The BCR/ABL hybrid gene plays a central role in the pathogenesis of the chronic phase of chronic myeloid leukemia (CML). We used a very sensitive quantitative reverse transcriptase-polymerase chain reaction to investigate the levels of hybrid BCR/ABL mRNA in bone marrow cells of 20 patients with Philadelphia positive (Ph+) CML treated with interferon- (IFN-) as a single agent. Bone marrow samples were collected at diagnosis and at hematologic remission induced by IFN-, or by hydroxyurea in case of resistance to IFN-. The mean levels of BCR/ABL transcripts in bone marrow mononuclear cells of patients who showed a complete hematologic response to IFN- were significantly reduced with respect to those at diagnosis (48 × 103v168 × 103; P < .001), whereas no difference was detected between the values at diagnosis and at hematologic remission in patients resistant to IFN-. In cell culture experiments, IFN- priming significantly reduced the levels of BCR/ABL hybrid transcripts in a dose-dependent manner in Ph+ bone marrow precursors obtained at diagnosis from patients who subsequently responded to IFN- treatment (P < .005). No downmodulation was observed in bone marrow precursors from patients who subsequently proved to be IFN-resistant. These results indicate that downmodulation of BCR/ABL gene expression could be one of the mechanisms involved in the response of CML patients to IFN- treatment.
THE BCR/ABL hybrid gene and its resulting chimeric P210BCR/ABL protein play a crucial role in chronic myeloid leukemia (CML).1 Experimental evidence indicates that this hybrid gene may be the only genetic lesion in the chronic phase of CML because its introduction into primitive murine cells results in a syndrome similar to human CML.2 The BCR/ABL hybrid gene sustains the abnormal expansion of myeloid committed precursors3,4 that characterizes the chronic phase of CML. The mechanisms of transformation by the P210BCR/ABL protein have yet to be fully clarified. In vitro, the P210BCR/ABLprotein (1) increases the survival of hematopoietic precursors and cell lines by blocking apoptosis5-7; and (2) enhances cell proliferation by MYC upregulation8; and (3) reduces differentiation capacity by RAS activation.9 However, it is not clear how these pathways affect the in vivo clinical situation.10
Interferon-α (IFN-α) belongs to a cytokine family that exhibits antiviral properties, immunomodulating effects, and antiproliferative activity on normal and transformed cells.11 These diverse properties are mediated through binding to a high-affinity cell surface receptor12 which, in turn, activates a variety of intracellular signals and modulates gene expression.13 At present, IFN-α is the drug of choice in the treatment of CML patients who cannot undergo allogeneic bone marrow transplantation.14-16 However, the response to IFN-α treatment is variable. The drug induces complete hematologic remission in up to 70% to 80% of patients after 6 to 12 months, but is ineffective in 20% to 30% of cases. Moreover, in 40% to 60% of patients, IFN-α treatment significantly reduces the percentage of Ph+ cells, an effect termed cytogenetic response,which is maximal after 12 to 24 months of treatment.15,17,18 However, the mechanism by which IFN-α controls the growth of Ph+ CML precursors is still unclear. There is evidence that enhancement of immunosurveillance through the induction of the expression of leukocyte function associated antigen-3 (LFA-3)19 and the reversal of deficient adhesion of CML cells to stromal elements might be involved in the therapeutic effects of IFN-α.20 However, recent studies failed to reveal LFA-3 induction by IFN-α21 or to document any antiproliferative effect of CML precursor binding to marrow stroma.3
We and others showed that IFN-α upmodulates membrane expression of FAS receptor.22-24 Recently we have also shown that, in CML, FAS-mediated apoptosis is restricted to IFN-α responder patients.25 Furthermore, in KT-1 cells (a human cell line derived from a CML patient), IFN-α reduces the expression of the BCR/ABL gene.26
In this report we show that in vivo and in vitro IFN-α treatment reduces the intracellular amounts of the BCR/ABL hybrid transcript and of the corresponding P210BCR/ABL protein, and that, as in the case of induction of FAS receptor expression,26 this effect is restricted to IFN-α responder patients.
MATERIALS AND METHODS
Patients.
After informed consent, 20 patients affected by CML were enrolled in the study (Table 1). The diagnosis was confirmed by the cytogenetic finding of the Ph chromosome and by the molecular finding of the hybrid BCR/ABL gene. All patients were treated with IFN-α as single agent according to the Italian Cooperative Study Group (ICSG) CML protocol.15
Clinical Features of CML Patients Enrolled in the Study
| . | Total . | IFN Responders* . | IFN Nonresponders* . |
|---|---|---|---|
| No. of cases | 20 | 14 | 6 |
| Follow-up (mo) | |||
| Median | 7.5 | 7 | 8 |
| Range | 6-24 | 6-24 | 6-19 |
| WBC at Diagnosis (×103/μL) | |||
| Median | 119 | 112 | 126 |
| Range | 41-363 | 41-363 | 94-264 |
| Platelets at Diagnosis (No. of cases) | |||
| <400,000/μL | 6 | 4 | 2 |
| >400,000/μL | 14 | 10 | 4 |
| Splenomegaly at Diagnosis (No. of cases) | |||
| Absent | 5 | 3 | 2 |
| <5 cm | 8 | 5 | 3 |
| >5 cm | 7 | 6 | 1 |
| . | Total . | IFN Responders* . | IFN Nonresponders* . |
|---|---|---|---|
| No. of cases | 20 | 14 | 6 |
| Follow-up (mo) | |||
| Median | 7.5 | 7 | 8 |
| Range | 6-24 | 6-24 | 6-19 |
| WBC at Diagnosis (×103/μL) | |||
| Median | 119 | 112 | 126 |
| Range | 41-363 | 41-363 | 94-264 |
| Platelets at Diagnosis (No. of cases) | |||
| <400,000/μL | 6 | 4 | 2 |
| >400,000/μL | 14 | 10 | 4 |
| Splenomegaly at Diagnosis (No. of cases) | |||
| Absent | 5 | 3 | 2 |
| <5 cm | 8 | 5 | 3 |
| >5 cm | 7 | 6 | 1 |
See text for the operational definition of hematologic response to IFN-α treatment.
Bone marrow samples were collected from the patients before the start of treatment and when the patients were in complete hematologic remission because of either IFN-α (responders) or hydroxyurea after IFN-α withdrawal (nonresponders). The molecular analysis was performed within a few days of sampling, without cell cryopreservation.
Our operational definition of the response to IFN-α treatment after 6 months of the maximum tolerated daily dosage (6 or 9 million units) was: (1) complete hematologic response: if white blood cell (WBC) <10 × 109/L, circulating immature hematopoietic precursors <5% in the differential, platelet <500 × 109/L, and spleen not palpable; (2) insufficient hematologic response (NR) if WBC were >10 × 109/L, immature hematopoietic precursors >5% in the differential, platelet >500 × 109/L, or persistent splenomegaly.
Bone marrow cell separation.
Bone marrow was aspirated from the posterior iliac crest into syringes containing 1:10 EDTA solution. Bone marrow mononuclear cells (MNC) were isolated by Ficoll density gradient centrifugation at 800g for 30 minutes (Ficoll-Paque; Pharmacia Biotech, Sweden), and washed in phosphate buffer saline (PBS).
Hematopoietic suspension cultures.
Bone marrow MNC isolated from 9 CML untreated patients were resuspended within a few hours from withdrawal in Iscove’s modified Dulbecco’s medium (IMDM) containing 20% fetal calf serum (FCS), and aliquots of 2 million cells were incubated for 24 hours in 24-well plates at 37°C with increasing concentrations of IFN-α, ranging from 20 to 1,000 U/mL, which encompasses the levels of IFN-α reached in blood (50 to 100 U/mL) during the treatment of CML patients. A control culture without IFN-α was also prepared for each set of cells. IMDM and FCS were purchased from Life Technologies Italia (Milan, Italy) and IFN-α from Hoffman La Roche (Basel, Switzerland).
Detection and quantitation of hybrid BCR/ABL mRNA.
Total RNA was extracted from bone marrow MNC using the acid guanidinium thiocyanate and phenol-chloroform method.27 Any contaminating DNA detected was digested using 5 U of RNAse-free DNAse I (Boehringer Mannheim Italia, Monza, Italy), and purified RNA was re-extracted using the phenol-chloroform method.27 At least 40 μg of RNA was extracted from each sample, corresponding to 4 to 8 million cells. The reverse transcriptase-polymerase chain reaction (RT-PCR) analysis was performed as described elsewhere.28Briefly, 1 μg of total RNA extracted was prewarmed for 10 minutes at 60°C and then incubated for 60 minutes at 37°C in a 40 μL reaction mixture containing 10 mmol/L Tris HCl (pH 8.3), 50 mmol/L KCl, 5 mmol/L MgCl2, 1 mmol/L of each deoxyribonucleotide, 40 U of RNAsin (Promega, Madison, WI), 2.5 mmol/L of an antisense primer, 5′-TGTGATTATAGCCTAAGACCCGGAG-3′, that hybridizes to sequences of the second ABL exon, and 100 U of MoMLV reverse transcriptase (BRL, Bethesda, MD). A 20-μL aliquot of this solution was amplified by PCR in a 100 μL final volume of a mixture containing 10 mmol/L Tris HCl (pH 8.3), 50 mmol/L KCl, 2 mmol/L MgCl2, 0.2 mmol/L of each deoxyribonucleotide, 2.5 U of Taq polymerase, 0.5 μmol/L of the same antisense ABL primer, 5′-TGTGATTATAGCCTAAGACCCGGAG-3′, and 0.5 μmol/L of a sense primer, 5′-GAAGAAGTGTTTCAGAAGCTTCTCCC-3′, complementary to sequences of BCR exons b1 and b2. The ABL sequences were amplified in the remaining 20-μL aliquot of reverse transcriptase reaction mixture, as a control of both RNA integrity and the PCR reaction. Both amplification reactions consisted of 30 cycles with the following temperature cycle: 95°C for 30 seconds; 55°C for 30 seconds; 72°C for 30 seconds. PCR products were analyzed on a 2% agarose gel containing ethidium bromide.
The absolute amount of BCR/ABL transcript was quantitated as previously described.29 30 Briefly, 2 aliquots (500 and 250 ng, respectively) of total RNA extracted from each sample were reverse transcribed and the 2 cDNAs were PCR-amplified independently in 2 tubes, using a radiolabeled deoxynucleotide. The PCR conditions, ie, the total number of amplification cycles and the reaction mixture, were standardized to ensure a constant amplification efficiency throughout all amplification cycles and to terminate the reaction during the exponential phase of the amplification. Under these conditions there is a log-log linear relation between the number of starting molecules of BCR/ABL mRNA contained in the sample and the amount of PCR products estimated by the radioactivity of the amplified bands, measured by densitometric analysis using a computerized apparatus, the Molecular Imager (Bio-Rad Laboratories, Hercules, CA), which is equipped with a phosphor crystal screen and a laser screen-scanner. The absolute number of BCR/ABL mRNA molecules was calculated by interpolating the amount of PCR products with the titration curve obtained by amplifying, in parallel with the samples, known amounts of a “synthetic” RNA molecule whose sequence is identical to that of the BCR/ABL mRNA being quantitated. The reduction (approximately 2-fold) of the number of molecules calculated in the 250-ng aliquot of total RNA analyzed as compared with the 500-ng sample is a control that the amplification efficiency is constant in the samples being analyzed, and, hence, that the assay is accurate. All samples that differed by more than 15% from the results obtained with the 500-ng and 250-ng aliquots were reanalyzed.
The limiting number of amplification cycles up to which amplification proceeds with a constant efficiency was previously calculated by amplifying various sets of scalar dilutions from 500 to 5 ng of total RNA by RT-PCR using a decreasing number of cycles (data not shown). After each experiment, logarithmically transformed data were analyzed by linear regression, and the highest number of cycles that ensured a log-log linear relationship between RNA sample dilutions and amplified products was used in the assays. The synthetic RNA used as a standard was synthesized in vitro using a 2-step procedure, as previously described.29
The RT and PCR assay conditions differed slightly from those used for the qualitative assay: the aliquots of total RNA were incubated for 60 minutes at 37°C in a 20-μL reaction mixture containing 20 mmol/L Tris HCl (pH 8.3), 50 mmol/L KCl, 5 mmol/L MgCl2, 0.5 mmol/L of each deoxyribonucleotide, 2.5 U of RNAsin, 0.75 μmol/L of antisense ABL primer (see above), and 50 U of MoMLV reverse transcriptase; the reaction was stopped by heating at 95°C for 10 minutes. The PCR amplification of the cDNA obtained was performed in a reaction mixture containing 20 mmol/L Tris HCl (pH 8.3), 50 mmol/L KCl, 2.5 mmol/L MgCl2, 0.1 μmol/L of both sense and antisense primers (see above), 0.1 mmol/L of each deoxyribonucleotide, 2 mCi of [α32P] dCTP, and 2.5 U of Taq DNA polymerase. The reaction was performed using a total of 20 cycles and the quantitative frame, ie, the linearity range of the assay under these conditions was between 1.2 × 106 and 6 × 103molecules.
Southern analysis of the M-BCR region.
High-molecular-weight genomic DNA was extracted, using the standard phenol/chloroform method,31 and the relative proportion of Ph+ and Ph− cells was assessed by densitometric evaluation of Southern analysis of M-BCR.32Briefly, for the identification of the BCR/ABL fused gene, 15 μg of genomic DNA were digested BglII (New England Biolabs, Beverly, MA), electrophoresed on 0.8% agarose gel, blotted on positively charged nylon membranes (Hybond N-plus, Amersham, UK), and hybridized to 50 ng of α32P-labeled 0.7-kbHindIII-BamHI genomic probe encompassing exons 2 and 3 of the M-BCR region of the BCR gene.33 Densitometric analysis of autoradiograph bands was performed with the Molecular Imager. The ratio of rearranged band to germline band was used to assess the relative proportion of Ph+ cells in the various bone marrow samples.
Western blotting of the P210BCR/ABL protein.
Cell-culture pellets were lysed by incubation for 10 minutes at 4°C with PBS-TDS buffer (PBS buffer containing 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS). Samples were then centrifuged at 13,000g to remove cellular debris, and the protein concentration of supernatants was determined by colorimetric assay (micro-BCA; Pierce, Rockford, IL) according to the manufacturer’s specifications. Seventy milligrams of each whole-cell lysate, together with molecular weight markers (Amersham, Little Chalfont, UK) were analyzed by sodium dodecyl sulfate-polyacrylamide gel 7.5% electrophoresis. After electrophoresis migration, the gel was soaked for 30 minutes in a transfer buffer (125 mmol/L Tris, 960 mmol/L glycine, 20% methanol) and the proteins were transferred on a 0.45-mm nitrocellulose membrane using a semidry apparatus (Protean Mini-gel; Bio-Rad Laboratories) for 40 minutes at 5.5 mA/cm2.
Block treatment of membranes was performed by overnight incubation in TST buffer (10 mmol/L Tris pH 8.0, 0.15 mmol/L NaCl, 0.05% Tween 20) containing 1% nonfat dry milk.
Specific proteins blotted onto membranes were detected with a 2-step procedure. First, the membrane was incubated overnight at 4°C with 10 μg/mL of anti–BCR/ABL antibody (Calbiochem, Cambridge, MA) and then for 1 hour with a 1:1,000 dilution of anti-mouse horse radish peroxidase-labeled secondary antibody (Amersham, Buckinghamshire, UK). Visualization of the protein bands was obtained using a chemoluminescent detection method (ECL Western Blotting Detection System, Amersham) following the manufacturer’s directions.
Statistical analysis.
The 2-tailed Student’s t-test for paired data was used to analyze the results of the assays of BCR/ABL transcripts in the samples drawn at diagnosis and at remission in the responder and nonresponder groups of CML patients. The results of the analysis of BCR/ABL hybrid transcript levels in cell cultures were analyzed by the Friedman test, a 2-way analysis of variance by ranks for matched samples testing the hypothesis that the effects of the different concentrations of IFN are the same in all sets of cultures. The statistical analyses were performed using the StatView 4.51 software for the PowerPc Macintosh computer (Abacus Concepts Inc, Berkeley, CA).
RESULTS
Intracellular amount of hybrid BCR/ABL mRNA after in vivo treatment.
The number of molecules of hybrid BCR/ABL mRNA was calculated by quantitative RT-PCR of total RNA extracted from the bone marrow MNC fraction. Because cytogenetic conversion, particularly at remission, could lead to reduction in the number of BCR/ABL mRNA molecules due to decreases in the proportion of Ph+ cells in the samples, the relative proportion of Ph+ clonal cells was verified by Southern blot, and the number of hybrid mRNA molecules for each sample was adjusted to the proportion of clonal cells evaluated by Southern blot analysis. Hence, the results are expressed as number of transcripts per microgram of Ph+ bone marrow MNC total RNA.
Fourteen of 20 (70%) patients obtained a complete hematologic response with IFN-α treatment in a median time of 7 months (range, 5 to 24), and were considered responders (Table 1). In the other 6 patients, IFN-α treatment was discontinued because of insufficient hematologic response, and the patients were induced into hematologic remission by hydroxyurea treatment. The number of hybrid mRNA molecules, and the relative proportion of Ph+ cells were measured at diagnosis and at remission, in both groups of patients.
Given the relatively short period of IFN-α treatment (median, 7 months; Table 1), none of the responder patients had, at the end of the study, a major or minor cytogenetic response (ie, relative proportion of Ph+ cells <33% or between 66% and 33%, respectively), which was evaluated by densitometric analysis of the M-BCR; a slight reduction of Ph+ cells was detected in only 3 cases (Table 2; cases 1, 8, and 10). This indicated that, at the end of the study, most of the hematopoietic tissue was still clonal in both responder and nonresponder patients.
Hybrid BCR/ABL Molecules in Ph+ Bone Marrow Mononuclear Cells of IFN- Responder and Nonresponder CML Patients Enrolled in the Study
| Case . | Hybrid BCR/ABL mRNA Junction . | At Diagnosis . | At Hematologic Remission . | Hematologic Response to IFN . | ||
|---|---|---|---|---|---|---|
| % of Ph+ Cells . | Molecules of BCR/ABL mRNA* . | % of Ph+ Cells . | Molecules of BCR/ABL mRNA* . | |||
| 1 | b3/a2 | 90 | 425 × 103 | 80 | 228 × 103 | Complete |
| 2 | b3/a2 | 100 | 151 × 103 | 100 | 12 × 103 | Complete |
| 3 | b2/a2 | 100 | 179 × 103 | 100 | 18 × 103 | Complete |
| 4 | b2/a2 | 100 | 57 × 103 | 100 | 17 × 103 | Complete |
| 5 | b2/a2 | 100 | 74 × 103 | 100 | 24 × 103 | Complete |
| 6 | b2/a2 | 100 | 135 × 103 | 100 | 65 × 103 | Complete |
| 7 | b3/a2 | 100 | 59 × 103 | 100 | 15 × 103 | Complete |
| 8 | b2/a2 | 100 | 438 × 103 | 95 | 47 × 103 | Complete |
| 9 | b3/a2 | 100 | 122 × 103 | 100 | 57 × 103 | Complete |
| 10 | b3/a2 | 85 | 157 × 103 | 75 | 60 × 103 | Complete |
| 11 | b3/a2 | 100 | 58 × 103 | 100 | 12 × 103 | Complete |
| 12 | b2/a2 | 100 | 145 × 103 | 100 | 72 × 103 | Complete |
| 13 | b3/a2 | 100 | 198 × 103 | 100 | 42 × 103 | Complete |
| 14 | b2/a2 | 100 | 122 × 103 | 100 | 37 × 103 | Complete |
| x ± SD | 166 × 103 ± 117 × 103 | 50 × 103 ± 53 × 103 | ||||
| 15 | b2/a2 | 100 | 94 × 103 | 100 | 1,093 × 103 | No response |
| 16 | b3/a2 | 100 | 357 × 103 | 100 | 984 × 103 | No response |
| 17 | b2/a2 | 100 | 112 × 103 | 100 | 205 × 103 | No response |
| 18 | b3/a2 | 100 | 110 × 103 | 100 | 470 × 103 | No response |
| 19 | b2/a2 | 100 | 81 × 103 | 100 | 164 × 103 | No response |
| 20 | b3/a2 | 100 | 144 × 103 | 100 | 178 × 103 | No response |
| x ± SD | 150 × 103 ± 95 × 103 | 516 × 103 ± 385 × 103 | ||||
| Case . | Hybrid BCR/ABL mRNA Junction . | At Diagnosis . | At Hematologic Remission . | Hematologic Response to IFN . | ||
|---|---|---|---|---|---|---|
| % of Ph+ Cells . | Molecules of BCR/ABL mRNA* . | % of Ph+ Cells . | Molecules of BCR/ABL mRNA* . | |||
| 1 | b3/a2 | 90 | 425 × 103 | 80 | 228 × 103 | Complete |
| 2 | b3/a2 | 100 | 151 × 103 | 100 | 12 × 103 | Complete |
| 3 | b2/a2 | 100 | 179 × 103 | 100 | 18 × 103 | Complete |
| 4 | b2/a2 | 100 | 57 × 103 | 100 | 17 × 103 | Complete |
| 5 | b2/a2 | 100 | 74 × 103 | 100 | 24 × 103 | Complete |
| 6 | b2/a2 | 100 | 135 × 103 | 100 | 65 × 103 | Complete |
| 7 | b3/a2 | 100 | 59 × 103 | 100 | 15 × 103 | Complete |
| 8 | b2/a2 | 100 | 438 × 103 | 95 | 47 × 103 | Complete |
| 9 | b3/a2 | 100 | 122 × 103 | 100 | 57 × 103 | Complete |
| 10 | b3/a2 | 85 | 157 × 103 | 75 | 60 × 103 | Complete |
| 11 | b3/a2 | 100 | 58 × 103 | 100 | 12 × 103 | Complete |
| 12 | b2/a2 | 100 | 145 × 103 | 100 | 72 × 103 | Complete |
| 13 | b3/a2 | 100 | 198 × 103 | 100 | 42 × 103 | Complete |
| 14 | b2/a2 | 100 | 122 × 103 | 100 | 37 × 103 | Complete |
| x ± SD | 166 × 103 ± 117 × 103 | 50 × 103 ± 53 × 103 | ||||
| 15 | b2/a2 | 100 | 94 × 103 | 100 | 1,093 × 103 | No response |
| 16 | b3/a2 | 100 | 357 × 103 | 100 | 984 × 103 | No response |
| 17 | b2/a2 | 100 | 112 × 103 | 100 | 205 × 103 | No response |
| 18 | b3/a2 | 100 | 110 × 103 | 100 | 470 × 103 | No response |
| 19 | b2/a2 | 100 | 81 × 103 | 100 | 164 × 103 | No response |
| 20 | b3/a2 | 100 | 144 × 103 | 100 | 178 × 103 | No response |
| x ± SD | 150 × 103 ± 95 × 103 | 516 × 103 ± 385 × 103 | ||||
The number of BCR/ABL molecules are expressed per microgram of total RNA and are adjusted to the relative proportion of Ph+ cells, as evaluated by Southern analysis.
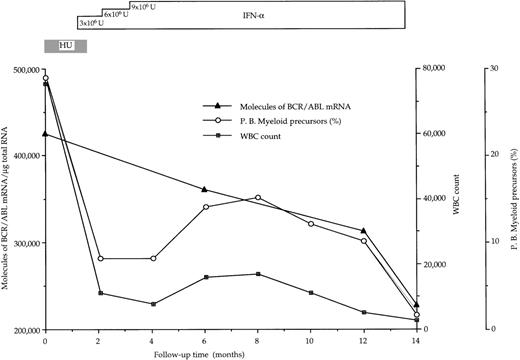
At diagnosis, the number of BCR/ABL hybrid transcripts/μg of total RNA varied from patient to patient; however, there was no difference between the mean values in the 2 groups of patients (168 × 103 and 144 × 103 molecules/μg of total RNA, respectively; Table 2 and Fig1). At the end of the study, in responder patients the number of hybrid transcripts was significantly lower (48 × 103 vs 168 × 103;P < .01 at the Student’s t-test for paired data), whereas in IFN-α-resistant patients hybrid transcripts tended to increase (Table 2 and Fig 1). During the course of treatment, 3 sequential bone marrow samples were collected from the same responder patient (no. 1): the results of quantitative RT-PCR assay of the number of BCR/ABL transcripts paralleled the decrease of WBC and of peripheral blood myeloid precursors (Fig 2).
Effect of IFN- treatment on the levels of hybrid BCR/ABL mRNA in bone marrow mononuclear cells from CML patients with complete or poor hematologic responses to therapy. For each patient, values have been adjusted for the percentage of Ph+ cells. At diagnosis, the amount of transcript was similar in the 2 groups of patients; there was a significant difference (P < .01, paired t-test) between the values at diagnosis and those at the end of the study in the group of responder patients, whereas in patients resistant to IFN-, there was a trend toward higher values at the end of the study.
Effect of IFN- treatment on the levels of hybrid BCR/ABL mRNA in bone marrow mononuclear cells from CML patients with complete or poor hematologic responses to therapy. For each patient, values have been adjusted for the percentage of Ph+ cells. At diagnosis, the amount of transcript was similar in the 2 groups of patients; there was a significant difference (P < .01, paired t-test) between the values at diagnosis and those at the end of the study in the group of responder patients, whereas in patients resistant to IFN-, there was a trend toward higher values at the end of the study.
Progressive decline of hybrid BCR/ABL mRNA molecules in bone marrow mononuclear cells from a CML patient (no. 1 in Table 2) who had a complete hematologic response after IFN- therapy. BCR/ABL values are corrected for the percentage of Ph+cells.
Progressive decline of hybrid BCR/ABL mRNA molecules in bone marrow mononuclear cells from a CML patient (no. 1 in Table 2) who had a complete hematologic response after IFN- therapy. BCR/ABL values are corrected for the percentage of Ph+cells.
Differential effects of IFN-α on suspension cultures of Ph+ bone marrow MNC. Comparison with hematologic response to treatment.
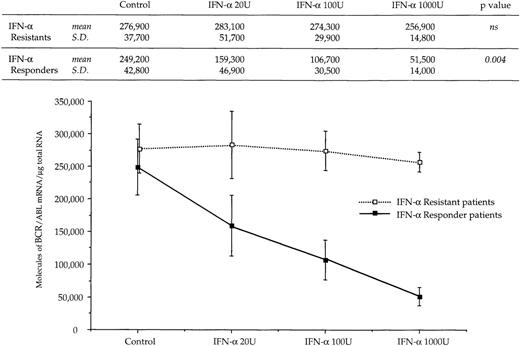
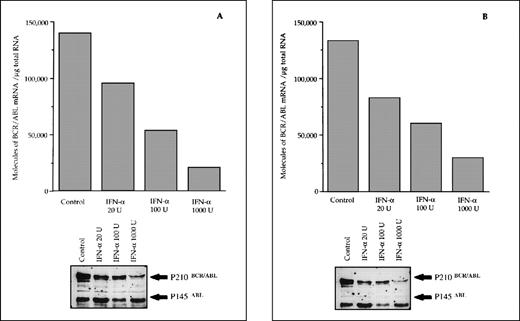
The reduced number of hybrid transcripts in Ph+ bone marrow MNC observed in responder patients could be because of either a direct or an indirect effect of in vivo IFN-α treatment. To clarify this issue, Ph+ bone marrow MNC isolated from 9 untreated CML patients were incubated for 24 hours in suspension culture with increasing amounts of IFN-α (from 20 to 1,000 UI/mL). At the end of incubation, we used quantitative RT-PCR to determine the number of transcripts in the total RNA fraction extracted from the cells. In 6 patients who subsequently had a complete hematologic response to IFN-α treatment, there was a decrease of BCR/ABL transcript number per μg of total RNA (P = .004 at Friedman test; Fig 3). In an attempt to obtain a rough evaluation of the corresponding intracellular P210BCR/ABL protein content, we analyzed the cell lysates from this experiment with the Western blot technique. The decrease of the P210BCR/ABL proteins was less pronounced than that of mRNA levels, it was dose-dependent in 3 of 6 responder patients studied and the greatest reductions occurred in the MNCs cultured with 1,000 U of IFN-α (Fig 4A and B). The differential effect of IFN-α on the BCR/ABL transcript and on the P210BCR/ABL protein level could be explained by their different half-lives.
Dose response relationship between levels of BCR/ABL transcripts and concentration of IFN- used in culture experiments. Symbols and vertical lines are mean ± SD, respectively. Numerical values and the results of the Friedman statistical analysis (see Materials and Methods for details) are reported in the upper part of the figure.
Dose response relationship between levels of BCR/ABL transcripts and concentration of IFN- used in culture experiments. Symbols and vertical lines are mean ± SD, respectively. Numerical values and the results of the Friedman statistical analysis (see Materials and Methods for details) are reported in the upper part of the figure.
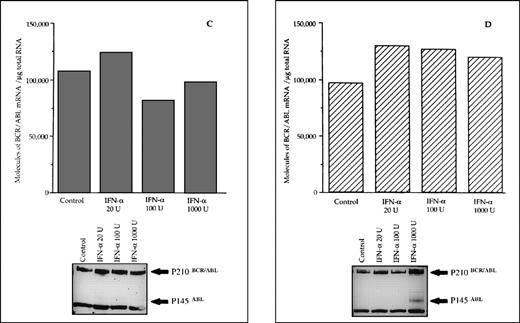
Differential effects of IFN- priming on BCR/ABL expression in bone marrow mononuclear cell cultures from CML patients (A and B, IFN- responder patients; C, IFN-–resistant patient; D, a patient presenting in initial blastic transformation). Bone marrow mononuclear cells were cultured for 24 hours with the indicated concentrations (U/mL) of IFN-. Lower panels: Western blot for intracellular P210BCR/ABLcontent, including P145ABL determination used as an internal control.
Differential effects of IFN- priming on BCR/ABL expression in bone marrow mononuclear cell cultures from CML patients (A and B, IFN- responder patients; C, IFN-–resistant patient; D, a patient presenting in initial blastic transformation). Bone marrow mononuclear cells were cultured for 24 hours with the indicated concentrations (U/mL) of IFN-. Lower panels: Western blot for intracellular P210BCR/ABLcontent, including P145ABL determination used as an internal control.
When the same set of experiments was performed on bone marrow MNC isolated from 3 CML patients who subsequently proved to be resistant to IFN-α treatment, we found no variation in the intracellular levels of both the hybrid transcript and the P210BCR/ABL protein between untreated and IFN-α–primed cells (Figs 3 and 4C). No in vitro IFN-α–induced downmodulation of BCR/ABL mRNA and of P210BCR/ABL protein was observed in cultured bone marrow MNC from an untreated patient who presented with CML in initial blastic transformation (Fig 4D).
DISCUSSION
IFN-α is a first-line drug in the treatment of CML and induces hematologic remission in the majority of patients after 3 to 6 months of treatment.14-16 Despite intensive efforts, the molecular mechanisms of the antiproliferative effects of IFN-α in CML have not yet been clarified.3 We and others have shown that IFN-α upmodulates FAS-R expression on cell membrane of bone marrow CD34+ cells from CML patients,22-24 and that FAS triggering in the presence of IFN-α inhibits the growth of these cells.22 We also showed that only CD34+ cells isolated from patients who were responsive to in vivo IFN-α treatment had a dose-dependent decrease of proliferation in methylcellulose colony assays when exposed to IFN-α in the presence of a FAS agonist.25 Interestingly, growth inhibition correlated to increased apoptotic rate of these cells, and to posttranscriptional downmodulation of the P210BCR/ABL protein. On the other hand, growth of colony-forming units and apoptosis rate of Ph+ progenitors isolated from CML patients who were resistant to IFN-α treatment remained unchanged.25Moreover, a new cell line, KT-1, established from a Ph+ CML patient in blastic crisis, is reported to be sensitive to both IFN-α and IFN-γ in vitro.26 Indeed, IFN-α dose-dependently suppressed the growth of KT-1 cells both in suspension and in semisolid cultures, inducing G1 cell-cycle arrest and apoptotic death. In addition, Northern blot analysis showed that IFN-α significantly reduces the expression of the BCR/ABL hybrid gene within 6 hours and that this effect is more pronounced after 24 hours of IFN-α exposure.26 Downmodulation of the BCR/ABL gene in the KT-1 cell line seems to be a specific effect of IFN-α. When IFN-γ was added to cultures, it suppressed the growth of KT-1, as did IFN-α, thus exerting a synergistic effect with IFN-α. However, the growth arrest of KT-1 cells induced by IFN-γ was not associated with downmodulation of the BCR/ABL gene.26 These findings suggested that IFN-α and IFN-γ control the proliferation of KT-1 cells through different signal-transduction pathways, and that the antiproliferative effect of IFN-α may depend on downmodulation of BCR/ABL expression. The latter mechanism could be the most plausible in vivo, given the limited efficacy of IFN-γ in the clinical setting.34-36
In the present study, we investigated the intracellular content of BCR/ABL hybrid mRNA in Ph+ precursors during in vivo and in vitro exposure to IFN-α. The data regarding bone marrow MNC of CML patients at diagnosis and at hematologic remission were normalized to the percentage of Ph+ cells to avoid a bias because of cytogenetic conversion. However, in only 3 of 14 cases that achieved hematologic remission was there a limited reduction of Ph+cell percentage. On complete hematologic response (after a median of 8 months of IFN-α treatment), the number of intracellular BCR/ABL mRNA molecules was reduced by a mean of 72% with respect to the number at diagnosis. Interestingly, there was a clear correlation with the hematologic response to IFN-α: at variance with IFN-α responders, nonresponder patients did not show BCR/ABL downmodulation, rather, they showed a trend toward enhanced intracellular levels of BCR/ABL mRNA. It is unlikely that changes in cell composition of the bone marrow MNC samples caused the decreases in transcript number in responder patients, because both responders and nonresponders were in hematologic remission at the end of the the study. The correlation between variation of intracellular levels of BCR/ABL mRNA and hematologic response to IFN-α treatment suggests that downmodulation of the BCR/ABL gene is an important mechanism of in vivo IFN-α activity.
In principle, the decreased BCR/ABL expression observed in vivo in responder patients could be attributed to an indirect effect of IFN-α treatment; however, we found a dose-dependent decrease of the number of BCR/ABL mRNA molecules and of the corresponding P210BCR/ABLprotein in bone marrow MNC isolated from CML patients after 24 hours of exposure to IFN-α in suspension cultures. Thus, in an experimental system closer to physiological conditions than the KT-1 cell line, we provide evidence that the decrease in intracellular levels of BCR/ABL mRNA during in vivo treatment is because of a direct interaction between IFN-α and Ph+ precursors. Remarkably, in vitro effects of IFN-α on BCR/ABL mRNA in bone marrow MNC correlated with subsequent hematologic response. This suggests that response to IFN-α treatment is an intrinsic property of the Ph+ clonal precursors.
The data reported here indicate that the mechanism of action of IFN-α could rely on multiple and, probably, synergistic effects. The P210BCR/ABL protein plays a crucial role in the pathogenesis of the chronic phase of CML given its ability to reduce, at least in vitro, the differentiation capacity8,37-40 and the adhesion to the stroma of the Ph+precursors,41-43 and to increase their proliferative potential.44,45 Therefore, it is possible that the IFN-α–mediated downmodulation of BCR/ABL expression exerts an initial antiproliferative effect on Ph+ stem cells directly, or alternatively indirectly, by restoring the adhesion to stroma. Decreased intracellular levels of the P210BCR/ABL protein could enhance the responsiveness of Ph+ progenitors to apoptosis and specifically to FAS-related apoptosis. In fact, decreased intracellular amounts of P210BCR/ABL protein results in reduced resistance of CML cells to apoptosis induced by a variety of agents.5-7 These mechanisms could account for the hematologic response of CML patients to IFN-α treatment, which is necessary, although not sufficient, for a significant cytogenetic conversion. It is still unknown whether the same mechanisms are also important for complete reversion to Ph− hematopoiesis, which is induced by IFN-α only in a small proportion of patients.
This study highlights the potential role of BCR/ABL downmodulation induced by IFN-α in the framework of the complex mechanisms by which IFN-α controls the chronic phase of CML. It also provides a potential tool by which in vivo responsiveness to IFN-α treatment can be predicted. Recent articles have dealt with this issue using different approaches.46,47 It is of considerable clinical relevance in terms of optimizing the therapeutic strategy (including bone marrow transplantation timing) and in terms of cost effectiveness of the treatment to be planned. Further experiments are necessary to identify the intracellular signals involved in the downmodulation described, which, in turn, may be important for the development of new therapeutic strategies for CML.
ACKNOWLEDGMENT
We are grateful to Jean Gilder for editorial assistance.
Supported in part by grants from AIRC (Milan), CNR—P.F. Biotecnologie (Rome), “Biogem” and “PRIN”—MURST (Rome), and Regione Campania.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Fabrizio Pane, MD, Dipartimento di Biochimica e Biotecnologie Mediche, Facoltà di Medicina, Università di Napoli Federico II, via S Pansini 5—80131 Napoli, Italy; e-mail: fabpane@unina.it.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal