TCRBV (T-cell receptor B variable) gene usage and CDR3 size distribution were analyzed using reverse transcription polymerase chain reaction (RT-PCR) to assess the T-cell repertoire of 10 patients with B-cell chronic lymphocytic leukemia (B-CLL) and in nine age-matched healthy control donors. When the usage of each TCRBV gene within the CD8+ T cells of the patients was compared with that of the controls, no statistically significant difference was noted except for BV 6S1-3. In contrast, within the CD4+ T cells of the CLL patients, a statistically significant overexpression for four BV families (2, 3, 5S1, 6S1-3) was seen while an underrepresentation was noted for five BV families (10, 11, 15, 16, 19). Based on the criterion that a value of any BV higher than the mean + 3 standard deviation (SD) of healthy controls indicated an overexpression, individual patients were shown to overexpress several TCRBV genes compared with the controls. Analyses of the CDR3 length polymorphism showed a significantly higher degree of restriction within CD4+ and CD8+ T cells of the patients, as compared with the corresponding control T-cell population. There was a significant difference in the CDR3 size distribution pattern with a more polymorphic CDR3 length pattern in the age-matched controls as compared with CLL patients, suggesting different mechanisms driving the T cells towards a clonal/oligoclonal TCRBV usage in patients and controls, respectively. The results show major perturbations of T cells in CLL patients, more frequently seen in the CD4+ T-cell subset, indicating that nonmalignant CD4+ T cells may be involved in the pathogenesis of CLL, but also CD8+ T cells.
B-CELL CHRONIC lymphocytic leukemia (B-CLL) is characterized by proliferation and accumulation of clonal CD5+ B cells expressing surface membrane immunoglobulins that share idiotypic determinants attesting to the monoclonal origin of the cells.1,2 In these patients, who are mainly above the age of 50, abnormal T-cell functions have been reported.3-7
Most circulating mature T cells use the α/β heterodimeric T-cell receptors (TCR) for specific recognition of antigenic peptides in context of major histocompatibility complex (MHC) molecules.8,9 Genes encoding the variable domains of the α and β polypeptide chains of the TCR are assembled by somatic recombination of one of each of variable (V), diversity (D, only for β chain), and joining (J) exons. Three hypervariable or complementarity determining regions (CDR1, CDR2, CDR3) have been defined for the two TCR chains. The portion of the TCR mainly responsible for the specific interaction with the antigenic peptide is the CDR3 loops of the variable domain.10 Specific recognition of peptide might result in a clonal expansion of T cells exhibiting TCR V gene products with identical CDR3 regions and CDR3 size distribution. Analyses of CDR3 size distribution has been used to define the degree of clonality of T cells in complex cell populations.11
An important issue to consider when studying TCR repertoires in normal and pathological conditions is that expanded oligoclonal T-cell populations are also found in normal adults, mainly elderly individuals.12-15 This phenomenon might cause misinterpretation in diseases, especially in those that mainly affect elderly people, as B-CLL.
The functional significance of nonmalignant T cells in CLL is an important issue to establish. A deeper understanding of the specificity of T cells in B-CLL may be of paramount importance for the future development of new therapeutic strategies. Oligoclonal TCR V gene usage in CLL patients has been reported earlier.16 17 In the present study, we have used reverse transcription polymerase chain reaction (RT-PCR) and the CDR3 size distribution pattern to analyze the TCRBV gene usage in CD4+ and CD8+ T cells of CLL patients and compared the results with those of age-matched control donors. The data indicated a preferentially perturbed CD4 T-cell population in CLL patients.
MATERIALS AND METHODS
Patients and controls.
Ten CLL patients (three males and seven females) with a mean age of 69 years (range, 38 to 82 years) were studied. The diagnostic criteria have been described earlier.18 The staging system according to Rai et al was applied.19 Clinical characteristics are shown in Table 1. Patients were considered to have progressive disease (PD),20 if there was a progression during the preceding 3 months in disease-related anemia (Hb < 100 g/L), in thrombocytopenia (platelet count < 100 × 109/L), and/or in spleen/liver/lymph node size (evaluated by both clinical examination and computed tomography of the abdomen), and/or in more than a doubling of the blood lymphocyte count and/or appearance of constitutional symptoms. Otherwise, B-CLL patients were defined as having nonprogressive disease (NPD). No patient had received any treatment for at least 3 months before testing, except for patient CLL 13 who received chlorambucil one month before sampling. Nine healthy control donors (one male and eight females) with an average age of 60 years (range, 54 to 67 years) were included.
Characteristics of the CLL Patients
| Patient . | Age (yr) . | Sex . | Rai Stage . | Disease Activity . | WBC Count (×109/L) . | Total T-cell Counts (×109/L)* . | ||
|---|---|---|---|---|---|---|---|---|
| CD3+ . | CD4+ . | CD8+ . | ||||||
| CLL 2 | 67 | F | III | PD | 212.0 | 2.12 | 1.21 | 0.74 |
| CLL 3 | 38 | F | 0 | NPD | 30.0 | 4.20 | 2.39 | 1.81 |
| CLL 7 | 81 | M | I | PD | 30.0 | 0.90 | 0.31 | 0.55 |
| CLL 9 | 63 | M | 0 | NPD | 14.0 | 2.24 | 0.90 | 1.34 |
| CLL 10 | 62 | F | I | NPD | 24.1 | 1.70 | 1.05 | 0.63 |
| CLL 11 | 68 | F | I | NPD | 83.2 | 17.47 | 12.41 | 4.72 |
| CLL 13 | 75 | F | II | PD | 71.2 | 1.42 | 0.80 | 0.62 |
| CLL 14 | 82 | F | III | NPD | 37.1 | 0.74 | 0.27 | 0.33 |
| CLL 15 | 77 | F | III | PD | 137.9 | 2.76 | 1.23 | 1.53 |
| CLL 16 | 81 | M | II | PD | 326.0 | 4.89 | 1.86 | 3.00 |
| Patient . | Age (yr) . | Sex . | Rai Stage . | Disease Activity . | WBC Count (×109/L) . | Total T-cell Counts (×109/L)* . | ||
|---|---|---|---|---|---|---|---|---|
| CD3+ . | CD4+ . | CD8+ . | ||||||
| CLL 2 | 67 | F | III | PD | 212.0 | 2.12 | 1.21 | 0.74 |
| CLL 3 | 38 | F | 0 | NPD | 30.0 | 4.20 | 2.39 | 1.81 |
| CLL 7 | 81 | M | I | PD | 30.0 | 0.90 | 0.31 | 0.55 |
| CLL 9 | 63 | M | 0 | NPD | 14.0 | 2.24 | 0.90 | 1.34 |
| CLL 10 | 62 | F | I | NPD | 24.1 | 1.70 | 1.05 | 0.63 |
| CLL 11 | 68 | F | I | NPD | 83.2 | 17.47 | 12.41 | 4.72 |
| CLL 13 | 75 | F | II | PD | 71.2 | 1.42 | 0.80 | 0.62 |
| CLL 14 | 82 | F | III | NPD | 37.1 | 0.74 | 0.27 | 0.33 |
| CLL 15 | 77 | F | III | PD | 137.9 | 2.76 | 1.23 | 1.53 |
| CLL 16 | 81 | M | II | PD | 326.0 | 4.89 | 1.86 | 3.00 |
Abbreviation: WBC, white blood cell.
At the time when the T cell repertoire was analyzed.
Purification of CD4+ and CD8+ T cells.
Peripheral blood mononuclear cells (PBMC) were isolated from heparinized venous blood by Ficoll-Paque (Amersham Pharmacia Biotech, Uppsala, Sweden) gradient centrifugation. T cells were enriched by nylon wool purification. Furthermore, a positive selection of CD4 and CD8 T cells, respectively, was performed by collecting from a column the effluent cells using anti-CD4 and anti-CD8 monoclonal antibodies (MoAb) coupled to magnetic beads (Dynal A/S, Oslo, Norway), according to the manufacturer’s instructions.
Immunofluorescent (IFL) staining and flow cytometry.
The purity of the cell populations was determined in triple staining using flow cytometry. Briefly, 0.5 × 106 purified T cells or PBMC were stained with fluorochrome-conjugated MoAbs. Fluorescein isothiocyanate (FITC)–labeled anti-CD3, phycoerythrin (PE)-conjugated anti-CD4 or anti-CD8, and peridinin chlorophyll (PerCP)-conjugated anti-CD19 MoAb (Becton Dickinson, Mountain View, CA) were added simultaneously, and the cells were incubated on ice for 20 minutes. The cells were then washed twice with ice-cold phosphate-buffered saline (PBS) and analyzed by FACScan using the software program Cellquest (Becton Dickinson) and a Power Macintosh 7300 computer (Apple computer Inc, Silicon Valley, CA).
RNA extraction and cDNA synthesis.
Total RNA was extracted from 1 to 2 × 106CD4+ or CD8+ T cells, using RNAzol B (ams, Biotechnology Europe, Stockholm, Sweden) based on the guanidine thiocyanate phenol-chloroform extraction method.21 RNA was denaturated at 90°C for 5 minutes and immediately chilled on ice. First-strand cDNA synthesis was performed in a 20-μL reaction mixture containing 3 μg RNA in 10 mL volume, 4 μL 5× Bethesda Research Laboratories (BRL) buffer, 1.5 μL dithiothreitol (DTT; 100 mmol/L, BRL), 2 μL dNTP (5 mmol/L each; Amersham Pharmacia Biotech), 1.0 μL random hexamer primers (100 pmol/μL; Amersham Pharmacia Biotech), 0.5 μL RNasin (20 U/μL; Promega, Madison, WI), and 1.0 μL of murine Molony leukemia virus reverse transcriptase (RT; 200 U/μL, BRL). The reaction mixture was incubated at 40°C for 45 minutes, followed by 5 minutes at 95°C to inactivate RT, and then stored at −20°C.
Normalization of TCR β chain–specific cDNA concentration.
The TCR β chain–specific cDNA concentration was normalized using a TCR beta chain constant region (BC)–specific polymerase chain reaction (PCR) by amplifying part of the TCR BC gene.22 23 Briefly, a 5′ sense, “5′ BC” primer, and a 3′ anti-sense, “3′ BC” primer (Table 2), both specific for TCR BC1 and TCR BC2 genes, were used in a 30-cycle PCR with serial twofold dilutions of cDNA. The PCR products were then electrophoresed on 1.5% ethidium bromide–stained agarose gels and photographed using Polaroid 665 films (Polaroid AB, Stockholm, Sweden). The TCR BC–specific bands (∼140 bp) on the negative films were quantified by gel scanning (2400 Gel Scan XL; Amersham Pharmacia Biotech). Based on the scanned data, equal amounts of TCR β chain–specific cDNA were used in the subsequent TCR BV PCR amplifications.
Sequences of Oligonucleotide Primers and Probes Used for PCR Amplification and Detection of TCR β Chain Specific Transcripts
| Primers | |
| TCRBV 1 | 5′-GCA CAA CAG TTC CCT GAC TTG CAC-3′ |
| TCRBV 2 | 5′-TCA TCA ACC ATG CAA GCC TGA CCT-3′ |
| TCRBV 3 | 5′-GGG GTA CAG TGT CTC TAG AGA GA-3′ |
| TCRBV 4 | 5′-ACA TAT GAG AGT GGA TTT GTC ATT-3′ |
| TCRBV 5S1 | 5′-ATA CTT CAG TGA GAC ACA GAG AAA C-3′ |
| TCRBV 5S2-3 | 5′-TTC CCT AAC TAT AGC TCT GAG CTG-3′ |
| TCRBV 6S1-3 | 5′-AGG CCT GAG GGA TCC GTC TC-3′ |
| TCRBV 7 | 5′-CCT GAA TGC CCC AAC AGC TCT C-3′ |
| TCRBV 8 | 5′-ATT TAC TTT AAC AAC AAC GTT CCG-3′ |
| TCRBV 9 | 5′-CCT AAA TCT CCA GAC AAA GCT CAC-3′ |
| TCRBV 10 | 5′-CTC CAA AAA CTC ATC CTG TAC CTT-3′ |
| TCRBV 11 | 5′-TCA ACA GTC TCC AGA ATA AGG ACG-3′ |
| TCRBV 12 | 5′-AAA GGA GAA GTC TCA GAT-3′ |
| TCRBV 13S1 | 5′-CAA GGA GAA GTC CCC AAT-3′ |
| TCRBV 13S2 | 5′-GGT GAG GGT ACA ACT GCC-3′ |
| TCRBV 14 | 5′-GTC TCT CGA AAA GAG AAG AGG AAT-3′ |
| TCRBV 15 | 5′-AGT GTC TCT CGA CAG GCA CAG GCT-3′ |
| TCRBV 16 | 5′-AAA GAG TCT AAA CAG GAT GAG TCC-3′ |
| TCRBV 17 | 5′-CAG ATA GTA AAT GAC TTT CAG-3′ |
| TCRBV 18 | 5′-GAT GAG TCA GGA ATG CCA AAG GAA-3′ |
| TCRBV 19 | 5′-CAA TGC CCC AAG AAC GCA CCC TGC-3′ |
| TCRBV 20 | 5′-AGC TCT GAG GTG CCC CAG AAT CTC-3′ |
| BC primer | 5′-TTC TGA TGG CTC AAA CAC-3′ |
| 3′ BC | 5′-GTG CAC CTC CTT CCC ATT-3′ |
| 5′ BC | 5′-GTC GCT GTG TTT GAG CCA TCA GAA-3′ |
| BC reporter | 5′-CAC AGC GAC CTC GGG TGG GAA CAC-3′ |
| Primers | |
| TCRBV 1 | 5′-GCA CAA CAG TTC CCT GAC TTG CAC-3′ |
| TCRBV 2 | 5′-TCA TCA ACC ATG CAA GCC TGA CCT-3′ |
| TCRBV 3 | 5′-GGG GTA CAG TGT CTC TAG AGA GA-3′ |
| TCRBV 4 | 5′-ACA TAT GAG AGT GGA TTT GTC ATT-3′ |
| TCRBV 5S1 | 5′-ATA CTT CAG TGA GAC ACA GAG AAA C-3′ |
| TCRBV 5S2-3 | 5′-TTC CCT AAC TAT AGC TCT GAG CTG-3′ |
| TCRBV 6S1-3 | 5′-AGG CCT GAG GGA TCC GTC TC-3′ |
| TCRBV 7 | 5′-CCT GAA TGC CCC AAC AGC TCT C-3′ |
| TCRBV 8 | 5′-ATT TAC TTT AAC AAC AAC GTT CCG-3′ |
| TCRBV 9 | 5′-CCT AAA TCT CCA GAC AAA GCT CAC-3′ |
| TCRBV 10 | 5′-CTC CAA AAA CTC ATC CTG TAC CTT-3′ |
| TCRBV 11 | 5′-TCA ACA GTC TCC AGA ATA AGG ACG-3′ |
| TCRBV 12 | 5′-AAA GGA GAA GTC TCA GAT-3′ |
| TCRBV 13S1 | 5′-CAA GGA GAA GTC CCC AAT-3′ |
| TCRBV 13S2 | 5′-GGT GAG GGT ACA ACT GCC-3′ |
| TCRBV 14 | 5′-GTC TCT CGA AAA GAG AAG AGG AAT-3′ |
| TCRBV 15 | 5′-AGT GTC TCT CGA CAG GCA CAG GCT-3′ |
| TCRBV 16 | 5′-AAA GAG TCT AAA CAG GAT GAG TCC-3′ |
| TCRBV 17 | 5′-CAG ATA GTA AAT GAC TTT CAG-3′ |
| TCRBV 18 | 5′-GAT GAG TCA GGA ATG CCA AAG GAA-3′ |
| TCRBV 19 | 5′-CAA TGC CCC AAG AAC GCA CCC TGC-3′ |
| TCRBV 20 | 5′-AGC TCT GAG GTG CCC CAG AAT CTC-3′ |
| BC primer | 5′-TTC TGA TGG CTC AAA CAC-3′ |
| 3′ BC | 5′-GTG CAC CTC CTT CCC ATT-3′ |
| 5′ BC | 5′-GTC GCT GTG TTT GAG CCA TCA GAA-3′ |
| BC reporter | 5′-CAC AGC GAC CTC GGG TGG GAA CAC-3′ |
Analysis of TCRBV gene usage.
PCR amplifications of BV-specific cDNA were performed using a panel of 22 TCR BV–specific 5′ primers and a TCR BC–specific 3′ primer, “BC (B constant) primer” (Table 2). This BC primer also recognizes sequences in both BC1 and BC2 genes.22 PCR mixtures contained 10× concentrated buffer (100 mmol/L Tris-HCl, 15 mmol/L MgCl2, 500 mmol/L KCl, 1 mg/mL gelatine, pH 8.3; Boehringer Mannheim Scandinavia AB, Stockholm, Sweden); 0.2 μmol/L final concentration of each of dNTPs (Amersham Pharmacia Biotech); 0.1 μL DNA Taq polymerase (5 U/μL; Boehringer Mannheim Scandinavia AB); and 0.5 μmol/L of each primer. Samples were overlayed with mineral oil and amplified in a Perkin-Elmer thermocycler (Perkin-Elmer Applied Biosystems Division, Norwalk, CT) for 32 cycles with a temperature profile of 94°C for denaturation, 55°C for annealing, and 72°C for extension. Each step lasted for 30 seconds, except for the last extension that continued for an additional 9 minutes to ensure complete extension of the products. To ensure the expected size of the amplified BV-BC PCR products, 8 μL of each of the products was electrophoresed on 1.5% ethidium bromide–stained agarose gels and photographed.
Southern blot analysis of TCRBV-BC PCR products.
PCR products were subjected to electrophoresis on 1.5% agarose gels. The gels were denatured using a solution containing 0.5 mol/L NaOH and 1.5 mol/L NaCl for 15 minutes and then neutralized in 0.5 mol/L Tris-HCl and 1.5 mol/L NaCl for 15 minutes, followed by incubation for 20 minutes in 10× SSC. The products were transferred onto nylon membranes (Amersham Pharmacia Biotech).24 Membranes were air dried and baked for 2 hours at 80°C.
5′ end-labeling of BC oligonucleotide probe and hybridization to PCR products.
A TCR BC–specific oligonucleotide probe, “BC reporter” (Table2), recognizing both BC1 and BC223 was designed, synthesized (Scandinavian Gene Synthesis AB, Köping, Sweden), and 5′-end labeled24 using T4 polynucleotide kinase enzyme (Amersham Pharmacia Biotech) and γ-32P-adenosine triphosphate (ATP) (Amersham Pharmacia Biotech). The baked nylon membranes were first prehybridized in a buffer containing 2× SSPE, 5× Denhardt’s, and 0.5% sodium dodecyl sulfate (SDS) for 3 hours at 42°C followed by hybridization in the same buffer with 1 × 106 cpm/mL of the 5′-end labeled BC reporter overnight at 42°C. They were then washed twice at 42°C in a washing solution containing 0.2 × SSPE and 0.5% SDS and were subsequently exposed to Hyper Film MP (Amersham Pharmacia Biotech) for 6 hours at −70°C.
Quantification of the relative usage of BV genes.
The intensity of the signals exerted by specific probing of the TCR BV-BC amplified products on the films was quantified by gel scanning. The relative usage of each BV gene was determined using a standard curve and a computer program.13
Analysis of CDR3 length polymorphism.
The CDR3 length polymorphism analysis was performed as described.11,25 Briefly, cDNA samples were amplified in 40 cycles of PCR using BV 2, 3, 4, 5S1, 6S1-3, 7, 8, and 9 as 5′ primers and 3′ BC-FITC as the 3′ primer. The products were then checked on 1.5% ethidium bromide–stained agarose gels. An aliquot was loaded onto 6% denaturing polyacrylamide sequencing gels, and the electrophoresis was run in the presence of fluorescent size markers (Amersham Pharmacia Biotech), in an ALF-DNA sequencing machine (Amersham Pharmacia Biotech). The data were collected by a computer and analyzed by the “Fragment Manager” software program (Amersham Pharmacia Biotech). A dominant CDR3 peak (or peaks) was defined as a high-intensity signal with a dramatic reduction of other CDR3 signals within the particular TCRBV family, ie, when the counts in a single peak were ≥50% of the total counts.11 17
Statistical analysis.
The nonparametric Wilcoxon-Mann-Whitney two-tailed rank sum test was used for comparison of independent samples. For comparison of dependent observations, the two-tailed nonparametric Wilcoxon signed rank test was applied. For frequency comparisons, Fisher’s exact two-tailed test was applied. A perturbed T-cell subset was defined as a subset statistically significantly different from that of the corresponding control population.
RESULTS
TCRBV gene usage in CD4+ and CD8+ T cells.
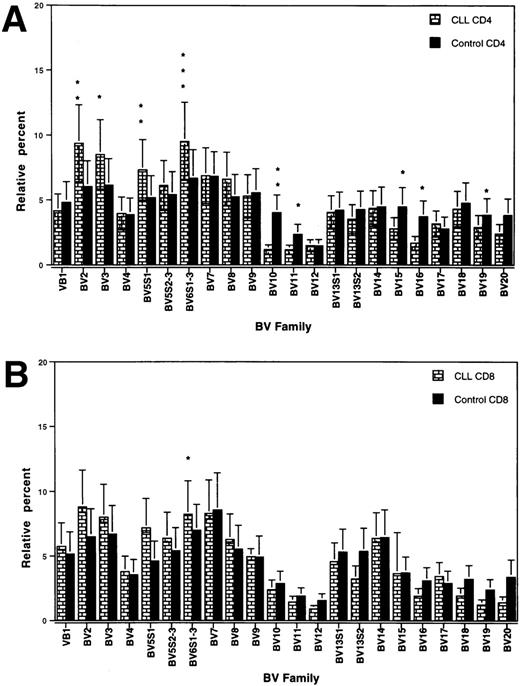
The total T-cell counts of the patients at test are shown in Table 1. The purification procedure yielded >95% enriched T cells of the respective population. The relative usage of different BV genes in CD4+ and CD8+ T cells, respectively, of patients and control donors, is shown in Fig1. In the CD4 population, a significant bias towards a higher usage of BV 2, 3, 5S1, and 6S1-3 and a lower usage of BV 10, 11, 15, 16, and 19 was observed in the patients as compared with controls (Fig 1A). In the CD8+ T-cell subset, a bias towards a higher usage in the patients was only confined to BV 6S1-3 (Fig 1B).
TCRBV gene segment usage (mean ± SEM) in CD4 (A) and CD8 (B) T cells of CLL patients and control donors. ∗, ∗∗, and ∗∗∗ indicate statistically significant differences atP values of <0.05, <0.01, and <0.001, respectively.
TCRBV gene segment usage (mean ± SEM) in CD4 (A) and CD8 (B) T cells of CLL patients and control donors. ∗, ∗∗, and ∗∗∗ indicate statistically significant differences atP values of <0.05, <0.01, and <0.001, respectively.
BV gene overexpression in individual patients.
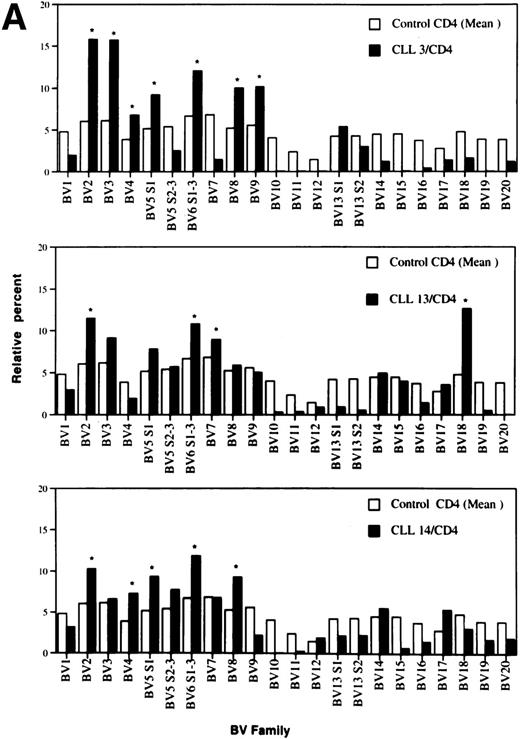
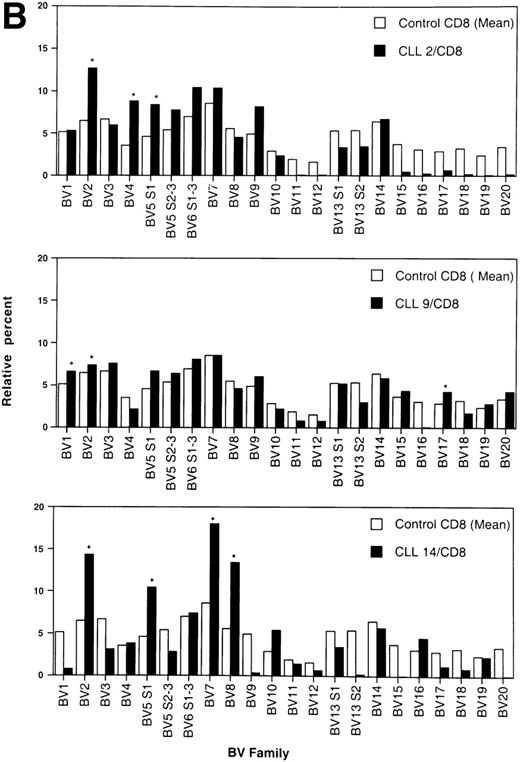
By convention, a value of any BV higher than the mean + 3 standard deviation (SD) of healthy age-matched controls was considered an overexpression.26 BV overexpressions in the CD4+ and CD8+ T-cell populations of individual patients are shown in Table 3. In total, there were overexpressions of 20 different TCRBV families in the CD4 population as compared with 15 in the CD8 T-cell subset. Only in three patients could no BV overexpression in the CD4 subpopulation be detected. All these patients had PD. In contrast, three of four patients with no TCRBV overexpression within the CD8+ subset had NPD. None of the age-matched controls showed any overexpression, neither in the CD4 nor in the CD8 T-cell subset. Representative examples of the TCRBV usage patterns in three CLL patients who showed overexpressions within the CD4+ T-cell population of more than 3 BV families are shown in Fig 2A. The pattern of three representative patients with overexpression of 3 or more BV families within the CD8+ T subset is depicted in Fig 2B.
Relative TCRBV gene segment usage of three representative CLL patients compared with the mean control values for CD4 (A) and CD8 (B) T-cell subsets. *Indicates a statistically significant overexpression as defined in Table 4.
Relative TCRBV gene segment usage of three representative CLL patients compared with the mean control values for CD4 (A) and CD8 (B) T-cell subsets. *Indicates a statistically significant overexpression as defined in Table 4.
TCRBV transcript CDR3 length polymorphism.
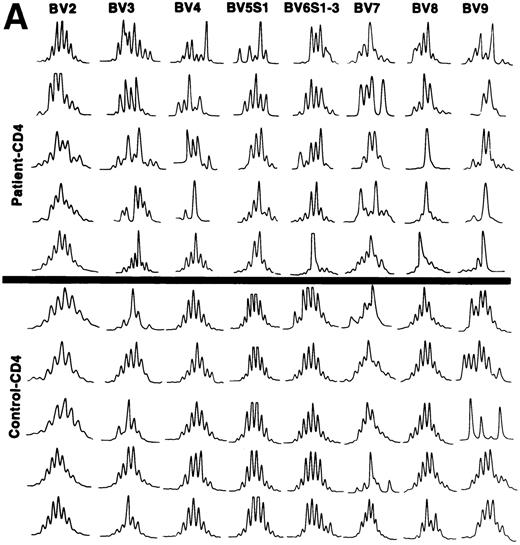
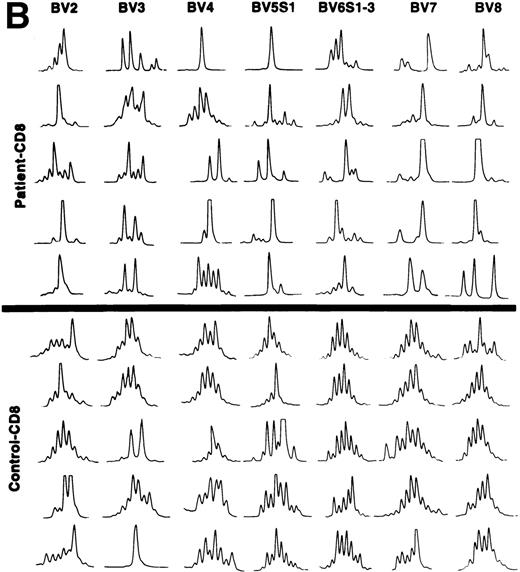
The CDR3 size distribution pattern of a defined BV transcript might give information with regard to an antigen-driven restriction of the BV gene usage. A monoclonal BV pattern represented by one single peak of the CDR3 distribution pattern indicates recognition of one single conventional antigen.11 A superantigen-driven process is expected to show a polyclonal CDR3 size pattern. To define the CDR3 size distribution of TCRBV transcripts, we used the method described by Pannetier et al.11 BV genes included in the analyses were selected from those showing overexpression in individual patients (Table 3). The tests were performed on the same T-cell samples as the BV gene usage analyses. In general, a restricted pattern of the CDR3 size distribution in the patients was observed (data not shown). All our patients were elderly individuals, except for patient no. 3, who was 38 years old. Several reports have shown a more restricted CDR3 length pattern, mainly within CD8+ T cells, in older compared with young, normal individuals.12,15 27 A key question is thus whether the restricted pattern of CDR3 length polymorphism in our patients was related to age or could be caused by other factors, maybe a response against the tumor cells. To address this issue, we compared the numbers of different CDR3 lengths for each BV gene of patients and age-matched controls, respectively. The results are shown in Table4. In 9 of 15 analyses, a statistically significant difference was observed comparing the number of CDR3 peaks with a more restricted CDR3 length pattern (fewer peaks) used by the patients as compared with the controls. This indicates that in addition to age, other mechanism(s) might be involved in shaping the TCRBV repertoire of the CLL patients. Examples of the CDR3 size distributions of different BV transcripts in CD4+ and CD8+ T cells of patients and controls are shown in Fig 3A and B. Although a few cases of restricted usage of BV genes with dominant CDR3 lengths were detected in control samples, such cases and also cases with only one dominant peak (suggesting monoclonality) were much more abundant in the CLL patients (P < .01 for CD4+ T cells andP < .05 for CD8+ T cells).
Overexpression of Individual TCRBV Gene Segments in CD4+ and CD8+ T Cells, Respectively, of 10 CLL Patients Compared With Age-Matched Healthy Controls
| Patients . | BV-CD4 . | BV-CD8 . |
|---|---|---|
| CLL 2 | 7 | |
| CLL 3 | 2, 3, 4, 5S1, 6S1-3, 8, 9 | 2, 4, 5S1 |
| CLL 7 | None | None |
| CLL 9 | 4 | 2, 5S1 |
| CLL 10 | 7 | 1, 2, 17 |
| CLL 11 | 7 | None |
| CLL 13 | 2, 6S1-3, 7, 18 | None |
| CLL 14 | 2, 4, 5S1, 6S1-3, 8 | None |
| CLL 15 | None | 2, 5S1, 7, 8 |
| CLL 16 | None | 5S1, 8 |
| 11 |
| Patients . | BV-CD4 . | BV-CD8 . |
|---|---|---|
| CLL 2 | 7 | |
| CLL 3 | 2, 3, 4, 5S1, 6S1-3, 8, 9 | 2, 4, 5S1 |
| CLL 7 | None | None |
| CLL 9 | 4 | 2, 5S1 |
| CLL 10 | 7 | 1, 2, 17 |
| CLL 11 | 7 | None |
| CLL 13 | 2, 6S1-3, 7, 18 | None |
| CLL 14 | 2, 4, 5S1, 6S1-3, 8 | None |
| CLL 15 | None | 2, 5S1, 7, 8 |
| CLL 16 | None | 5S1, 8 |
| 11 |
A TCRBV gene segment was regarded as overexpressed when the relative frequency was more than the mean + 3 SD for that of the control donors (n = 9).
Statistical Comparison (P values) of CDR3 Lengths of TCR β Chains Using Different V Genes in CLL Patients (n = 10) and Age-Matched Healthy Controls (n = 9)
| BV Genes . | CD4+ Subset . | CD8+ Subset . |
|---|---|---|
| BV2 | .2352 | .1768 |
| BV3 | .2758 | .1819 |
| BV4 | .00414-150 | .00624-150 |
| BV5S1 | .0616 | .02604-150 |
| BV6S1-3 | .00214-150 | .00604-150 |
| BV7 | .8340 | .00534-150 |
| BV8 | .00124-150 | .04644-150 |
| BV9 | .01684-150 | 4-151 |
| BV Genes . | CD4+ Subset . | CD8+ Subset . |
|---|---|---|
| BV2 | .2352 | .1768 |
| BV3 | .2758 | .1819 |
| BV4 | .00414-150 | .00624-150 |
| BV5S1 | .0616 | .02604-150 |
| BV6S1-3 | .00214-150 | .00604-150 |
| BV7 | .8340 | .00534-150 |
| BV8 | .00124-150 | .04644-150 |
| BV9 | .01684-150 | 4-151 |
BV genes included in the calculations were selected based on the data in Table 3.
Statistically significant differences comparing the CDR3 lengths of the CLL patients and the control donors, with a skew towards a more polymorphic CDR3 length pattern in the control donors.
No TCRBV overexpression was found.
(A) CDR3 length polymorphism in CD4 T cells of CLL patients expressing TCRBV 2, 3, 4, 5 S1, 6 S1-3, 7, 8, and 9 genes and control donors. Results of five individuals are shown. (B) CDR3 length polymorphism in CD8 T cells of CLL patients expressing TCRBV 2, 3, 4, 5 S1, 6 S1-3, 7, and 8 genes and control donors. Results of five individuals are shown.
(A) CDR3 length polymorphism in CD4 T cells of CLL patients expressing TCRBV 2, 3, 4, 5 S1, 6 S1-3, 7, 8, and 9 genes and control donors. Results of five individuals are shown. (B) CDR3 length polymorphism in CD8 T cells of CLL patients expressing TCRBV 2, 3, 4, 5 S1, 6 S1-3, 7, and 8 genes and control donors. Results of five individuals are shown.
DISCUSSION
There have been a few attempts to characterize the TCR usage in CLL patients. In the first report, clonally expanded TCR beta chain rearrangements were detected by Southern blot analysis.28Analyses of TCRAV and TCRBV gene usages of unfractionated T cells showed overrepresentation of unique TCRAV or BV gene segment transcripts by PCR, as well as expansions of TCRBV-expressing T cells using flow cytometry and mAbs. A CD8+/BV19+T-cell clone from one CLL patient recognized specifically the autologous tumor cells.16 A recent study addressed the question of clonality of T cells within the CD4+ and CD8+ subsets by using a multiplex PCR assay for CDR3 length. Oligoclonal expansions were predominantly found within the CD57+ subset of CD4+ T cells as well as of CD8+ T cells in CLL patients.17
In the present study, we used RT-PCR and CDR3 size distribution technologies to study in detail the TCR repertoire of B-CLL patients and healthy age-matched control donors. A statistically significant overexpression within the CD4+ T-cell subset of four BV families (BV 2, 3, 5S1, 6S1-3) was noted in the CLL patients, whereas five families (BV 10, 11, 15, 16, 19) showed underrepresentation compared with control donors. In contrast, only one BV family was overexpressed (BV 6S1-3) within the CD8+ T cells of the patients. In normal individuals, clonal expansions are more frequently seen within the CD8+ T-cell subset than within the CD4+ T cells.29 Thus, our results indicated a perturbed CD4+ population in CLL patients. The oligoclonal/monoclonal pattern of the CDR3 length pattern of CD4+ T cells in the CLL patients was in contrast with the Gaussian-like CDR3 distribution of control CD4+ T cells, which may further strengthen the assumption of a perturbed CD4+ population in the CLL patients. The CD8+subset of the CLL patients also showed an increased frequency of clonal T-cell expansions measured by TCR-CDR3 length analyses as compared with control donors. The relatively more often noted clonally or restricted pattern of TCR usage within CD8+ cells of older individuals12,15,27 30 was overwhelmed by the pattern seen in the CLL patients. When the degree of clonality was analyzed and comparisons were made between patients and controls, the difference for CD4+ T cells was, however, even more obvious than for CD8+ cells (P < .01 for CD4+ T cells and P < .05 for CD8+ T cells). The clonally perturbed CD4+ population in the CLL patients might imply that these expanded cells may have been involved in an MHC class II–restricted recognition. CLL B cells express MHC class II molecules, which are restriction elements for antigen presentation to CD4+ T cells.
The specificity and functional properties of the clonal T cells in B-CLL patients are not yet known. They are unlikely to be induced by superantigens, but may rather recognize peptide antigens in a conventional manner. Do these T cells recognize structures on the tumor clone, or are they driven by other antigens as might be the case for a normal old population? In a recent study, a multiple myeloma patient was shown to have three major expanded BV T-cell populations. None of the expansions reacted with the tumor-derived idiotypic immunoglobulin. However, a nonexpanded T-cell population (BV22) reacted specifically with the tumor-associated idiotype, the population of which was shown to be highly restricted by CDR3 size distribution analysis.26 In a CLL patient, it was shown that a clonally expanded BV19 T-cell population recognized specifically the tumor cells.16 Overexpression of restricted TCRBV gene segments is also a common feature of tumor-infiltrating lymphocytes. Mostly, 1 to 3 TCRAV or TCRBV regions have been shown to be overexpressed, and the pattern differed for each patient.31 In melanoma patients matched for HLA-A2, known to be a dominant restriction element for several melanoma-associated antigenic peptides, TCRBV14 was overexpressed in all studied lesions.32 33 TCRBV14 overexpression was not observed in HLA-A2–negative patients. The overexpression was specific for the neoplastic tissue.
The mechanisms responsible for driving these T cells towards clonality and maintaining them as clonal expansions in CLL are not yet understood. Whether such expanded T cells might be involved in a specific recognition of the tumor cells or may just be part of a general immune dysfunction in CLL are important issues to be addressed.
In conclusion, this study shows that TCRBV overexpressions in CLL are more frequently seen within the CD4 T-cell population than within the CD8 T-cell subset. CDR3 size analyses revealed a higher degree of clonality within CD4 T cells than within CD8 T cells. These are novel observations. Taken together, the TCRBV overexpressions and the monoclonal/oligoclonal pattern of TCRBV CDR3 length may suggest that CD4 T cells might be involved in the disease process in CLL but maybe also CD8 T cells. CD4 T cells may recognize MHC class II–restricted peptides presented on the leukemic B cells. MHC class II–restricted T cells specifically recognizing the CDR3 region of the tumor-derived immunoglobulin heavy chain have been identified in the blood of CLL patients (own unpublished data).
ACKNOWLEDGMENT
The excellent secretarial help of Gunilla Buren and Gerd Ståhlberg is highly appreciated.
Supported by grants from the Cancer Society in Stockholm, the King Gustav Vth Jubilee Fund, the Swedish Cancer Society, the Åke Wiberg Foundation, the Cancer and Allergy Foundation, and the Gunnar Nilsson Foundation.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Mahmood Jeddi-Tehrani, PhD, Immune and Gene Therapy Lab, Cancer Center Karolinska (CCK), Karolinska Hospital, S-17176 Stockholm, Sweden.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal