Abstract
Within hematopoiesis, C/EBP is expressed only in myeloid cells, and PU.1 is expressed mainly in myeloid and B-lymphoid cells. C/EBP-deficient mice lack the neutrophil lineage and retain monocytes, whereas PU.1-deficient mice lack monocytes and have severely reduced neutrophils. We expressed a C/EBP-estrogen receptor ligand-binding domain fusion protein, C/EBPWT-ER, in 32D cl3 myeloblasts. 32D cl3 cells proliferate in interleukin-3 (IL-3) and differentiate to neutrophils in granulocyte colony-stimulating factor (G-CSF). In the presence of estradiol, C/EBPWT-ER induced morphologic differentiation and the expression of the myeloperoxidase, lactoferrin, and G-CSF receptor mRNAs. C/EBPWT-ER also induced a G1/S cell cycle block, with induction of p27 and Rb hypophosphorylation. bcr-ablp210 prevented 32D cl3 cell differentiation. Activation of C/EBP-ER in 32D-bcr-ablp210 or Ba/F3 B-lymphoid cells induced cell cycle arrest independent of terminal differentiation. C/EBPWT-ER induced endogenous PU.1 mRNA within 8 hours in both 32D cl3 and Ba/F3 cells, even in the presence of cycloheximide, indicating that C/EBP directly activates the PU.1 gene. However, activation of a PU.1-ER fusion protein in 32D cl3 cells induced myeloperoxidase (MPO) RNA but not terminal differentiation. Thus, C/EBP acts downstream of G-CSF and upstream of PU.1, p27, and potentially other factors to induce myeloblasts to undergo granulocytic differentiation and cell cycle arrest.
GENES EXPRESSED predominantly in immature myeloid cells are often activated by a combination of the C/EBPα, PU.1, c-Myb, and AML1 transcription factors.1-12c-Myb–null or AML1-null mice do not develop myeloid, lymphoid, or erythroid cells, consistent with the expression of these factors in multipotent hematopoietic stem cells and in the immature cells that give rise to these mature blood elements.13-15
PU.1 is expressed in myeloid cells and in B cells.16 One PU.1-deficient murine line lacks B and T lymphocytes, monocytes, and neutrophils,17 although in the presence of interleukin-3 (IL-3) and granulocyte colony-stimulating factor (G-CSF), progenitors from these mice generate immature granulocytes with myeloperoxidase-positive granules.18 A second PU.1-deficient line lacks monocytes and B lymphocytes, but retains neutrophils, although these neutrophils are reduced in number and do not express some markers of terminal differentiation.19
The C/EBP family of transcription factors are expressed in multiple cell types, including hepatocytes, adipocytes, enterocytes, and pulmonary epethilium.20-22 Within hematopoiesis, C/EBPα, C/EBPβ, and C/EBPδ are expressed predominantly in myeloid cells and in eosinophils.23-26 C/EBPβ is the most prominent isoform in monocytic cells, and C/EBPβ-null mice have dysfunctional monocytes and normal granulocytes.27,28 C/EBPα expression is prominent in immature myeloid cells,3,25,26 and C/EBPα-null mice lack the entire granulocytic lineage, but develop normal monocytes.29 Also, ectopic expression of C/EBPα in U937 monocytic leukemia cells induces granulocytic differentiation over a 2-week period and inhibits monocytic differentiation.25Thus, hematopoietic progenitors require PU.1 to initiate monocyte differentiation and C/EBPα to initiate granulopoiesis. C/EBPβ contributes to monocyte activation, and PU.1 contributes to granulocyte differentiation.
Colony-forming unit–granulocyte-macrophage (CFU-GM) is a bipotent progenitor that can give rise to granulocytes and monocytes. C/EBPα-null mice lack the entire granulocytic lineage, including colony-forming unit-granulocyte (CFU-G), a unipotent progenitor, but retain CFU-GM, monocytes, and the other blood elements.29 CFU-G are dependent on G-CSF for survival and proliferation, and the G-CSFR promoter is activated by C/EBPα and PU.1.9 Therefore, decreased expression of the G-CSFR might in part account for the loss of the granulocytic lineage in C/EBPα or PU.1 null mice, as proposed.29 However, G-CSFR- and G-CSF-null mice retain neutrophils, albeit at reduced numbers.30,31 C/EBPα and PU.1 also regulate the GM-CSF receptor α gene,3 but mice lacking both G-CSF and GM-CSF also retain neutrophils.32 These results indicate that C/EBPα and PU.1 regulate additional genes besides cytokine receptors required for granulopoiesis.
To gain further insight into the transcriptional program regulating myelopoiesis, we have expressed here C/EBPα and PU.1 inducibly in 32D cl3 myeloblasts.33,34 32D cl3 cells are diploid and are nonleukemic in syngeneic murine recipients. They require IL-3 for proliferation and differentiate to mature granulocytes in response to G-CSF, with a concomitant cell cycle arrest, just as normal progenitors do. 32D cl3 cells also retain monocytic potential.35Differentiation occurs only if IL-3 is removed. The membrane distal segment of the G-CSF receptor is required for G-CSF to induce differentiation of 32D cl3 and other myeloblastic cell lines.36-38
We demonstrate here that activation of a C/EBPα-estrogen receptor ligand-binding domain fusion protein, C/EBPαWT-ER, by estradiol is sufficient to induce terminal granulocytic differentiation and a G1 cell cycle arrest in 32D cl3 cells despite the continued presence of IL-3. bcr-ablp210 prevented 32D cl3 cell differentiation, including myeloperoxidase (MPO) RNA induction. Inhibition of cell growth by C/EBPαWT-ER occured even in 32D cl3 cells expressing bcr-ablp210 or in Ba/F3 B-lymphoid cells, without induction of differentiation. Cell cycle arrest was assoicated with elevated p27Kip1 levels. PU.1 protein and mRNA levels were increased within 4 hours of C/EBPαWT-ER activation, in 32D cl3, 32D-bcr-ablp210, or Ba/F3 cells, and induction of PU.1 mRNA occured even in the presence of cycloheximide, suggesting that induction of endogenous PU.1 RNA by C/EBPαWT-ER results from direct transcriptional activation. However, activation of PU.1-ER(T) in 32D cl3 cells induced MPO RNA but not cell cycle arrest or terminal differentiation.
Thus, in 32D cl3 myeloblasts, C/EBPα acts independent of G-CSF signals, directly upstream of PU.1, and upstream of p27Kip1and additional factors to limit proliferation and induce granulocytic differentiation.
MATERIALS AND METHODS
Cell culture and proliferation assays.
32D cl3 cells34 were maintained in Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 10% heat-inactivated fetal calf serum (HI-FCS) and 1 ng/mL IL-3 (R&D Systems, Minneapolis, MN). Where indicated, 32D cl3 lines were washed twice with phosphate-buffered saline (PBS) and placed in IMDM-10% HI-FCS with 1,000 U of G-CSF (Amgen, Thousand Oaks, CA) per milliliter. Ba/F3 cells39 were maintained in RPMI 1640 medium with 10% HI-FCS and 1 ng/mL IL-3. Estradiol was added to 1 μmol/L from a 1,000× stock in ethanol, and 4-hydroxytamoxifen (4HT; Sigma, St Louis, MO) was added to 200 nmol/L from a 1,000× stock in ethanol. Viable cell numbers were determined by enumerated cells that excluded Trypan Blue Dye using a hemocytometer. BrdU/PI staining and FACScan analysis were performed as described.40 Morphologic differentiation was assessed by cytospin followed by Wright’s-Giemsa staining. Cycloheximide was used at 50 μg/mL, Actinomycin D was used at 10 μg/mL, and each was added to cell cultures 30 minutes before estradiol. ϕCRE packaging cells41 and NIH-3T3 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 10% heat-inactivated calf serum. 293T cells were maintained in DMEM with 10% HI-FCS.
Retroviral transduction.
Retroviral constructs were transfected into ϕCRE cells by calcium-phosphate precipitation, and pooled transfectants were then selected in 2 μg/mL puromycin. For retroviral transduction, ϕCRE packaging lines were irradiated to 3,000 cGy and cocultured with 32D cl3 or Ba/F3 cells for 48 hours with 8 μg/mL Polybrene. Stable transfectants were then selected by limiting dilution in 96-well dishes with 2 μg/mL puromycin. 293T cells were used to package and transduce a bcr-ablp210 retroviral vector as described.42
Plasmids and transient tranfection.
The C/EBPαWT-ER cDNA has been described.43,44 The PU.1-ER(T) construct was prepared by linking the hormone-binding domain, amino acids 281 to 599, from a modified murine estrogen receptor that responds to 4HT but not estradiol,45 to the full-length murine PU.1 cDNA by polymerase chain reaction. Each of these cDNAs was then inserted into the polylinker of the pBabePuro retroviral vector.46 The pSRαMSVtkNeo-bcr-ablp210 retroviral vector and ϕ2 ecotropic packaging DNA have been described.47 The PU.1-ER(T) cDNA was also ligated downstream of the phosphoglycerol kinase (PGK) promoter to construct pPGK-PU.1-ER(T). cDNAs encoding PU.1-ER(T), PU.1, or antisense PU.1 (AS-PU.1) were linked to the pPGK promoter and analyzed for their ability to activate pB4TKCAT in NIH 3T3 cells, in cooperation with CMV-Pip, as described.48
Western and Northern blotting.
Preparation of total cellular protein and RNA, Western blotting, and Northern blotting were performed as described.40C/EBPα, c-Myb, and p27Kip1 antisera and Rb and p21WAF1/CIP1 monoclonal antibodies have been described.6,26 40 C/EBPβ (C19), PU.1 (T21), and ER(MC-20) rabbit antisera (Santa Cruz, Santa Cruz, CA) were used at 1:200. C-abl (Ab3) antibody (Calbiochem, San Diego, CA) was used at 1:200. Blots were stained with Fast Green as a loading control and were stripped between probes in 2% sodium dodecyl sulfate (SDS), 62.5 mmol/L Tris, pH 6.8, 100 mmol/L 2-mercaptoethanol at 50°C for 30 minutes.
The murine MPO and lactoferrin cDNA probes have been described,33 as has the PU.1 cDNA.6 The murine G-CSF receptor cDNA37 was kindly provided by S. Nagata (Osaka Biosciences Institute, Osaka, Japan). The blots were stripped between probes in 1 mmol/L Tris, pH 8.0, 1 mmol/L EDTA, 0.1× Denhardt’s solution at 75°C for 2 hours and were probed with an 18S oligonucleotide, 5′-GTGCGTACTTAGACATGCATG-3′, as a loading control. This oligonucleotide was radiolabeled with T4 kinase and hybridized in 6× SSC, 0.01 mol/L NaHPO4, pH 6.8, 1 mmol/L EDTA, 100 μg/mL sonicated salmon sperm DNA, and 0.1% nonfat dried milk at 45°C. The blots were then washed with 4× SSC, 0.1% SDS at 50°C and subjected to autoradiography.
RESULTS
C/EBPα induces granulocytic differentiation of 32D cl3 cells in IL-3.
C/EBPαWT-ER, a fusion protein containing the entire C/EBPα polypeptide and the ligand-binding domain of the estrogen receptor, was shown to activate transcription from a C/EBPα-binding site in the presence of estradiol.44 32D cl3 cells were transduced with a pBabePuro or pBabePuro-C/EBPαWT-ER retroviral vector, and clonal, puromycin-resistant lines were obtained by limiting dilution. Expression of C/EBPαWT-ER in two 32D cl3 lines proliferating in IL-3 was verified by Western blotting (Fig 1A). 32D cl3 cells express little endogenous C/EBPα in IL-3.26Material detected at 46 kD likely represents degradation of C/EBPαWT-ER. Thus, in IL-3, these two lines express C/EBPαWT-ER in large excess to endogenous C/EBPα. As these lines differentiated to granulocytes, C/EBPαWT-ER remained severalfold higher than endogenous C/EBPα (data not shown).
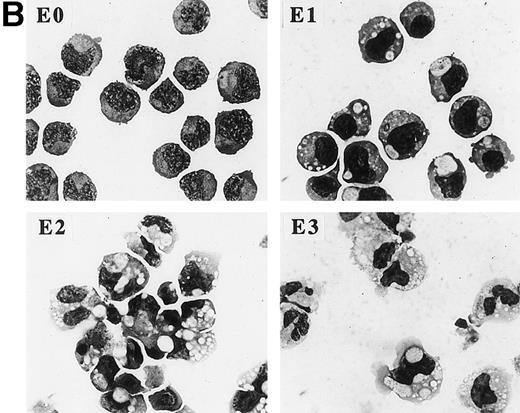
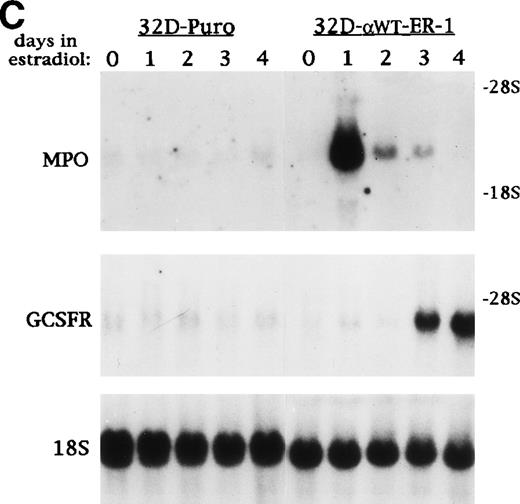
C/EBP induces terminal differentiation of 32D cl3 myeloblasts in IL-3. (A) Total cellular protein extracts derived from a 32D cl3 subclone transduced with a control retrovirus (32D-Puro) and from two 32D cl3 subclones transduced with a retrovirus expressing C/EBPWT-ER (32D-WT-ER-1 and 32D-WT-ER-2) were subjected to Western blotting using an antisera specific for C/EBP. The location of C/EBPWT-ER is indicated. (B) 32D-WT-ER-1 cells in IL-3 were exposed to 1 μmol/L estradiol. Cytospins were prepared before estradiol addition (E0) and 1, 2, and 3 days later (E1, E2, and E3) and were subjected to Wright’s-Giemsa staining. (C) 32D-Puro and 32D-WT-ER-1 cells in IL-3 were exposed to estradiol. For each culture, total cellular RNA was prepared after 0, 1, 2, 3, or 4 days in estradiol. These RNAs were then subjected to Northern blotting for MPO, G-CSF receptor (GCSFR), and 18S RNAs. (D) 32D-WT-ER-2 cells in IL-3 were exposed to estradiol, and total cellular RNA was prepared after 0 hours, 4 hours, 8 hours, 1 day, 2 days, 3 days, or 4 days in estradiol. These RNAs were then subjected to Northern blotting for MPO, lactoferrin (LF), or 18S RNAs.
C/EBP induces terminal differentiation of 32D cl3 myeloblasts in IL-3. (A) Total cellular protein extracts derived from a 32D cl3 subclone transduced with a control retrovirus (32D-Puro) and from two 32D cl3 subclones transduced with a retrovirus expressing C/EBPWT-ER (32D-WT-ER-1 and 32D-WT-ER-2) were subjected to Western blotting using an antisera specific for C/EBP. The location of C/EBPWT-ER is indicated. (B) 32D-WT-ER-1 cells in IL-3 were exposed to 1 μmol/L estradiol. Cytospins were prepared before estradiol addition (E0) and 1, 2, and 3 days later (E1, E2, and E3) and were subjected to Wright’s-Giemsa staining. (C) 32D-Puro and 32D-WT-ER-1 cells in IL-3 were exposed to estradiol. For each culture, total cellular RNA was prepared after 0, 1, 2, 3, or 4 days in estradiol. These RNAs were then subjected to Northern blotting for MPO, G-CSF receptor (GCSFR), and 18S RNAs. (D) 32D-WT-ER-2 cells in IL-3 were exposed to estradiol, and total cellular RNA was prepared after 0 hours, 4 hours, 8 hours, 1 day, 2 days, 3 days, or 4 days in estradiol. These RNAs were then subjected to Northern blotting for MPO, lactoferrin (LF), or 18S RNAs.
32D-αWTER-1 cells were exposed to estradiol for 0, 1, 2, or 3 days, and the morphology of the cells was assessed daily by cytospin and Wright’s-Giemsa staining (Fig 1B). After 1 day (E1), chromatin condensation and increased phagocytosis of dust particles is evident. The cell in the lower right of the panel exhibits cytoplasmic blebbing, an early sign of apoptosis. By the second day (E2), the nuclei in some cells have become more eccentric and indented. A cell with a bi-lobed nucleus is evident in the panel, as are several very small cells that are likely in a later stage of apoptosis (as demonstrated by BrdU/PI staining below). By the third day (E3), several cells with tri-lobed nuclei are seen, reminiscent of mature granulocytes. 32D-αWTER-2 cells underwent similar morphologic changes in estradiol, as did several additional subclones, including one expressing severalfold less C/EBPαWT-ER (data not shown).
32D-Puro and 32D-αWTER-1 cells were exposed to estradiol for 0, 1, 2, 3, or 4 days, and total cellular RNA was prepared daily. These RNAs were subjected to Northern blotting for MPO, G-CSF receptor (GCSFR), and 18S RNAs (Fig 1C). Estradiol did not affect MPO or GCSFR RNA expression in the 32D-Puro cells, but MPO RNA levels increased dramatically by day 1 in the 32D-αWTER-1 cells and then diminished, as seen when 32D cl3 cells differentiate in G-CSF33,34 (see Fig 4B). G-CSFR RNA levels also increased on days 3 and 4, again consistent with increased G-CSFR RNA levels observed during G-CSF–induced differentiation (see Fig 4B). Total cellular RNAs were then prepared from 32D-αWTER-2 cells exposed to estradiol for 0 hours, 4 hours, 8 hours, 1 day, 2 days, 3 days, or 4 days. These RNAs were then subjected to Northern blotting and probed for MPO, Lactoferrin, and 18S RNAs (Fig 1D). MPO RNA was increased by 4 hours, was maximal by 8 hours, and was again diminished on days 2 through 4. Lactoferrin RNA increased on days 2 to 3, as MPO RNA levels decreased, as seen when 32D cl3 cells differentiate in G-CSF.33 34Thus, activation of C/EBPαWT-ER in 32D cl3 myeloblasts proliferating in IL-3 induced differentiation to neutrophils, as assessed morphologically and by induction of early and late RNA markers.
Induction of MPO RNA by C/EBPα requires new protein and RNA synthesis.
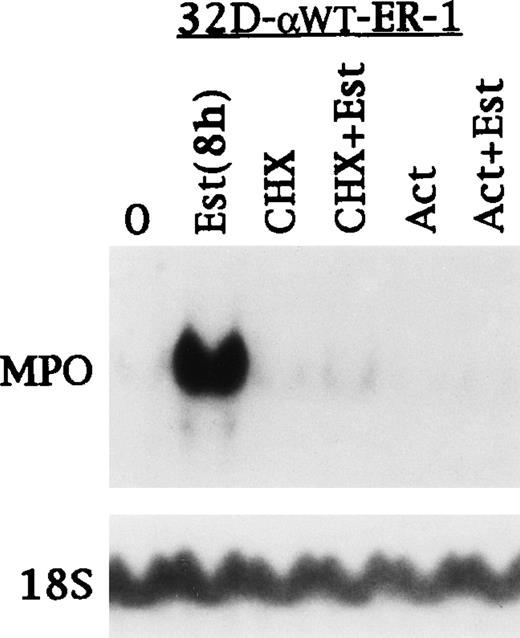
32D-αWTER-1 cells were cultured 0 or 8 hours in estradiol or for 8 hours in cycloheximide, estradiol with cycloheximide, Actinomycin D, or estradiol with Actinomycin D (Fig 2). Again, MPO RNA was rapidly induced, and this induction was prevented by both cycloheximide and Actinomycin D. Attempts to perform similar experiments at 4 hours of estradiol exposure were unsuccessful, because MPO was not induced or only weakly induced (data not shown). The effect of Actinomycin D indicates that induction of MPO RNA is not simply due to RNA stabilization. The effect of cycloheximide and the delayed maximal induction of MPO RNA is consistent with a requirement for C/EBPα to induce another factor, which then induces MPO transcription, either alone or with C/EBPα.
Induction of MPO RNA by C/EBP requires new protein and RNA synthesis. 32D-WT-ER-1 cells in IL-3 were exposed to estradiol for 0 or 8 hours. A third culture was exposed to cycloheximide (CHX) for 8 hours. A fourth culture was exposed to CHX for 30 minutes and then to CHX with estradiol for 8 hours. A fifth culture was exposed to Actinomycin D (Act) for 8 hours. A sixth culture was exposed to Act for 30 minutes and then to Act with estradiol for 8 hours. Total cellular RNAs prepared from each culture were then subjected to Northern blotting for MPO and 18S RNAs.
Induction of MPO RNA by C/EBP requires new protein and RNA synthesis. 32D-WT-ER-1 cells in IL-3 were exposed to estradiol for 0 or 8 hours. A third culture was exposed to cycloheximide (CHX) for 8 hours. A fourth culture was exposed to CHX for 30 minutes and then to CHX with estradiol for 8 hours. A fifth culture was exposed to Actinomycin D (Act) for 8 hours. A sixth culture was exposed to Act for 30 minutes and then to Act with estradiol for 8 hours. Total cellular RNAs prepared from each culture were then subjected to Northern blotting for MPO and 18S RNAs.
C/EBPα directly slows hematopoietic cell proliferation.
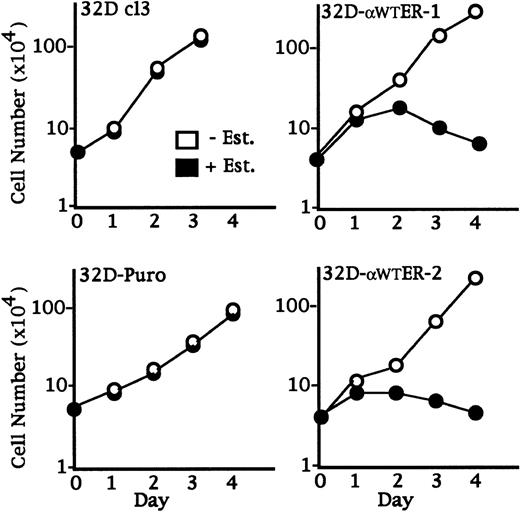
32D cl3, 32D-Puro, 32D-αWTER-1, and 32D-αWTER-2 cells were cultured in the absence or presence of estradiol, and viable cell numbers were enumerated daily (Fig 3). Estradiol did not affect the proliferation of 32D cl3 or 32D-Puro cells, but dramatically slowed the accumulation of lines expressing C/EBPαWT-ER. Continuous activation of C/EBPαWT-ER was required for growth inhibition, because removal of estradiol after 24 hours allowed the 32D-αWTER cells to resume proliferating at control rates (data not shown).
C/EBP slows 32D cl3 cell proliferation. 32D cl3 32D cl3, 32D-Puro, 32D-WT-ER-1, and 32D-WT-ER-2 cells in IL-3 were cultured in the absence or presence of estradiol (− Est. or + Est.). Viable cell numbers were enumerated daily with a hemocytometer and Trypan Blue Dye. Results shown are the mean of three determinations. Standard errors (SEs) were less than 15% of each mean.
C/EBP slows 32D cl3 cell proliferation. 32D cl3 32D cl3, 32D-Puro, 32D-WT-ER-1, and 32D-WT-ER-2 cells in IL-3 were cultured in the absence or presence of estradiol (− Est. or + Est.). Viable cell numbers were enumerated daily with a hemocytometer and Trypan Blue Dye. Results shown are the mean of three determinations. Standard errors (SEs) were less than 15% of each mean.
Inhibition of 32D cl3 cell proliferation might either be a primary effect of C/EBPα or a secondary effect due to induction of terminal differentiation. To distinguish between these possibilities, we sought to coexpress C/EBPαWT-ER with bcr-ablp210, which we hypothesized would prevent 32D cl3 cell differentiation in response to G-CSF or C/EBPαWT-ER. 32D cl3 cells were transduced with the pBabeNeo or SRαMSVtkNeo-bcr-ablp210 retroviral vectors and subjected to G418 selection. Expression of bcr-ablp210 was assessed by Western blotting in the resulting cell pools, in parental 32D cl3 cells, and in K562 cells, which are known to express bcr-ablp210 (Fig 4A, left panel). bcr-ablp210 was detected at high levels, relative to endogenous c-abl, in transduced 32D cl3 cells. The 32D-bcr/abl cells proliferated in the absence of IL-3 (data not shown). A 32D cl3 subclone expressing bcr-ablp210 was isolated by limiting dilution and transduced with pBabePuro-C/EBPαWT-ER. Expression of C/EBPαWT-ER in this 32D-bcr/abl line was documented by Western blotting (Fig 4A, right panel).
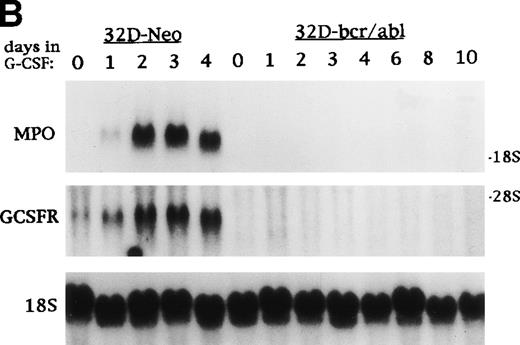
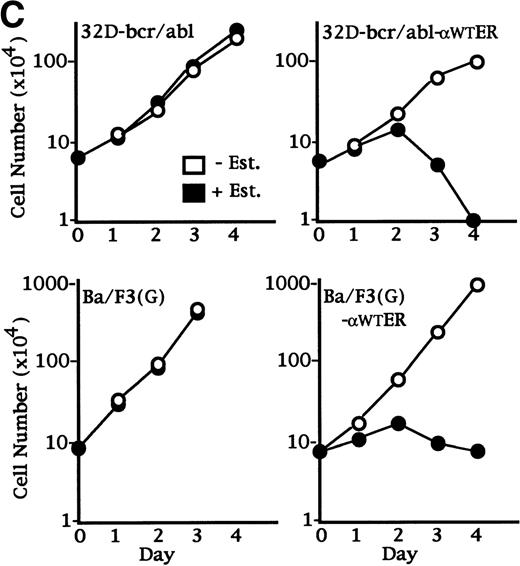
C/EBP slows the proliferation of 32D cl3 myeloblasts that cannot differentiate due to the expression of bcr-ablp210 and of Ba/F3 lymphoid cells. (A) Total cellular proteins were prepared from 32D cl3 cells (32D), from a cell line known to express bcr-ablp210 (K562), from a pool of 32D cl3 cells transduced with a control retrovirus (32D-Neo), and from a pool of 32D cl3 cells transduced with a retrovirus expressing bcr-ablp210 and G418 resistance (32D-bcr/abl). These extracts were then subjected to Western blotting with a c-abl antisera (left panel). The locations of c-abl and bcr-ablp210 are indicated. A G418-resistant subclone was derived the 32D-bcr/abl pool by limiting dilution and was shown to express a similar level of bcr-ablp210 (not shown). This line was then transduced with the pBabePuro-C/EBPWT-ER retroviral vector. A subclone resistant to G418 and puromycin was selected. A total cellular protein extract derived from this line was subjected to Western blotting for C/EBP (right panel). (B) Total cellular RNAs were prepared from 32D-Neo and 32D-bcr/abl cells in IL-3 or after 0, 1, 2, 3, or 4 days in G-CSF. RNA samples were also prepared from 32D-bcr/abl cells cultured in G-CSF for 6, 8, or 10 days. These RNAs were then subjected to Northern blotting for MPO, GCSFR, and 18S RNAs. (C) 32D-bcr/abl, 32D-bcr/abl-WT-ER, Ba/F3(G), and Ba/F3(G)-WTER cells in IL-3 were cultured in the absence or presence of estradiol. Ba/F3(G) cells are a derivative of Ba/F3 cells which express the G-CSFR. Ba/F3(G)-WTER cells express C/EBPWT-ER in addition. Cell counts were enumerated daily with a hemocytometer and Trypan Blue Dye. Results shown are the mean of three determinations. SEs were less than 10% of each mean.
C/EBP slows the proliferation of 32D cl3 myeloblasts that cannot differentiate due to the expression of bcr-ablp210 and of Ba/F3 lymphoid cells. (A) Total cellular proteins were prepared from 32D cl3 cells (32D), from a cell line known to express bcr-ablp210 (K562), from a pool of 32D cl3 cells transduced with a control retrovirus (32D-Neo), and from a pool of 32D cl3 cells transduced with a retrovirus expressing bcr-ablp210 and G418 resistance (32D-bcr/abl). These extracts were then subjected to Western blotting with a c-abl antisera (left panel). The locations of c-abl and bcr-ablp210 are indicated. A G418-resistant subclone was derived the 32D-bcr/abl pool by limiting dilution and was shown to express a similar level of bcr-ablp210 (not shown). This line was then transduced with the pBabePuro-C/EBPWT-ER retroviral vector. A subclone resistant to G418 and puromycin was selected. A total cellular protein extract derived from this line was subjected to Western blotting for C/EBP (right panel). (B) Total cellular RNAs were prepared from 32D-Neo and 32D-bcr/abl cells in IL-3 or after 0, 1, 2, 3, or 4 days in G-CSF. RNA samples were also prepared from 32D-bcr/abl cells cultured in G-CSF for 6, 8, or 10 days. These RNAs were then subjected to Northern blotting for MPO, GCSFR, and 18S RNAs. (C) 32D-bcr/abl, 32D-bcr/abl-WT-ER, Ba/F3(G), and Ba/F3(G)-WTER cells in IL-3 were cultured in the absence or presence of estradiol. Ba/F3(G) cells are a derivative of Ba/F3 cells which express the G-CSFR. Ba/F3(G)-WTER cells express C/EBPWT-ER in addition. Cell counts were enumerated daily with a hemocytometer and Trypan Blue Dye. Results shown are the mean of three determinations. SEs were less than 10% of each mean.
Total cellular RNAs were prepared from 32D-Neo cells cultured in IL-3 or in G-CSF for 1, 2, 3, or 4 days and from the 32D-bcr/abl subclone cultured in IL-3 or in G-CSF for 1, 2, 3, 4, 6, 8, or 10 days. The RNAs were subjected to Northern blotting for MPO, GCSFR, and 18S RNAs (Fig4B). bcr-ablp210 prevented induction of MPO and GCSFR RNA by G-CSF. bcr-ablp210 also prevented morphologic differentiation, alone or when coexpressed with exogenous G-CSFR, and prevented induction of PU.1, c-Myb, C/EBPα, and C/EBPβ by G-CSF (data not shown).
32D-bcr/abl and 32D-bcr/abl-αWT-ER cells were cultured in IL-3 in the presence or absence of estradiol, and viable cell counts were enumerated daily (Fig 4C, top panels). C/EBPα inhibited proliferation of 32D-bcr/abl cells, although MPO and LF RNAs were not induced (data not shown). We also transduced the C/EBPαWT-ER into Ba/F3(G) cells, a B-lymphoid cell line that required IL-3 for proliferation and that also expresses the G-CSFR. Ba/F3(G) and a Ba/F3(G) subclone expressing C/EBPαWT-ER were also cultured in IL-3 in the presence or absence of estradiol, and viable cell counts were enumerated daily (Fig4C, bottom panels). Activation of C/EBPαWT-ER markedly inhibited proliferation in this hematopoietic cell line, although MPO RNA was only slightly induced (data not shown). Thus, C/EBPα can slow the proliferation of myeloid or lymphoid cells without inducing granulocytic differentiation.
C/EBPα inhibits the G1/S transition in 32D cl3 cells.
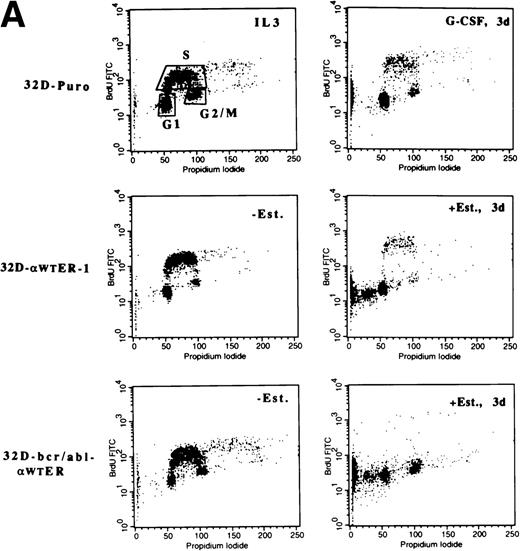
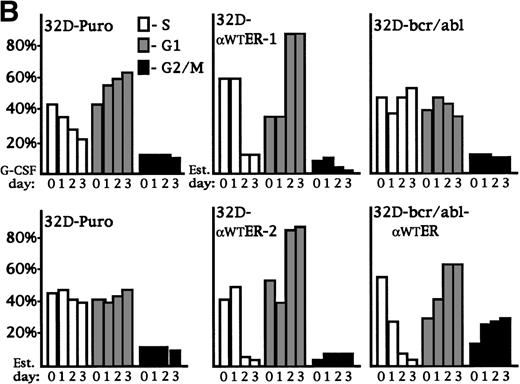
32D-Puro cells were cultured in IL-3 or in G-CSF for 1, 2, or 3 days. The proportion of cells in the G1, S, and G2/M cell cycle phases was determined by BrdU/PI staining (Fig 5A, top panels; Fig 5B, top left panel). As the 32D-Puro cells differentiated, they underwent a G1/S cell cycle arrest. 32D-Puro, 32D-αWTER-1, and 32D-bcr/abl-αWTER cells were cultured in IL-3 in the presence or absence of estradiol for 0, 1, 2, or 3 days. The proportion of cells in each cell cycle phase was determined similarly (Fig 5A and B). Activation of C/EBPαWT-ER in 32D cl3 or 32D-bcr/abl cells also induced a G1/S arrest. A G2/M arrest may also be present in 32D-bcr/abl-αWTER cells. In addition, G-CSF induced apoptosis in 10% of 32D-Puro cells over 3 days, and C/EBPαWT-ER induced apoptosis in 30% of 32D-αWTER-1 cells and in 50% of 32D-bcr/abl cells after 3 days (Fig 5A). Apoptosis may result from excessive levels of C/EBPα-ER in a subset of cells.
C/EBP inhibits the G1 to S transition and induces p27Kip1 levels and Rb hypophosphorylation in 32D cl3 cells. (A) 32D-Puro cells were cultured in IL-3 or in G-CSF for 1, 2, or 3 days. 32D-Puro, 32D-WT-ER-1, 32D-WT-ER-2, and 32D-bcr/abl, 32D-bcr/abl-WTER cells in IL-3 were exposed to estradiol for 0, 1, 2, or 3 days. The proportion of cells in the G1, S, and G2/M cell cycle phases was determined daily by BrdU/PI staining. Representative FACScan data from days 0 and 3 are shown for 32D-Puro cells in IL-3 or G-CSF and from 32D-WT-ER-1 and 32D-bcr/abl-WTER cells in IL-3 and estradiol. The location of G1, S, and G2/M cells is shown in the upper left panel. Signals to the left of the G1 population have less than 2n DNA content and represent cells undergoing apoptosis. (B) The proportion of cells in S, G1, or G2/M on each day for these six cultures is shown from a representative experiment. Cells with less than 2n DNA content were excluded from these calculations. (C) Total cellular protein extracts were prepared from 32D-Puro, 32D-WT-ER-1, and 32D-WT-ER-2 cells exposed in IL-3 to estradiol for 0, 1, 2, or 3 days. These extracts were then subjected to Western blotting for Rb, p21WAF1/CIP1, and p27Kip1. Retinoblastoma protein was detected as a doublet (Rb-P, Rb).
C/EBP inhibits the G1 to S transition and induces p27Kip1 levels and Rb hypophosphorylation in 32D cl3 cells. (A) 32D-Puro cells were cultured in IL-3 or in G-CSF for 1, 2, or 3 days. 32D-Puro, 32D-WT-ER-1, 32D-WT-ER-2, and 32D-bcr/abl, 32D-bcr/abl-WTER cells in IL-3 were exposed to estradiol for 0, 1, 2, or 3 days. The proportion of cells in the G1, S, and G2/M cell cycle phases was determined daily by BrdU/PI staining. Representative FACScan data from days 0 and 3 are shown for 32D-Puro cells in IL-3 or G-CSF and from 32D-WT-ER-1 and 32D-bcr/abl-WTER cells in IL-3 and estradiol. The location of G1, S, and G2/M cells is shown in the upper left panel. Signals to the left of the G1 population have less than 2n DNA content and represent cells undergoing apoptosis. (B) The proportion of cells in S, G1, or G2/M on each day for these six cultures is shown from a representative experiment. Cells with less than 2n DNA content were excluded from these calculations. (C) Total cellular protein extracts were prepared from 32D-Puro, 32D-WT-ER-1, and 32D-WT-ER-2 cells exposed in IL-3 to estradiol for 0, 1, 2, or 3 days. These extracts were then subjected to Western blotting for Rb, p21WAF1/CIP1, and p27Kip1. Retinoblastoma protein was detected as a doublet (Rb-P, Rb).
To investigate the mechanism of cell cycle arrest induced by C/EBPα, 32D-Puro, 32D-αWTER-1, and 32D-αWTER-2 cells were cultured in estradiol for 0, 1, 2, or 3 days. Total cellular proteins were prepared daily and were subjected to Western blotting for Retinoblastoma protein (Rb), p21WAF1/CIP1, and p27Kip1 (Fig 5C). Activation of C/EBPα led to Rb hypophosphorylation; the Rb-P:Rb ratio was equal to or greater than 1 for the 32D-Puro extracts, but became much less than 1 for the 32D-αWTER extracts on days 2 and 3. Estradiol induced either minimal or no change in p21 levels in the 32D-αWTER lines, but markedly induced p27 by day 2 in these lines. A mild increase in p27 was also evident on day 3 in the 32D-Puro cells, and basal levels of p21 and p27 differed between the three lines. Induction of p27Kip1 is also seen when 32D cl3 cells are exposed to G-CSF.49 Thus, C/EBPα slows cell cycle progression in 32D cl3 myeloblasts at or before the restriction point in G1 and may do so by inducing p27, either directly or indirectly.
C/EBPα rapidly induces PU.1 in 32D cl3 cells.
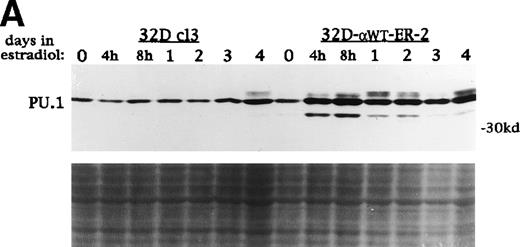
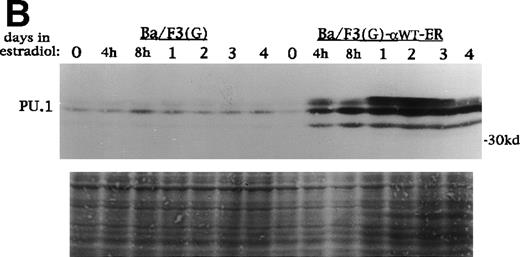
PU.1, c-Myb, C/EBPα, and C/EBPβ levels increase during G-CSF-induced 32D cl3 cell differentiation.1,6 26 To examine the effect of C/EBPαWT-ER on these factors, total cellular protein extracts were prepared from 32D cl3 and 32D-αWTER cells cultured in estradiol for 0 hours, 4 hours, 8 hours, 1 day, 2 days, 3 days, or 4 days. These extracts were subjected to Western blotting for PU.1, c-Myb, C/EBPα, and C/EBPβ. Activation of C/EBPαWT-ER resulted in increases in c-Myb, endogenous C/EBPα, and C/EBPβ, although these increases were delayed by at least 24 hours (data not shown). In contrast, PU.1 levels increased dramatically by 4 hours in 32D-αWTER-2 cells (Fig 6A) and in 32D-αWTER-1 cells (data not shown). Estradiol did not affect PU.1 expression in 32D cl3 cells (Fig 6A). A similar set of extracts from Ba/F3(G) and Ba/F3(G)-αWTER cells were also subjected to Western blotting for PU.1 (Fig 6B). Activation of C/EBP-αWTER rapidly induced expression of PU.1 in Ba/F3(G)-αWTER cells. In 32D-bcr/abl cells, C/EBP-αWTER also rapidly induced PU.1, but did not induce c-Myb, C/EBPβ, or endogenous C/EBPα (data not shown).
C/EBP rapidly induces PU.1 protein in 32D cl3 and Ba/F3 cells. (A) Protein extracts were prepared from 32D cl3 and 32D-WT-ER-2 cells in IL-3 exposed to estradiol for 0 hours, 4 hours, 8 hours, 1 day, 2 days, 3 days, or 4 days. These extracts were subjected to Western blotting for PU.1. Fast Green staining of the blot is shown as a control for protein loading. (B) Protein extracts were prepared from Ba/F3(G) and Ba/F3(G)-WT-ER cells cultured in IL-3 and exposed to estradiol for 0 hours, 4 hours, 8 hours, 1 day, 2 days, 3 days, or 4 days. These extracts were also subjected to Western blotting for PU.1.
C/EBP rapidly induces PU.1 protein in 32D cl3 and Ba/F3 cells. (A) Protein extracts were prepared from 32D cl3 and 32D-WT-ER-2 cells in IL-3 exposed to estradiol for 0 hours, 4 hours, 8 hours, 1 day, 2 days, 3 days, or 4 days. These extracts were subjected to Western blotting for PU.1. Fast Green staining of the blot is shown as a control for protein loading. (B) Protein extracts were prepared from Ba/F3(G) and Ba/F3(G)-WT-ER cells cultured in IL-3 and exposed to estradiol for 0 hours, 4 hours, 8 hours, 1 day, 2 days, 3 days, or 4 days. These extracts were also subjected to Western blotting for PU.1.
C/EBPα activates the endogenous PU.1 gene.
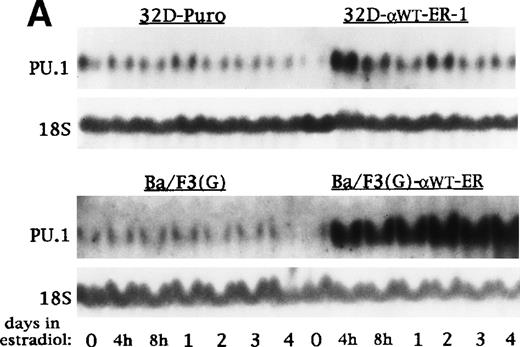
RNA samples were prepared from 32D-Puro, 32D-αWTER-1, Ba/F3(G), and Ba/F3(G)-αWTER cells cultured in estradiol for 0 hours, 4 hours, 8 hours, 1 day, 2 days, 3 days, or 4 days. These RNAs were subjected to Northern blotting for PU.1 and 18S RNA (Fig7A). Activation of C/EBPαWTER led to a marked increase in PU.1 RNA levels within 4 hours. RNA samples were then prepared from 32D-αWTER-1 and Ba/F3(G)-αWTER cells cultured in the presence of estradiol for 0 or 8 hours or in the presence of cycloheximide with or without estradiol for 8 hours. RNA samples were also prepared from 32D-αWTER-1 cells cultured with Actinomycin D with or without estradiol for 8 hours. These samples were subjected to Northern blotting for PU.1 and 18S RNA (Fig 7B). Estradiol induced PU.1 RNA despite the presence of cycloheximide, but not in the presence of Actinomycin D. Similar results were obtained in an additional experiment as well (data not shown). These data suggest that C/EBPα directly activates the PU.1 gene.
C/EBP induces transcription of the endogenous PU.1 gene in the absence of protein synthesis. (A) Total cellular RNAs were prepared from 32D-Puro, 32D-WT-ER-1, Ba/F3(G), and Ba/F3(G)-WT-ER cells proliferating in IL-3 with estradiol for 0 hours, 4 hours, 8 hours, 1 day, 2 days, 3 days, or 4 days. These RNAs were then subjected to Northern blotting for PU.1 and 18S RNAs. (B) The Northern blot described in Fig 2 was probed also for PU.1 RNA. The 18S RNA levels shown in Fig 2 are again shown (top panels). Total cellular RNAs were also prepared from Ba/F3(G)-WT-ER cells cultured in IL-3 with estradiol for 0 or 8 hours, from a third culture exposed to CHX for 8 hours, and from a fourth culture exposed to CHX for 30 minutes followed by CHX with estradiol for 8 hours. These RNAs were subjected to Northern blotting for PU.1 and 18S RNAs (bottom panels).
C/EBP induces transcription of the endogenous PU.1 gene in the absence of protein synthesis. (A) Total cellular RNAs were prepared from 32D-Puro, 32D-WT-ER-1, Ba/F3(G), and Ba/F3(G)-WT-ER cells proliferating in IL-3 with estradiol for 0 hours, 4 hours, 8 hours, 1 day, 2 days, 3 days, or 4 days. These RNAs were then subjected to Northern blotting for PU.1 and 18S RNAs. (B) The Northern blot described in Fig 2 was probed also for PU.1 RNA. The 18S RNA levels shown in Fig 2 are again shown (top panels). Total cellular RNAs were also prepared from Ba/F3(G)-WT-ER cells cultured in IL-3 with estradiol for 0 or 8 hours, from a third culture exposed to CHX for 8 hours, and from a fourth culture exposed to CHX for 30 minutes followed by CHX with estradiol for 8 hours. These RNAs were subjected to Northern blotting for PU.1 and 18S RNAs (bottom panels).
PU.1 is not sufficient to induce terminal granulopoiesis in 32D cl3 cells.
Rapid induction of PU.1 RNA by C/EBPαWT-ER raises the possibility that increased levels of PU.1 might be sufficient to induce terminal granulocytic differentiation and cell cycle arrest in 32D cl3 cells. To test this possibility, we expressed PU.1-ER(T), a fusion protein containing the entire PU.1 polypeptide and a modified estrogen receptor ligand-binding domain responsive to 4HT but not estradiol. pB4TKCAT contains four binding sites for the PU.1:Pip heterodimer, derived from the λ2-4 enhancer, adjacent to the thymidine kinase promoter and the chloramphenicol acyl-transferase (CAT) cDNA.48 PU.1-ER(T) activated transcription from pB4TKCAT with Pip only in the presence of 4HT (Fig 8A). Antisense PU.1 did not stimulate pB4TKCAT in the presence of 4HT (Fig 8A). PU.1-ER(T) also induced the endogenous M-CSFR gene in hematopoietic cells derived from PU.1−/− ES cells (D. Weil and E.S., unpublished data).
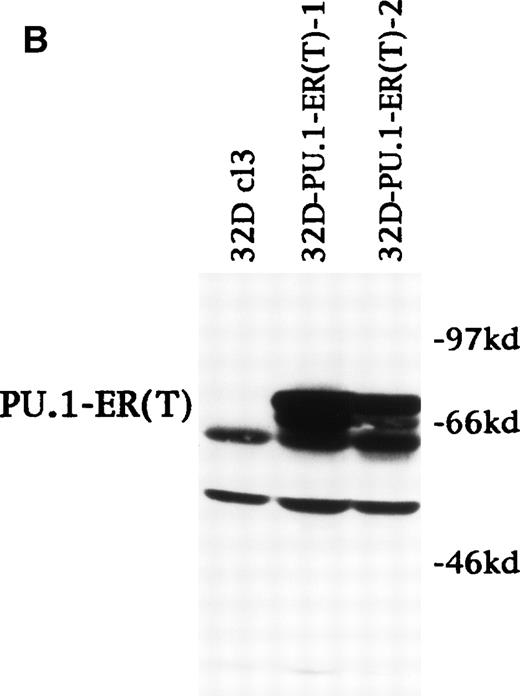
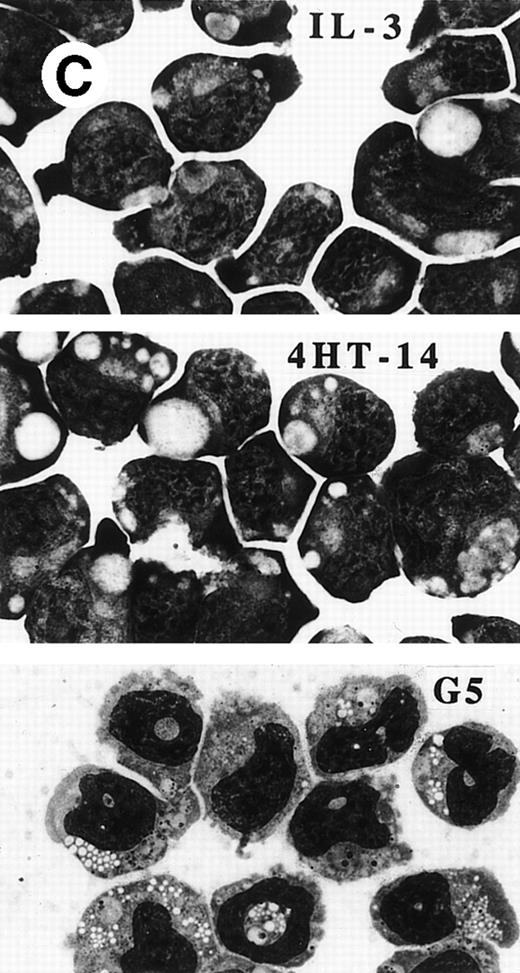
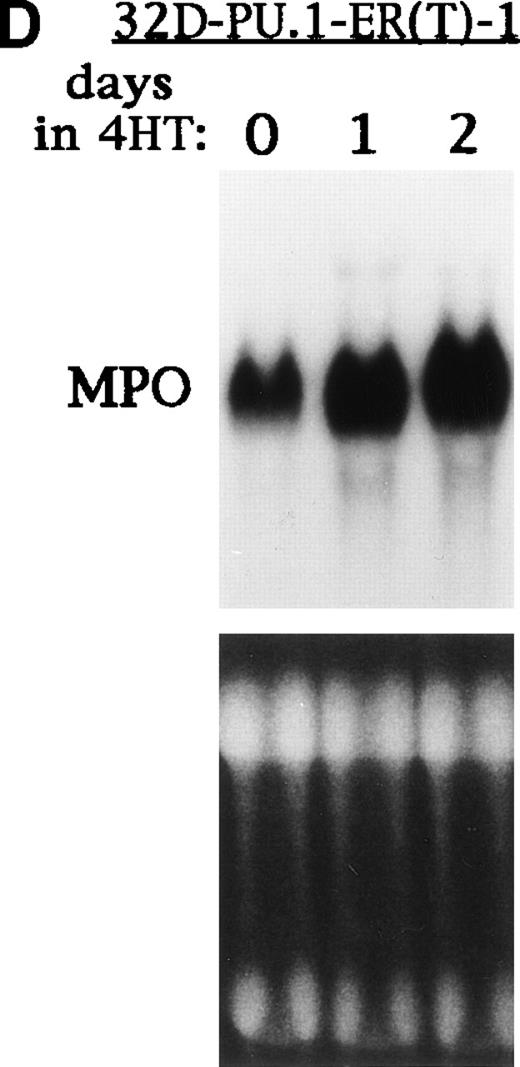
PU.1-ER(T) induces MPO RNA, but does not induce terminal granulopoiesis in 32D cl3 myeloblasts. (A) Ten micrograms of pB4TKCAT was transfected into NIH 3T3 cells with 10 μg pCMV-Pip and 10 μg of pPGK-PU.1 or pPGK-PU.1-ER(T) and was cultured with or without 100 nmol/L 4HT (lanes 1 through 4). pB4TKCAT was similarly transfected with pCMV-Pip and pPGK-AS-PU.1, which expresses an antisense PU.1 RNA, and cultured with 4HT (lane 5). CAT assays performed 48 hours after transfection are shown. (B) Total cellular protein extracts prepared from 32D cl3 cells and from two puromycin-resistant subclones transduced with the BabePuro-PU.1-ER(T) retroviral vector were subjected to Western blotting using an ER antisera. The location of PU.1-ER(T) is indicated. (C) Cytospins were prepared from 32D-PU.1-ER(T) cells in IL-3, in IL-3 and after exposure to 4HT for 14 days, and after transfer from IL-3– to G-CSF–containing media for 5 days. The cytospins were subjected to Wright’s-Giemsa staining. (D) Total cellular RNAs were prepared from 32D-PU.1-ER(T) cells in IL-3 after exposure to 4HT for 0, 1, or 2 days. These RNAs were subjected to Northern blotting for MPO RNA (top panel). The integrity of the RNA samples was assessed by ethidium bromide staining (bottom panel).
PU.1-ER(T) induces MPO RNA, but does not induce terminal granulopoiesis in 32D cl3 myeloblasts. (A) Ten micrograms of pB4TKCAT was transfected into NIH 3T3 cells with 10 μg pCMV-Pip and 10 μg of pPGK-PU.1 or pPGK-PU.1-ER(T) and was cultured with or without 100 nmol/L 4HT (lanes 1 through 4). pB4TKCAT was similarly transfected with pCMV-Pip and pPGK-AS-PU.1, which expresses an antisense PU.1 RNA, and cultured with 4HT (lane 5). CAT assays performed 48 hours after transfection are shown. (B) Total cellular protein extracts prepared from 32D cl3 cells and from two puromycin-resistant subclones transduced with the BabePuro-PU.1-ER(T) retroviral vector were subjected to Western blotting using an ER antisera. The location of PU.1-ER(T) is indicated. (C) Cytospins were prepared from 32D-PU.1-ER(T) cells in IL-3, in IL-3 and after exposure to 4HT for 14 days, and after transfer from IL-3– to G-CSF–containing media for 5 days. The cytospins were subjected to Wright’s-Giemsa staining. (D) Total cellular RNAs were prepared from 32D-PU.1-ER(T) cells in IL-3 after exposure to 4HT for 0, 1, or 2 days. These RNAs were subjected to Northern blotting for MPO RNA (top panel). The integrity of the RNA samples was assessed by ethidium bromide staining (bottom panel).
Using retroviral transduction, we obtained 12 32D cl3 subclones expressing PU.1-ER(T). The two lines with the highest level of expression were selected for evaluation (Fig 8B). In IL-3, line 1 expressed PU.1-ER(T) at a level similar to endogenous PU.1 (data not shown). 32D cl3, 32D-PU.1-ER(T)-1, and 32D-PU.1-ER(T)-2 cells were cultured in 4HT for 14 days. Cell counts were obtained and cell morphology was assessed every 1 to 2 days. The ability of 32D-PU.1-ER(T)-1 and 32D-PU.1-ER(T)-2 cells to differentiate morphologically in G-CSF was also assessed. After 5 days in G-CSF, 32D-PU.1-ER(T)-1 cells displayed evidence of granulocytic differentiation, including cytoplasmic granulation and nuclear changes (Fig 8C, bottom panel). However, no evidence of differentiation was evident when these cells were exposed to 4HT in IL-3, even after 14 days (Fig 8C, center panel). Similar results were obtained with 32D-PU.1-ER(T)-2 cells (data not shown). Activation of PU.1-ER(T) led to increased MPO RNA levels in 32D-PU.1-ER(T)-1 cells after 24 hours (Fig 8D). In a second experiment with this cell line, activation of PU.1-ER(T) similarly induced MPO RNA by 24 hours, but no induction was seen at 8 hours (data not shown). PU.1-ER(T) inhibited 32D-PU.1-ER(T)-1 cell proliferation twofold over a 3-day period, whereas 4HT did not affect 32D cl3 cell proliferation during this time. During the subsequent 10 days, each of the lines proliferated at similar rates in the presence or absence of 4HT (data not shown). These data indicate that induction of PU.1 by C/EBPα is not sufficient to induce terminal differentiation or cell cycle arrest in 32D cl3 cells.
DISCUSSION
To gain further insight into the role of C/EBPα in neutrophil development, we first expressed an estradiol-inducible form of C/EBPα, C/EBPαWT-ER, in 32D cl3 cells. 32D cl3 cells resemble CFU-GM, although inhibition of G-CSF–induced differentiation of 32D cl3 cells by IL-3 is at odds with the properties of this myeloid progenitor. We demonstrate that C/EBPαWT-ER rapidly induced a full program of granulopoiesis in 32D cl3 cells proliferating in IL-3, including an early increase in MPO RNA, followed by a delayed cell cycle arrest in G1, an increase in LF and G-CSFR RNAs, and the development of a neutrophilic morphology. Moreover, both G-CSF–induced and C/EBPα-induced 32D cl3 cell differentiation resulted in increased expression of PU.1, c-Myb, C/EBPα, C/EBPβ, and p27Kip1.
Increased expression of PU.1 protein and RNA occured very rapidly, and PU.1 RNA was induced by C/EBPα in the presence of cycloheximide but not that of Actinomycin D. Thus, C/EBPα may directly activate transcription of the endogenous PU.1 gene. Of note, PU.1 mRNA can be detected in fetal liver cells derived from C/EBPα null mice.50 This result demonstrates that PU.1 transcription can occur in vivo in the absence of C/EBPα, as we also observe in Ba/F3 cells. The murine PU.1 promoter contains several sites, at −225, −175, −75, and −65, that resemble the consensus for C/EBPα DNA-binding.51 In future experiments we intend to determine whether C/EBPα binds and activates the PU.1 gene via these or other sites.
Having made these observations, we considered the possibility that expression of PU.1 inducibly in 32D cl3 cells might reproduce the effects observed with C/EBPαWT-ER. Activation of PU.1-ER(T) did lead to increased expression of MPO RNA, but did not lead to cell cycle arrest or terminal granulopoiesis. It is noteworthy that induction of MPO RNA by C/EBPα-WTER was sensitive to cycloheximide, indicating the requirement for an additional factor, perhaps PU.1.2Because induction of MPO RNA by PU.1-ER(T) was not evident at 8 hours, inhibition by cycloheximide could not be assessed. Also, whereas we readily obtained 32D cl3 lines that expressed C/EBPαWT-ER at levels that exceeded that of endogenous C/EBPα, PU.1-ER(T) could at best be expressed at a level that equalled that of endogenous PU.1. Nevertheless, our results are consistent with the reported phenotype of 32D cl3 cells stably overexpressing PU.1 at high levels: 32D/PU.1 cells proliferated as undifferentiated myeloblasts at a rate similar to control cells in IL-3 and differentiated more rapidly to neutrophils than control lines in G-CSF.52 Together with the observation that PU.1-deficient mice retain some neutrophilic potential,18,19 53 these results indicate that the PU.1 gene is not the only C/EBPα target required for granulopoiesis.
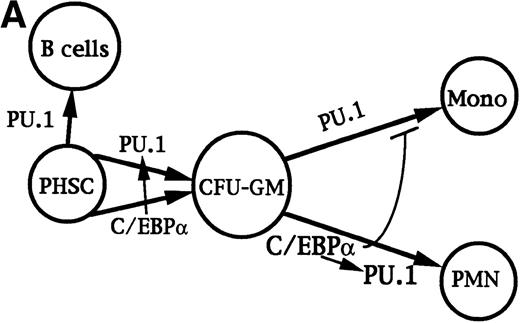
C/EBPα-deficient mice retained normal levels of CFU-GM and CFU-M, indicating that C/EBPα is not absolutely required for the development of these progenitors. Also, overexpression of either PU.1 or C/EBPβ in an avian multipotent progenitor induced myeloid lineage commitment.24,54 To account for these results and those discussed above, we propose a model in which either PU.1 or C/EBPα can commit multipotent cells to myelopoiesis. Perhaps C/EBPα stimulates PU.1 expression even in such early progenitors. Increased C/EBPα activity resulting from as yet unknown mechanisms then directs myeloid progenitors along the granulocytic lineage and in so doing further activates the PU.1 gene. Increased C/EBPα activity may also inhibit monocytic development.25 PU.1 is required for further development along the monocytic lineage, perhaps in cooperation with c-Jun,55 and also for B-lymphoid development. This model is depicted in Fig 9A.
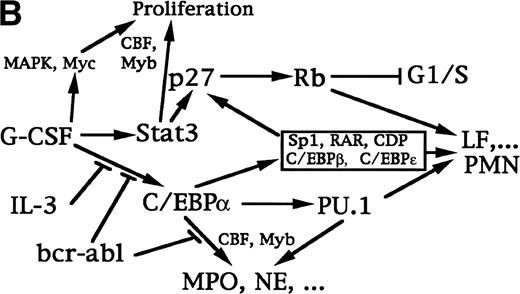
Transcriptional regulation of myelopoiesis. (A) Model for the transcriptional regulation of commitment to the granulocytic and monocytic lineages. Arrows represent stimulation and the perpendicular bar indicates inhibition. C/EBP or PU.1 commit pluripotent hematopoietic stem cells (PHSC) to CFU-GM. C/EBP may modestly induce PU.1 in these cells. Increased activity of C/EBP stimulates granulopoiesis, with further induction of PU.1, and may inhibit monocyte development.25 PU.1 is required for terminal monocyte differentiation and is also required for B-lymphoid development. (B) Model for the transcriptional program regulating granulopoiesis in committed progenitors. G-CSF activates several signal transduction pathways that allow cell proliferation and stimulate differentiation, including the Ras/MAPK pathway, the Jak/Stat pathway, and induction of c-Myc. The CBF and c-Myb transcription factors also stimulate proliferation. Removal of IL-3 signals, addition of G-CSF signals, or both lead to elevated C/EBP levels, a phenomenon inhibited by bcr/abl. C/EBP induces increased levels of PU.1 and, together with CBF and c-Myb, these factors then activate early markers of myeloid differentiation, including the MPO and NE genes. C/EBP also leads to a delayed increased in p27Kip1, in cooperation with Stat3 and other factors. p27 in turn induces Rb hypophosphorylation and a G1/S arrest. Hypophosphorylated Rb, PU.1, and several other transcription factors expressed, activated, or inactivated at a later stage in granulopoiesis (eg, C/EBPβ, C/EBPɛ, Sp1, RAR, and loss of CDP) may then induce late differentiation markers, such as the LF gene and genes required to induce the neutrophilic morphology.
Transcriptional regulation of myelopoiesis. (A) Model for the transcriptional regulation of commitment to the granulocytic and monocytic lineages. Arrows represent stimulation and the perpendicular bar indicates inhibition. C/EBP or PU.1 commit pluripotent hematopoietic stem cells (PHSC) to CFU-GM. C/EBP may modestly induce PU.1 in these cells. Increased activity of C/EBP stimulates granulopoiesis, with further induction of PU.1, and may inhibit monocyte development.25 PU.1 is required for terminal monocyte differentiation and is also required for B-lymphoid development. (B) Model for the transcriptional program regulating granulopoiesis in committed progenitors. G-CSF activates several signal transduction pathways that allow cell proliferation and stimulate differentiation, including the Ras/MAPK pathway, the Jak/Stat pathway, and induction of c-Myc. The CBF and c-Myb transcription factors also stimulate proliferation. Removal of IL-3 signals, addition of G-CSF signals, or both lead to elevated C/EBP levels, a phenomenon inhibited by bcr/abl. C/EBP induces increased levels of PU.1 and, together with CBF and c-Myb, these factors then activate early markers of myeloid differentiation, including the MPO and NE genes. C/EBP also leads to a delayed increased in p27Kip1, in cooperation with Stat3 and other factors. p27 in turn induces Rb hypophosphorylation and a G1/S arrest. Hypophosphorylated Rb, PU.1, and several other transcription factors expressed, activated, or inactivated at a later stage in granulopoiesis (eg, C/EBPβ, C/EBPɛ, Sp1, RAR, and loss of CDP) may then induce late differentiation markers, such as the LF gene and genes required to induce the neutrophilic morphology.
Because C/EBPα substituted for G-CSF signals to induce 32D cl3 cell differentiation, G-CSF may induce C/EBPα expression or stimulates its activity in normal myeloid progenitors. A proposed transcriptional program regulating granulopoiesis, beginning with G-CSF signals, which takes account of our observations and prior results (as reviewed56 and/or discussed herein), is shown in Fig 9B.
C/EBPα was first shown to induce cell cycle arrest in adipocytes.44 C/EBPα was later shown to induce a G1/S cell cycle arrest in hepatocytes and to induce p21WAF1/CIP1in fibrosarcoma cells.57,58 We observed that activation of C/EBPαWT-ER led to a delayed increase in p27Kip1, but did not increase p21WAF1/CIP1, as is also observed when 32D cl3 cells differentiate in G-CSF.49 Interestingly, dominant-negative STAT3 prevents G-CSF–induced growth arrest of myeloid cell lines,49,59 and STAT3 can directly activate the p27Kip1 gene.49 Perhaps C/EBPα induces the expression of a factor, such as C/EBPβ or C/EBPε, which cooperates with STAT3 to activate the p27Kip1 gene.
bcr-ablp210 prevented G-CSF–induced 32D cl3 cell differentiation. Interestingly, bcr-ablp210 also prevented C/EBPαWT-ER–induced 32D cl3 cell granulopoiesis. These effects of bcr-ablp210 might be relevant to the transition of CML from chronic phase to blast crisis. However, bcr-ablp210 did not prevent C/EBPαWT-ER from inducing PU.1, indicating that bcr-ablp210 does not inhibit granulopoiesis by inactivating C/EBPα via protein modification.
C/EBPα was previously shown to be capable of inducing granulopoiesis when expressed inducibly in U937 cells.25 We have extended those observations by showing that induction of granulopoiesis by C/EBPα can occur also in 32D cl3 cells, which more closely resemble normal myeloblasts. Moreover, C/EBPα-induced granulopoiesis in 32D cl3 cells occurs over a 4-day period, proceeds in the absence of G-CSF signals and despite the presence of IL-3 signals, is associated with rapid induction of PU.1 even in the presence of cycloheximide, and is associated with delayed induction of p27 and a G1/S cell cycle arrest.
ACKNOWLEDGMENT
The authors thank S. Nagata for the murine GCSFR cDNA; M. Britos-Bray, W. Wang, and D. Weil for technical assistance; and C. Civin for assistance with FACScan analysis.
Supported in part by National Institutes of Health Grants No. HL15388 (A.D.F.) and CA72769 (E.S.). A.D.F., E.S., and C.L.S. are Leukemia Society Scholars.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Alan D. Friedman, MD, Johns Hopkins Oncology Center, Room 3-109, 600 N Wolfe St, Baltimore, MD 21287; e-mail: adfrdman@jhmi.edu.







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal