinogen from the propositus showed no abnormalities in the band patterns relative to normal Aα-, Bβ-, and γ-chains (data not shown). We concluded that the patient has normal fibrinogen at an abnormally low concentration.
Characterization of the fibrinogen defect.
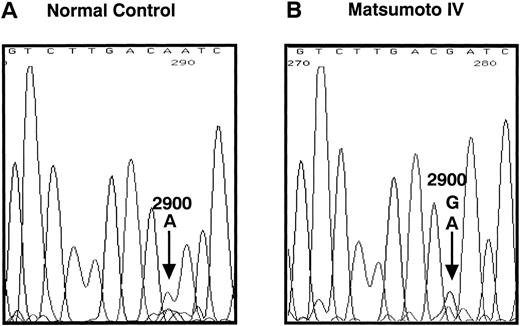
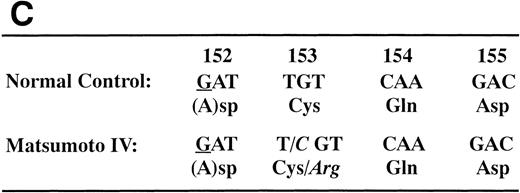
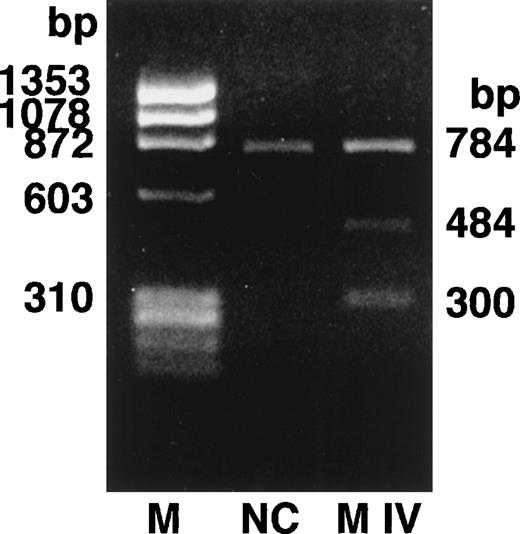
We determined the nucleotide sequence of the Aα-, Bβ-, and γ-chain genes by direct sequence analysis of PCR-amplified DNA fragments, as described in Materials and Methods. The primers used for PCR amplification and the fragments sequenced are presented in Table 1. The sequence determined for exon VI of the γ-chain is shown in Fig 1. At position 2900 (numbered according to Chung et al31) of the γ-chain gene, both T and C were present in DNA amplified from the patient, whereas only T was found at this position in the normal control. This T to C substitution encodes the replacement of Cys at residue 153 by Arg. Because the mixed nucleotide sequence at position 2900 of γ-chain was faint in several sequence reactions, we confirmed the mixed sequence by endonuclease restriction digestion of the PCR-amplified fragments. The substitution of T to C introduces an Mbo I site at this position of the γ-chain gene. The restriction digests of DNA amplified from the propositus and a normal control are shown in Fig 2. The control showed 1 band as predicted from the sequence (784 bp) and the propositus showed 3 bands of the predicted sizes of 784, 484, and 300 bp, indicating that the patient genotype was heterozygous for GATT and GATC. We also found a mix of residues, G and A, at position 4266 of the Aα-chain gene. This position has been reported as polymorphic, with either Ala (GCT) or Thr (ACT) at Aα-chain residue 312.32 We concluded that the heterozygous mutation at position 2900 of the γ-chain gene caused the reduced level of normal fibrinogen in the propositus.
Nucleotide sequence of the fibrinogen γ-chain gene exon VI. The PCR-amplified γ-chain genes of the normal control (A) and the propositus (B) were directly sequenced by dideoxy termination method using the reverse primer. The nucleotide position 2900, indicated by the arrow in (B), of the propositus' gene was heterozygous for A and G. This nucleotide substitution changed the γ153Cys→Arg as shown in (C). The underlined nucleotide is located in the intron region.
Nucleotide sequence of the fibrinogen γ-chain gene exon VI. The PCR-amplified γ-chain genes of the normal control (A) and the propositus (B) were directly sequenced by dideoxy termination method using the reverse primer. The nucleotide position 2900, indicated by the arrow in (B), of the propositus' gene was heterozygous for A and G. This nucleotide substitution changed the γ153Cys→Arg as shown in (C). The underlined nucleotide is located in the intron region.
Endonuclease restriction digestion of fibrinogen γ-chain gene exon VI. The PCR-amplified γ-chain genes of the normal control (NC) and the propositus (M IV) were digested by Mbo I. The normal control showed 1 band of 784 bp and the heterozygous propositus showed 3 bands of 784, 484, and 300 bp.
Endonuclease restriction digestion of fibrinogen γ-chain gene exon VI. The PCR-amplified γ-chain genes of the normal control (NC) and the propositus (M IV) were digested by Mbo I. The normal control showed 1 band of 784 bp and the heterozygous propositus showed 3 bands of 784, 484, and 300 bp.
We found a few other differences from the sequence reported by Chung et al.31 The nucleotide A at position 796 of the γ-chain gene differed from the reported T. However, PCR-amplified DNA fragments from 12 normal volunteers also showed the nucleotide A.22Our results are in agreement with those of Baumann and Henschen32 in that all of 110 healthy individuals had an A. We found 3 silent mutations: A to G at position 57 and T to C at position 60 in exon I of the Aα-chain gene, and G to C at position 5141 in exon V of the Aα-chain gene.31 Furthermore, 2 mutations were found within introns. One was a deletion of 2 nucleotides, A and G, at positions 6939-6944 in intron F of the Bβ-chain gene, which is a triplet repeat of A and G.31The other was an inversion of TC to CT at position 206 and 207 in intron A of the γ-chain gene.31 Both intron mutations were also found in PCR-amplified DNA fragments from 1 normal control. No other mutations were found in the entire coding region or the exon-intron boundaries.
Failed expression of the variant fibrinogen in CHO cells.
To determine the basis for the reduced levels of fibrinogen in the patient's plasma, we attempted expression of the variant protein in CHO cells. We prepared 2 mutant expression vectors, pMLP-γ153R or pMLP-γ153A, by oligonucleotide-directed mutagenesis of the γ-chain cDNA in the expression vector pMLP-γ,25 as described in Materials and Methods. We sequenced the entire modified γ-chain cDNA to identify the directed changes and to confirm that no unanticipated coding changes were incorporated (data not shown). We cotransfected pMLP-γ, pMLP-γ153R, or pMLP-γ153A with pMSVhis into CHO-AαBβ cell lines, which express the normal Aα- and Bβ-chains, as described.25 27 The transfected CHO cells were called γ153C, γ153R, and γ153A, respectively. Twenty days after transfection, histidinol-resistant colonies were picked and transferred to 24-well plates. Cells were grown to confluence within 12 days, and 29 of 36 γ153C clones, 41 of 48 γ153R clones, and 14 of 24 γ153A clones were split into three 60-mm dishes. Cells were again grown to confluence, and 12 γ153C clones, 13 γ153R clones, and 6 γ153A clones were selected for further experiments.
Fibrinogen concentrations in the culture media and cell lysates were determined by ELISA, as described in Materials and Methods. The results are presented in Table 2. Fibrinogen concentrations in the conditioned media from the 12 colonies expressing normal fibrinogen (γ153C) varied from 0.23 to 4.8 μg/mL. In contrast, conditioned media from all the plates with γ153R or γ153A clones contained less than 10 ng/mL fibrinogen. Moreover, fibrinogen concentrations in cell lysates from 12 clones of γ153C were 0.33 to 3.2 μg/mL, whereas fibrinogen concentrations in lysates from 13 clones of γ153R and 6 clones of γ153A were less than 10 to 92 ng/mL and less than 10 to 346 ng/mL, respectively. Analysis of the CHO-AαBβ cell line used for these transfections demonstrated that less than 10 ng/mL fibrinogen was present in either the culture medium or the cell lysates.
Fibrinogen Antigen Levels Determined by ELISA
| Cell Lines . | Cell Lysates (ng/mL) . | Media (ng/mL) . |
|---|---|---|
| γ153C (n = 12) | 1,134 ± 810 (330-3,200) | 1,995 ± 1,265 (230-4,800) |
| γ153R (n = 13) | 30 ± 31 (<10-92) | <10 (<10) |
| γ153R (n = 9*) | 41 ± 31 (<10-92) | <10 (<10) |
| γ153A (n = 6) | 107 ± 121 (<10-346) | <10 (<10) |
| Aα · Bβ (n = 3) | <10 (<10) | <10 (<10) |
| Cell Lines . | Cell Lysates (ng/mL) . | Media (ng/mL) . |
|---|---|---|
| γ153C (n = 12) | 1,134 ± 810 (330-3,200) | 1,995 ± 1,265 (230-4,800) |
| γ153R (n = 13) | 30 ± 31 (<10-92) | <10 (<10) |
| γ153R (n = 9*) | 41 ± 31 (<10-92) | <10 (<10) |
| γ153A (n = 6) | 107 ± 121 (<10-346) | <10 (<10) |
| Aα · Bβ (n = 3) | <10 (<10) | <10 (<10) |
The concentrations of fibrinogen in the cell lysates or the conditioned media from transfected CHO cells were measured by ELISA described in Materials and Methods. Data represent the mean ± SD in numbers of CHO cell lines. Numbers in parentheses mean the range of fibrinogen concentration.
Number of clones after deletion of γ-chain plasmid nonincorporated clones.
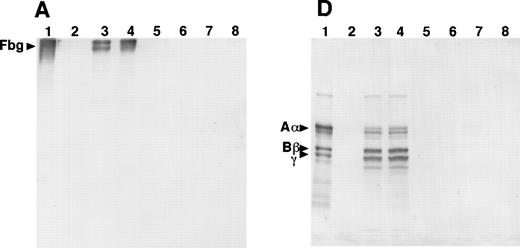
SDS-PAGE and immunoblot analysis of the culture media from the 12 γ153C clones showed the presence of fibrinogen or Aα-, Bβ-, and γ-chains under nonreducing or reducing conditions, respectively (Fig 3A and D). However, SDS-PAGE and immunoblot analysis of the culture media from all of the γ153R and all of the γ153A clones showed no cross-reacting bands under either nonreducing or reducing conditions (Fig 3A and D). Immunoblot analysis of cell lysates from all of the γ153C clones, all of the γ153A clones, and 9 of 13 γ153R clones showed several cross-reacting bands, including intact fibrinogen and the individual chains when SDS-PAGE was run under nonreducing conditions and blots were developed with an antifibrinogen antibody (Fig 3B). Similar immunoblots of gels run under reducing conditions showed Aα-chains, Bβ-chains, γ-chains, and several smaller species, presumably proteolytic degradation products, in all samples that contained fibrinogen (Fig 3E). When immunoblots were developed with an antibody to the fibrinogen γ-chain, several cross-reacting bands were seen with both nonreducing and reducing conditions (Fig 3C and F). Although this antibody apparently cross-reacted with both the β- and γ-chains, these immunoblots demonstrated that γ-chain was present in the cell lysates from all of the γ153C clones, all of the γ153A clones, and 9 of 13 γ153R clones. In contrast, SDS-PAGE and immunoblot analysis of the cell lysates from 4 clones of γ153R showed no γ-chain with either the antifibrinogen antibody or antifibrinogen γ-chain antibody (data not shown). Because these data suggest that γ-chains were not expressed in these 4 clones, we presumed that the expression plasmid pMLP-γ153R was not present. Therefore, we recalculated the mean fibrinogen concentration in cell lysates from our prior ELISA analysis including only 9 clones of γ153R (Table 2).
Western blot analysis of fibrinogen in media (A and D) or in CHO cell lysates (B, C, E, and F) that were transfected with mutant or normal fibrinogen γ-chain plasmid. The samples were subjected to 8% SDS-PAGE under nonreduced conditions (A, B, and C) or 10% SDS-PAGE under reduced conditions (D, E, and F). After transfer to nitrocellulose membrane, blots were developed with a rabbit antihuman fibrinogen antibody (A, B, D, and E) or a rabbit antihuman fibrinogen γ-chain antibody (C and F) that reacted to not only human fibrinogen γ-chain, but also to Bβ-chain. The samples in each figure were (1) purified fibrinogen from normal control plasma, (2) ABβ CHO cells, (3) γ153C-7, (4) γ153C-16, (5) γ153R-24, (6) γ153R-32, (7) γ153A-7, and (8) γ153A-11.
Western blot analysis of fibrinogen in media (A and D) or in CHO cell lysates (B, C, E, and F) that were transfected with mutant or normal fibrinogen γ-chain plasmid. The samples were subjected to 8% SDS-PAGE under nonreduced conditions (A, B, and C) or 10% SDS-PAGE under reduced conditions (D, E, and F). After transfer to nitrocellulose membrane, blots were developed with a rabbit antihuman fibrinogen antibody (A, B, D, and E) or a rabbit antihuman fibrinogen γ-chain antibody (C and F) that reacted to not only human fibrinogen γ-chain, but also to Bβ-chain. The samples in each figure were (1) purified fibrinogen from normal control plasma, (2) ABβ CHO cells, (3) γ153C-7, (4) γ153C-16, (5) γ153R-24, (6) γ153R-32, (7) γ153A-7, and (8) γ153A-11.
Analysis of cell lysates on immunoblots that were prepared from SDS-PAGE run under nonreducing conditions and developed with antifibrinogen antibody showed that fibrinogen, Aα-chains, Bβ-chains, γ-chains, and their degradation products were present in every γ153C clone and several clones of γ153A or γ153R. However, fibrinogen was not detected in the γ153A or γ153R clones in which the fibrinogen concentration was less than 10 ng/mL (data not shown). Cell lysates prepared from the CHO-AαBβ cells showed only Aα-chains, Bβ-chains, and their degradation products.
Level of expression with RT-PCR.
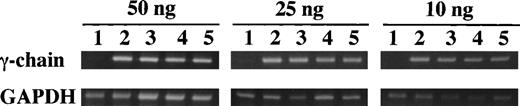
Semiquantitative RT-PCR was used to compare the level of expression of the γ-chain mRNA between the γ153C and the γ153R and the γ153A. Fibrinogen concentrations of culture media and cell lysates were as follows: γ153C-14, 3,600 and 3,200 ng/mL; γ153C-8, 230 and 390 ng/mL; γ153R-24, less than 10 and 90 ng/mL; and γ153A-10, less than 10 and 350 ng/mL. The amplified products for the γ-chain and GAPDH primers are 492 and 452 bp, respectively. GAPDH primers were used to show that similar amounts of RNA were added and that the efficiency of amplification was similar among the samples. As seen in Fig 4, we detected expression of γ-chain mRNA in not only γ153C cell lines, but also in γ153R and γ153A cell lines at all concentrations of input RNA. Furthermore, no γ-chain mRNA was detected in the parent CHO-AαBβ cell line. With 10 ng of input RNA, the quantity of PCR product from the γ153R and γ153A cell lines appeared slightly reduced as compared with that of the γ153C cell lines. Nevertheless, this semiquantitative analysis showed that variant messages were transcribed at levels comparable to the normal message.
Semiquantitative RT-PCR analysis of γ-chain mRNA expression in cell lines. Increasing amounts of total input RNA (10, 25, and 50 ng) from transfected CHO cell lines (γ153C-8 and -14, γ153R-24, and γ153A-10) were reverse transcribed and amplified for 25 cycles. The PCR products for the γ-chain (492 bp) and GAPDH primers (452 bp) were visualized by ethidium bromide staining with 2% agarose gels. The samples in the figure were (1) ABβ CHO cells, (2) γ153C-14, (3) γ153C-8, (4) γ153R-24, and (5) γ153A-10.
Semiquantitative RT-PCR analysis of γ-chain mRNA expression in cell lines. Increasing amounts of total input RNA (10, 25, and 50 ng) from transfected CHO cell lines (γ153C-8 and -14, γ153R-24, and γ153A-10) were reverse transcribed and amplified for 25 cycles. The PCR products for the γ-chain (492 bp) and GAPDH primers (452 bp) were visualized by ethidium bromide staining with 2% agarose gels. The samples in the figure were (1) ABβ CHO cells, (2) γ153C-14, (3) γ153C-8, (4) γ153R-24, and (5) γ153A-10.
Pulse-chase analysis of fibrinogen synthesis.
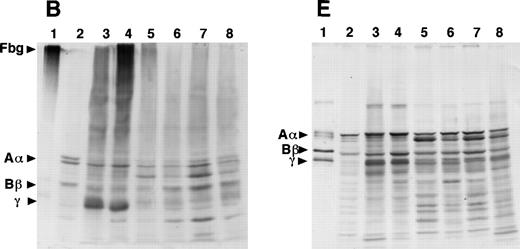
To examine the assembly and secretion of normal and variant fibrinogens, cells were grown in the presence of [35S]-methionine and the radiolabeled fibrinogen was analyzed by SDS-PAGE after immunoprecipitation from cell lysates or culture medium, as described in Materials and Methods. In 1 set of experiments, the cells were incubated with [35S]-methionine for 1 hour, the medium was replaced with unlabeled methionine and samples were immunoprecipitated at varying times up to 24 hours (Fig 5). The specificity of the immunoprecipitation was verified by the loss of labeled bands when excess plasma fibrinogen was added to samples before immunoprecipitation (Fig 5A, lane Inh). Three bands with relative mobilities of 195, 39, and 34 kD were seen in all samples, indicating that these species are not fibrinogen or fibrinogen fragments. Three immunospecific bands were seen in all the lysates from γ153C cells: the 340-kD band expected for fibrinogen and 2 additional high molecular weight bands of approximately 288 and 234 kD. These smaller bands may be intermediates formed during assembly of the fibrinogen chains into the intact molecule. All 3 immunospecific bands were present in samples prepared up to 8 hours after the radiolabel was removed, indicating that all 3 species were stable within the cell. After 24 hours, the intact fibrinogen and 234-kD band are seen, and the amounts of these are substantially reduced. Only 1 band, the 340-kD band of fibrinogen, was seen in immunoprecipitates of culture medium. This band became evident 1 hour after the addition of unlabeled medium and increased with successive incubation times, as expected for a complex protein that is assembled in the cell before secretion into medium. The presence of radiolabeled fibrinogen in the medium after 24 hours of incubation indicates that protein degradation did not exceed the rate of protein secretion from the cells. Under these labeling conditions, radiolabeled Aα-, Bβ-, or γ-chains were not seen in immunoprecipitates from either the γ153C cells or the culture medium.
Analysis of pulse-labeled fibrinogen in the transfected CHO cells. The cells were pulse-labeled for 1 hour with [35S]-methionine and chased for the indicated periods with an excess of unlabeled methionine. The recombinant fibrinogen and/or 3 polypeptides of fibrinogen were then immunoprecipitated from the cell lysates or the conditioned media with a rabbit antihuman fibrinogen antibody and protein A-Sepharose. The immunoprecipitates were subjected to electrophoresis on 4% to 12% gradient SDS-PAGE under nonreducing conditions and autoradiography. Lane Inh in (A) included the addition of purified plasma fibrinogen to the reaction mixtures of the 6-hour chase experiment to demonstrate the antibody specificity. Lane PC in (B) was the conditioned medium at the 6-hour chase of γ153C as a positive control.
Analysis of pulse-labeled fibrinogen in the transfected CHO cells. The cells were pulse-labeled for 1 hour with [35S]-methionine and chased for the indicated periods with an excess of unlabeled methionine. The recombinant fibrinogen and/or 3 polypeptides of fibrinogen were then immunoprecipitated from the cell lysates or the conditioned media with a rabbit antihuman fibrinogen antibody and protein A-Sepharose. The immunoprecipitates were subjected to electrophoresis on 4% to 12% gradient SDS-PAGE under nonreducing conditions and autoradiography. Lane Inh in (A) included the addition of purified plasma fibrinogen to the reaction mixtures of the 6-hour chase experiment to demonstrate the antibody specificity. Lane PC in (B) was the conditioned medium at the 6-hour chase of γ153C as a positive control.
In contrast, no immunospecific bands were seen with samples prepared from the γ153R cells or the culture medium (Fig 5B). Cell lysates showed 4 radiolabeled bands (195, 39, 37, and 34 kD), but 3 of these had the same mobilities (195, 39, and 34 kD) as the nonimmunospecific bands seen in the presence of plasma fibrinogen (lane Inh, Fig 5A). Furthermore, immunoprecipitates from the media contained no radiolabeled material. We noted that the autoradiogram from γ153R cell lysates showed significant radioactive material at the top of the lanes. When similar samples were analyzed by SDS-PAGE run under reducing conditions, bands corresponding to the Aα-, Bβ-, and γ-chains were seen (data not shown), suggesting that the radioactive material at the top of the lanes in Fig 5B represented large complexes that did not enter the gel. Pulse-chase analyses of γ153A cell lysates and media and CHO-AαBβ cell lysates and media showed results similar to those for γ153R cells (data not shown).
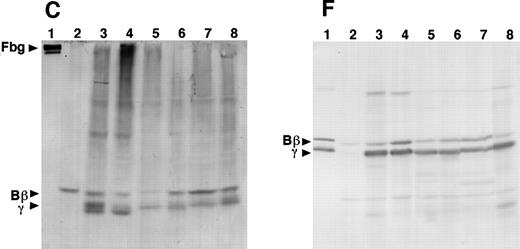
In a second set of experiments, the cells were incubated with [35S]-methionine for 3 minutes, the medium was replaced with unlabeled methionine, and cell lysates were immunoprecipitated at varying times up to 60 minutes. The samples were analyzed on 4% to 12% acrylamide gradient gels run under reducing and nonreducing conditions (Fig 6). Fully assembled fibrinogen (340 kD under nonreducing conditions) was detected in lysates of γ153C cells in samples prepared after a 10-minute chase, and the intensity of this band increased in both the 20- and 60-minute chase samples. These data indicate that radiolabeled chains continued to assemble into fibrinogen for at least 1 hour after the initial pulse. A 234-kD band, presumably a nascent form of fibrinogen, was seen 5 minutes after the initial pulse; the intensity of this band increased at both the 10- and 20-minute chase times and decreased at the 60-minute chase, as expected for an intermediate in the assembly process. SDS-PAGE analysis under reducing conditions showed that all 3 fibrinogen chains (Aα-chain was weak) were radiolabeled under these pulse-chase conditions, that the chains were first detected in the 5-minute chase, and that the label intensity increased with the chase interval. These data indicate that assembly was rapid relative to the time frame tested here. Furthermore, because labeled fibrinogen was not detected, but labeled 234-kD intermediate was detected in the 5-minute chase under nonreduced conditions, the data suggest that this intermediate was comprised of Aα-, Bβ-, and γ-chains. Finally, degradation of assembled fibrinogen or the individual chains was not detected under these conditions, indicating that, within the cell, assembled fibrinogen molecules were not degraded in the time frame of this experiment.
Analysis of pulse-labeled fibrinogen in the transfected CHO cells. The cells were pulse-labeled for 3 minutes with [35S]-methionine and chased for the indicated periods with an excess of unlabeled methionine. The immunoprecipitates from the cell lysates were subjected to electrophoresis on 4% to 12% gradient SDS-PAGE under nonreducing or reducing conditions and autoradiography.
Analysis of pulse-labeled fibrinogen in the transfected CHO cells. The cells were pulse-labeled for 3 minutes with [35S]-methionine and chased for the indicated periods with an excess of unlabeled methionine. The immunoprecipitates from the cell lysates were subjected to electrophoresis on 4% to 12% gradient SDS-PAGE under nonreducing or reducing conditions and autoradiography.
In contrast, under nonreduced conditions, no radiolabeled bands were detected in γ153R cell lysates. This result indicates that fibrinogen was not assembled from γ153R. Nevertheless, when these samples were analyzed under reducing conditions, radiolabeled bands equivalent to the Aα- and Bβ-chains were observed. These bands appeared in the 5-minute chase and increased in intensity after 10, 20, and 60 minutes. A radiolabeled species equivalent to the variant γ-chains was not observed. This result contrasted with that observed in the 1-hour pulse experiment, in which all 3 chains were seen in cell lysates under reduced conditions (data not shown). Together, these experiments suggest that the variant γ-chain was synthesized more slowly than the normal chain.
DISCUSSION
In these studies, we analyzed a case of hypofibrinogenemia to determine the etiology of this defect. By direct sequence analysis of a PCR-amplified γ-chain gene, we found a novel nucleotide substitution of T to C that changed a Cys to Arg at residue 153. This heterozygous substitution suggested that the variant chain was not assembled into fibrinogen such that the level of plasma fibrinogen was reduced in the patient. To confirm the causative role of this missense mutation for the fibrinogen deficiency, we prepared the mutant γ-chain expression vectors and transfected these into a CHO cell line that expressed normal human fibrinogen Aα- and Bβ-chains. The expression experiments indicated that the γ153R cell lines transcribed the variant transfected cDNA and synthesized the variant γ-chain but did not secrete fibrinogen into the culture media. In addition, metabolic pulse-chase analyses showed that fibrinogen was not assembled in these cells. Similar data were seen with γ153A cell lines. We concluded that the γ153Cys→Arg substitution led to the patient's hypofibrinogenemia.
High resolution structures of fibrinogen fragment D33 or a recombinant 30-kD C-terminal fragment of the fibrinogen γ-chain34 clearly showed the presence of an intrachain disulfide bond between γ153C and γ182C. Thus, the loss of Cys at residue 153 will necessarily disrupt this disulfide bond. Although the loss of a disulfide bond may cause profound changes in fibrinogen structure and associated function, there is no precedence to suggest that the loss of an individual Cys caused hypofibrinogenemia. In particular, fibrinogens Marburg (Aα1-460)18 and Milano III (Aα1-453)19 both lack Aα472Cys and therefore are missing the intrapeptide disulfide bond between Aα442Cys and Aα472Cys. Analysis of these patient plasmas demonstrated that the altered fibrinogen was present in both patients; furthermore, the variant plasma fibrinogens were linked by a novel disulfide bond to albumin.18,19 Even the loss of an interchain disulfide bond between Bβ65Cys and Aα36Cys in fibrinogen New York I (lacking Bβ residues 9-72) did not prohibit assembly of the variant chain into fibrinogen and its subsequent secretion into the patient's plasma.35 Thus, the impaired secretion of fibrinogen Matsumoto IV must reflect some characteristic of γCys153 and/or the intrapeptide disulfide bond between γ153Cys and γ182Cys.
The pulse-chase data presented here demonstrated that the variant chain was not assembled into fibrinogen within the CHO cells. In the short pulse (3 minutes) and chase analysis, no γ-chain was observed in the γ153R cell line. Because the variant γ-chain was not assembled into fibrinogen or nascent fibrinogen fragments in the cell, we think that the synthesis of the variant γ-chain was slower than the synthesis of the normal Aα- or Bβ-chains. Alternatively, the variant γ-chain may be unstable, because it cannot participate in the normal assembly process. In this case, the labeled variant γ-chains seen after a 1-hour pulse would reflect an accumulation of abnormal, high molecular weight complexes that contain normal Aα- and Bβ-chains and the variant γ-chain. These results suggest that our mutant cell lines, γ153R and γ153A, will be useful tools to investigate the transport of individual chains, posttranslational chain modification, and the assembly of the dimeric heterotrimer of 6 polypeptide chains within the cell, as well as the subsequent secretion of fibrinogen.
Our data suggest that the variant chains could not participate in the interchain interactions required for normal fibrinogen assembly. This conclusion is consistent with a previously proposed model for fibrinogen assembly that was based on data obtained from analysis of fibrinogen expression in cultured BHK or HepG2 cells.36 In this model, the initial steps in fibrinogen assembly were the formation of either an αγ or βγ dimer or both. The dimer complexes were detected in radiolabeled cell lysates and were shown to be linked by disulfide bonds. Our data suggest that these dimers could not be formed when γ153Cys was not present. Such a result would be found if either γCys153 has a direct role in complex formation or the free γCys182 impairs the normal formation of disulfide bonds in the 2 chain complexes. Because no unique higher molecular weight complex was seen in our radiolabel studies, our data support the proposed model in which γ-chain has a critical role in the initial stages of fibrinogen assembly.
In summary, fibrinogen Matsumoto IV with γ153Cys→Arg disrupted an intrapeptide disulfide bond between γ153Cys and γ182Cys and led to impaired secretion of fibrinogen. These results suggest that the fibrinogen γ-chain has an essential role in the assembly and/or secretion of fibrinogen.
ACKNOWLEDGMENT
The authors acknowledge Prof T. Katsuyama and M. Tozuka, PhD (Department of Laboratory Medicine, Shinshu University School of Medicine, Matsumoto, Japan) for helpful advice and encouragement and I. Ueno, PhD, and E. Hidaka for helpful advice about semiquantitative RT-PCR analysis.
Supported by grant from Kurozumi Medical Foundation.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Nobuo Okumura, PhD, Laboratory of Clinical Chemistry, Department of Medical Technology, School of Allied Medical Sciences, Shinshu University, 3-1-1 Asahi, Matsumoto 390-8621, Japan; e-mail: nobuoku@gipac.shinshu-u.ac.jp.

![Fig. 5. Analysis of pulse-labeled fibrinogen in the transfected CHO cells. The cells were pulse-labeled for 1 hour with [35S]-methionine and chased for the indicated periods with an excess of unlabeled methionine. The recombinant fibrinogen and/or 3 polypeptides of fibrinogen were then immunoprecipitated from the cell lysates or the conditioned media with a rabbit antihuman fibrinogen antibody and protein A-Sepharose. The immunoprecipitates were subjected to electrophoresis on 4% to 12% gradient SDS-PAGE under nonreducing conditions and autoradiography. Lane Inh in (A) included the addition of purified plasma fibrinogen to the reaction mixtures of the 6-hour chase experiment to demonstrate the antibody specificity. Lane PC in (B) was the conditioned medium at the 6-hour chase of γ153C as a positive control.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/12/10.1182_blood.v94.12.4122/5/m_blod42435005aw.jpeg?Expires=1763603741&Signature=haM5nsyvj46uA9T4OS5BiFpgYiLXdQEF2v1XXIA1C6SQMsjBy6-zzTbe2u9na4AnHYjw7b0pE0VzdjIey83XQ9dbkpMNHTytqFVNbkdpWR1K-wEROLnabVOtQKS9NI7W2M6GNxn9Yt-TmPjRk6UplCP9-4OpRvnQpBO~Ow6HqLl9Af7bDgX68UJa7WZek9abFk~txKncrXjbSCFMIsSYZGI-MPuqAscAjav9ywNNw3qi8zors6EjLedHHIDEdGrSSnRolUJ3leAPA0hNTIRDqV-n7wtXrM~sHUVyHv-ltze1HUYXOD-1D4Jl8FEXTxKSJa6IuR-yT5-WamYxwp0qaA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. Analysis of pulse-labeled fibrinogen in the transfected CHO cells. The cells were pulse-labeled for 1 hour with [35S]-methionine and chased for the indicated periods with an excess of unlabeled methionine. The recombinant fibrinogen and/or 3 polypeptides of fibrinogen were then immunoprecipitated from the cell lysates or the conditioned media with a rabbit antihuman fibrinogen antibody and protein A-Sepharose. The immunoprecipitates were subjected to electrophoresis on 4% to 12% gradient SDS-PAGE under nonreducing conditions and autoradiography. Lane Inh in (A) included the addition of purified plasma fibrinogen to the reaction mixtures of the 6-hour chase experiment to demonstrate the antibody specificity. Lane PC in (B) was the conditioned medium at the 6-hour chase of γ153C as a positive control.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/12/10.1182_blood.v94.12.4122/5/m_blod42435005bw.jpeg?Expires=1763603741&Signature=WlyN3JXHeJ2-Tx-73A9lHWLsZH2MGSt5DxRzh-2Ggo5BCPCAmoTPATeYBrk32MMu05sH32MmS6VshIfVk~RbbO8QIuStMKZCb0PNJUi9WguM9ooXZlo9jIcyotYhzZiBvGbgJf374Hvd4O5FikwUeaFi0J2eQoLJTucxFsfywEkf~5GKw6fb-NnueGpo5AbKLzqON7ws4hh5sKBTnNsC7GfM4PlpbMhi2EtXdK3l0EvjpN2wpCcPaFTc8TWFrf-xCXvP1~1ozeUtMlCXE-wJp8wgUxbC-m4bNGub0Cp0nswNIZ8v53f9VOa1p6eENY3B0a7RIboVBjY0v6yJLQrdCg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 6. Analysis of pulse-labeled fibrinogen in the transfected CHO cells. The cells were pulse-labeled for 3 minutes with [35S]-methionine and chased for the indicated periods with an excess of unlabeled methionine. The immunoprecipitates from the cell lysates were subjected to electrophoresis on 4% to 12% gradient SDS-PAGE under nonreducing or reducing conditions and autoradiography.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/12/10.1182_blood.v94.12.4122/5/m_blod42435006w.jpeg?Expires=1763603741&Signature=FdIiwcJfxMGEnxjDj-JrxtT~QxSx7y67BxJsceEEuT0MjCXLtPJp6awAYe4rKAr~POHPOyYQfGW~aEz7g0WnKkif8cAKxkWj2lsqiBsQJAwV2JgZwy39I-pFeqOEC83xwrm8iEOVeZCZ7LpxRqQ97wgueix1OzAgf6l1u8iUtRnIkx-u7LPo8b40m1RrRfFOjmdTmAtQgMjt6z-kM9~YI9pukoUj~Xtl0QjAjowYX99EP8ccTXrl6fFMxERoV4K2O5hAdWK6HI7r~6R4muLANnFKq2pt~DOThtUKXO-GtNe6rPolMxjtcVyV8WynmqLDN-uCD45-ZAs1-IgtHtlUUw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal