Familial Mediterranean fever (FMF) is a recessively inherited disorder characterized by recurrent, self-limited attacks of fever and serositis and by infiltration of affected tissues by large numbers of neutrophils. A candidate gene for FMF was identified by positional cloning and named “MEFV.” The corresponding protein was named “pyrin.” To elucidate the currently unknown function of pyrin, we characterized its tissue distribution, regulation of expression during hematopoietic differentiation, and subcellular localization. Reverse transcription-polymerase chain reaction analysis, followed by hybridization with an internal oligonucleotide, demonstrated expression of MEFV in different populations of peripheral blood cells. Among hematopoietic cell lines, MEFVwas almost exclusively expressed in cells of the myeloid lineage. Furthermore, MEFV messenger RNA was strongly expressed within 24 hours of dimethyl sulfoxide–induced granulocytic differentiation of HL-60 cells. Analysis of complementary DNA from human solid tumor–derived cell lines revealed expression of MEFV in several cell lines derived from colon and prostate cancers. Expression of MEFV fused to enhanced green fluorescent protein showed that pyrin localized in distinct patches in the cytoplasm, forming a perinuclear cap. Taken together, MEFV is predominantly expressed in myeloid cells and upregulated during myeloid differentiation, and the corresponding protein, pyrin, is expressed in the cytoplasm.
Familial Mediterranean fever (FMF) is a hereditary disease characterized by periodic episodes of fever accompanied by acute inflammatory attacks of serosal or synovial membranes.1,2 A massive influx of neutrophils into the affected tissues occurs during these attacks. This enormous invasion of neutrophils to the sites of inflammation plays a major role in the inflammatory process, although the regulating factors remain unknown. Recently, a gene linked to FMF was identified by positional cloning.3,4 The gene was named “MEFV,” and the predicted protein was called either “pyrin” or “marenostrin.” MEFV is located on chromosome 16p13.3 and consists of at least 10 exons. Analysis of the predicted amino acid sequence revealed the presence of 2 potential nuclear localization signals as well as a basic domain and a B-box–type zinc finger motif. Similarly, a coiled-coil domain and a B30.2 globular domain, which are implicated in dimerization and protein-protein interactions, respectively, were predicted from the MEFVsequence.3 4
Several missense mutations in the MEFV gene have been described in patients with FMF.3-5 FMF patients display decreased C5a-inhibitor activity,6,7 and an investigative group has hypothesized that pyrin might regulate expression of C5a-inhibitor activity.8 So far, this C5a-inhibitor activity has not been cloned, and a direct connection to pyrin awaits further investigation.
To date, the function of pyrin and its role in the pathogenesis of FMF remain unknown. Pyrin may act as a regulator of inflammatory stimuli in neutrophils in FMF as well as in other diseases.
Here, we investigated expression of MEFV messenger RNA (mRNA) in hematopoietic and nonhematopoietic tissues and cell lines. Induction of MEFV mRNA during granulocytic differentiation is demonstrated in HL-60 cells incubated with dimethyl sulfoxide (DMSO). Expression of pyrin as an EGFP (enhanced green fluorescent protein) fusion protein reveals cytoplasmic, perinuclear localization of the pyrin protein.
Materials and methods
Cell culture
Cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA). NB4 cells were generously provided by Dr M. Lanotte (St. Louis Hospital, Paris, France). ML-1 cells were a gift from Dr M. Kastan (Johns Hopkins University, Baltimore, MD). Kasumi1 and Kasumi3 cells were established by Dr Hiroya Asou (Hiroshima University, Hiroshima, Japan). Adherent and suspension cells were grown in DMEM containing 10% to 20% fetal calf serum (FCS) and in RPMI containing 10% FCS medium, respectively, in a humidified atmosphere with 5% carbon dioxide at 37°C. HL-60 cells were incubated in the presence of 1.25% (v/v) DMSO for up to 4 days to induce granulocytic differentiation.
Separation of peripheral blood neutrophils
Polymorphonuclear cells (neutrophils) were isolated from anticoagulated blood using PMN solution (Robbins Scientific Corp, Sunnyvale, CA) according to the 1-step density-gradient centrifugation method.9 10 Briefly, whole blood was layered over PMN solution and centrifuged for 25 minutes at 500g. Mononuclear cells and neutrophils were separated into 2 distinct bands, whereas erythrocytes pelleted to the bottom of the tube. Neutrophils were obtained from the lower band and washed twice with serum-free medium. Monocytes were separated from the mononuclear cell fraction by adherence to plastic cell-culture dishes. Viability of the cells was tested by trypan blue dye exclusion.
Sorting of peripheral blood leukocytes by flow cytometry
First, mononuclear cells were separated from heparinized blood by Ficoll-Paque (Pharmacia Biotech, Piscataway, NJ) density gradient centrifugation. Individual populations of peripheral blood leukocytes were further isolated by flow cytometry (FACStar, Becton Dickinson, Mountain View, CA) using monoclonal mouse antibodies against CD19 and CD3 conjugated to fluorescein isothiocyanate and PE, respectively (Dako, Carpinteria, CA).
Reverse transcription and polymerase chain reaction
Total RNA was isolated using TRIzol reagent (Gibco BRL, Gaithersburg, MD) according to the method described by Chomczynski and Sacchi11; 2 μg of total RNA were reverse transcribed using SuperscriptII (Gibco BRL) and random primers (Gibco BRL) according to the manufacturer's instructions. Polymerase chain reaction (PCR) was performed with Taq DNA polymerase (Gibco BRL) under standard conditions using MEFV-specific oligonucleotides (5′-ATCCAACTCCTCCACCAGAA-3′ and 5′-AGTGTTGGGC-ATTCAGTCAG-3′) that amplify a 690–base pair (bp) fragment. All PCR primer pairs span an exon/intron boundary to allow discrimination between complementary DNA (cDNA)- and DNA-generated fragments. Oligonucleotides specific for β-actin (5′-TACATGGCTGGGGTGTTGAA-3′ and 5′-AAGAGAGGCATCCTCACCCT-3′) amplify a 218-bp fragment, and oligonucleotides for lactoferrin (5′-GTTCAGTGGTGCGCCGTATC-3′ and 5′-CACCACAGCCACGGCATAAT-3′) generate a 279-bp fragment. Each cycle of PCR consisted of denaturation (1 minute at 95°C), annealing (1 minute at 60°C, 55°C, or 64°C forMEFV, lactoferrin, and β-actin, respectively), and extension (1 minute at 72°C) and was repeated 22 to 30 times, as indicated in “Results.” PCR-amplified DNA fragments were visualized by ethidium bromide staining after agarose gel electrophoresis.
Hybridization with an internal oligonucleotide
Gel-separated PCR products were blotted onto a positively charged nylon membrane (Boehringer Mannheim, Indianapolis, IN, or Amersham Life Science, Arlington Heights, IL) by capillary transfer in 20xSCC. Prehybridization (30-45 minutes) and hybridization (1-2 hours) occurred at 42°C in DIG (digoxigenin) Easy-Hyb solution (Boehringer Mannheim). As a hybridization probe, internal oligonucleotides specific for MEFV5′-GACAGC-ATGGATCCTGGGAGCCTGCAAG-3′, lactoferrin 5′-CCCTTGATGGTGGTTTCA-TATACGA-3′, or β-actin 5′-ATCGAGCACGGCATCGTCAC-3′ were labeled with DIG using the DIG 3′ End Labeling Kit (Boehringer Mannheim). Washes were repeated twice in 2xSCC for 5 minutes at room temperature and in 0.5xSCC for 5 minutes at 42°C. The membrane was blocked (1% blocking reagent in maleic acid buffer) for 30 minutes prior to incubation with anti-DIG antibodies coupled to alkaline phosphatase (30 minutes). After washing twice in maleic acid buffer, detection was performed by incubating with the chemiluminescent substrate CDP-Star (Tropix, Bedford, MA). Autoradiographic film (X-OMAT/AR, Eastman Kodak, Rochester, NY) was exposed to the blot for several seconds up to 30 minutes.
Cloning and expression of an EGFP-pyrin fusion protein
In a 2-step cloning procedure, the cDNAs of EGFP and pyrin were cloned into the pcDNA3.1(+) vector (Invitrogen, Carlsbad, CA). First, PCR-amplified EGFP cDNA (Clontech, Palo Alto, CA) was cloned into the vector using recognition sites for restriction endonucleasesHindIII and KpnI. Second, the full-length cDNA of pyrin was amplified by PCR using Pfu polymerase (Stratagene, La Jolla, CA) and the following primers that contain recognition sites forBglII and EcoRI, respectively: 5′-GCATATAGATCTATGGCTAAGACCCCTAG-3′ and 5′-GCATATGAATTCGGCAT-TCAGTCAGGCC-3′. PCR fragments were digested, gel-purified, and cloned in frame 3′ of EGFP cDNA into the EcoRI and BglII sites of a pCDNA3.1(+) expression vector. Transformants were tested for the correct insert by restriction enzyme digestion and DNA sequencing. Either EGFP or EGFP-pyrin plasmid (15 μg) were transfected into COS-1 cells using Superfect reagent (Qiagen, Valencia, CA). Transfected cells were plated on cover slips and analyzed 2 to 3 days after transfection by fluorescent microscopy using a 510-nm filter.
Western blot
Transfected cells were lysed in radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors on day 2 after transfection and separated by SDS-PAGE on a 4% to 15% acrylamide gradient gel. Proteins were transferred onto a nitrocellulose membrane and detected with a monoclonal anti-GFP antibody (Clontech). Incubation with a secondary goat anti-mouse antibody coupled to horseradish peroxidase and subsequent incubation with ECL1 and ECL2 substrate (Amersham Life Science) was followed by exposure to autoradiographic film (Biomax-MR, Eastman Kodak) for several seconds or minutes.
Results
Hematopoietic expression of MEFV
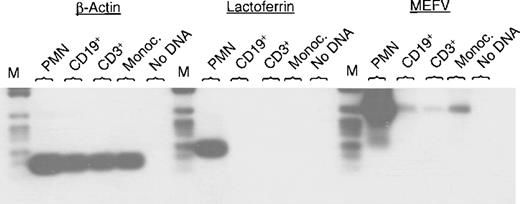
MEFV was recently identified as a disease-related gene in patients with FMF.3,4 So far, little is known about its function. To begin to address this issue, we studied the expression ofMEFV mRNA in different tissues and cell lines. Expression ofMEFV mRNA previously was described in peripheral blood leukocytes.3 4 In this report, we analyzed MEFVexpression in different populations of peripheral blood leukocytes. PCR was performed using specific oligonucleotides for MEFV, lactoferrin, and β-actin. Hybridization with gene-specific internal oligonucleotides confirmed the specificity of the PCR products.MEFV mRNA could be amplified in neutrophils, monocytes, and also weakly in CD19+ B cells and CD3+ T cells (Figure 1). Amplification of β-actin served as a control for cDNA quality (Figure 1). Lactoferrin is a specific granule marker and was chosen to exclude potential contamination of neutrophils to the other cell populations. Only neutrophils showed strong expression of lactoferrin mRNA, whereas CD19+ B lymphocytes, CD3+ T cells, and monocytes were negative for lactoferrin mRNA (Figure 1).
Expression of MEFV mRNA in different populations of peripheral blood cells.
Total RNAs were isolated from the indicated subsets of leukocytes and subjected to RT-PCR using specific oligonucleotides for MEFV, lactoferrin, and β-actin. Amplification occurred for 30, 25, and 22 cycles, respectively. PCR fragments were separated by agarose gel electrophoresis, transferred to a nylon membrane, and hybridized with a gene-specific internal oligonucleotide labeled with DIG. (PMN = polymorphonuclear cells (neutrophils); Monoc. = monocytes; M = DNA length standard VI: 154, 220/234, 298, 394, 453, 517, 653, 1033 bp, Boehringer Mannheim.)
Expression of MEFV mRNA in different populations of peripheral blood cells.
Total RNAs were isolated from the indicated subsets of leukocytes and subjected to RT-PCR using specific oligonucleotides for MEFV, lactoferrin, and β-actin. Amplification occurred for 30, 25, and 22 cycles, respectively. PCR fragments were separated by agarose gel electrophoresis, transferred to a nylon membrane, and hybridized with a gene-specific internal oligonucleotide labeled with DIG. (PMN = polymorphonuclear cells (neutrophils); Monoc. = monocytes; M = DNA length standard VI: 154, 220/234, 298, 394, 453, 517, 653, 1033 bp, Boehringer Mannheim.)
Next, we examined expression of MEFV in a panel of 22 leukemic cell lines. Interestingly, among the 11 myeloid leukemic cell lines, 8 cell lines showed MEFV expression after PCR amplification for 30 cycles followed by hybridization (Table1). Only 1 (Jurkat) of 11 lymphoid cell lines showed expression of MEFV mRNA.
Expression of MEFV in human cell lines
| Cell Type . | Cell Type . | Cell Lines . | Expression ofMEFV . |
|---|---|---|---|
| Hematopoietic | Myeloid and erythroid | THP-1 KG-1 | + + |
| Kasumi1 | − | ||
| Kasumi3 | + | ||
| ML-1 | + | ||
| SK-NO1 | − | ||
| NB4 | + | ||
| HL-60 | + | ||
| U937 | + | ||
| KCL22 | + | ||
| K-562 | − | ||
| Lymphoid | SLB-1 | − | |
| TALL-1 | − | ||
| TALL-104 | − | ||
| Jurkat | + | ||
| YTC3 | − | ||
| NALM-18 | − | ||
| JB38 | − | ||
| JB39 | − | ||
| Raji | − | ||
| BC-1 | − | ||
| BC-2 | − | ||
| Nonhematopoietic | Colon | HuTu 80 | + |
| SW837 | + | ||
| HT29 | − | ||
| SW 1463 | − | ||
| CaCo-2 | − | ||
| SK-CO-1 | − | ||
| LoVo | − | ||
| SW 480 | + | ||
| SW 48 | − | ||
| SW 403 | − | ||
| SW 1417 | + | ||
| Prostate | PC-3 | + | |
| LNCaP | + | ||
| DU 145 | + | ||
| Breast | SK-BR2 | − | |
| MDA-MB-231 | − | ||
| MCF7 | − | ||
| Others | HeLa | + | |
| SK-Hep-1 | − | ||
| IMR-32 | − | ||
| Saos-2 | − |
| Cell Type . | Cell Type . | Cell Lines . | Expression ofMEFV . |
|---|---|---|---|
| Hematopoietic | Myeloid and erythroid | THP-1 KG-1 | + + |
| Kasumi1 | − | ||
| Kasumi3 | + | ||
| ML-1 | + | ||
| SK-NO1 | − | ||
| NB4 | + | ||
| HL-60 | + | ||
| U937 | + | ||
| KCL22 | + | ||
| K-562 | − | ||
| Lymphoid | SLB-1 | − | |
| TALL-1 | − | ||
| TALL-104 | − | ||
| Jurkat | + | ||
| YTC3 | − | ||
| NALM-18 | − | ||
| JB38 | − | ||
| JB39 | − | ||
| Raji | − | ||
| BC-1 | − | ||
| BC-2 | − | ||
| Nonhematopoietic | Colon | HuTu 80 | + |
| SW837 | + | ||
| HT29 | − | ||
| SW 1463 | − | ||
| CaCo-2 | − | ||
| SK-CO-1 | − | ||
| LoVo | − | ||
| SW 480 | + | ||
| SW 48 | − | ||
| SW 403 | − | ||
| SW 1417 | + | ||
| Prostate | PC-3 | + | |
| LNCaP | + | ||
| DU 145 | + | ||
| Breast | SK-BR2 | − | |
| MDA-MB-231 | − | ||
| MCF7 | − | ||
| Others | HeLa | + | |
| SK-Hep-1 | − | ||
| IMR-32 | − | ||
| Saos-2 | − |
Induction of MEFV expression during granulocytic differentiation
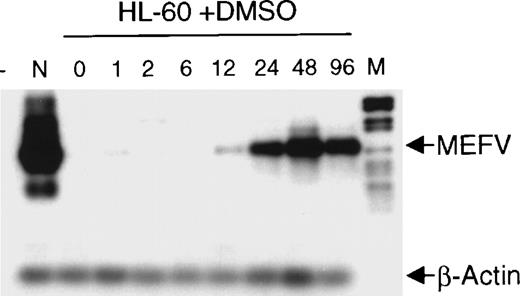
The finding that MEFV is preferentially expressed in myeloid cells and cell lines led us to analyze its expression during hematopoietic differentiation. HL-60 cells were induced to differentiate toward granulocytes by incubation with DMSO for up to 96 hours. Induction of MEFV mRNA was analyzed by semiquantitative reverse transcription (RT)-PCR with amplification for 25 instead of 30 cycles and subsequent hybridization with an MEFV-specific oligonucleotide (Figure 2). Expression of MEFVin unstimulated HL-60 cells was detected only after prolonged exposure of the film to the blot (30 minutes). Regular exposure times (1 minute) revealed MEFV mRNA beginning at 12 hours of incubation with DMSO and peaking by 48 hours. In contrast, the level of MEFVexpression was not altered in ML-1 cells incubated in the presence of tetradecanoyl phorbol acetate to induce monocytic differentiation (data not shown).
Induction of MEFV during granulocytic differentiation of HL-60 cells.
HL-60 cells were incubated with 1.25% (v/v) DMSO to induce granulocytic differentiation. Total RNAs were isolated at the indicated times (hours) and subjected to semiquantitative RT-PCR and hybridization with specific oligonucleotides for MEFV and β-actin. PCR products were amplified for 25 cycles (MEFV) and 20 cycles (β-actin) to stay within the linear range. (- = no DNA; n = neutrophils, M = DNA length standard VI: 298, 394, 453, 517, 653, 1033, 1230, 1760, 2176 bp.)
Induction of MEFV during granulocytic differentiation of HL-60 cells.
HL-60 cells were incubated with 1.25% (v/v) DMSO to induce granulocytic differentiation. Total RNAs were isolated at the indicated times (hours) and subjected to semiquantitative RT-PCR and hybridization with specific oligonucleotides for MEFV and β-actin. PCR products were amplified for 25 cycles (MEFV) and 20 cycles (β-actin) to stay within the linear range. (- = no DNA; n = neutrophils, M = DNA length standard VI: 298, 394, 453, 517, 653, 1033, 1230, 1760, 2176 bp.)
MEFV expression in nonhematopoietic cells and cell lines
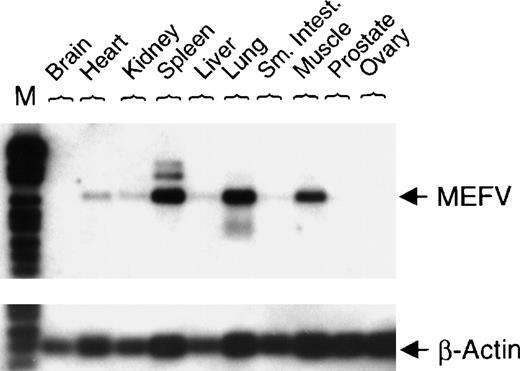
To investigate nonhematopoietic tissue distribution of MEFV mRNA, expression was analyzed in normal human tissues. Expression of MEFV was detected in human muscle, lung, and spleen (Figure 3).
Expression of MEFV in a collection of normal human tissue cDNAs.
cDNAs of normal human tissue were obtained (OriGene Technologies Inc, Rockville, MD), and PCR with specific primers for either MEFV(30 cycles) or β-actin (22 cycles) was performed. PCR fragments were blotted and hybridized with a gene-specific internal DIG-labeled oligonucleotide. (Sm. Intest. = small intestine; M = DNA length standard VI.)
Expression of MEFV in a collection of normal human tissue cDNAs.
cDNAs of normal human tissue were obtained (OriGene Technologies Inc, Rockville, MD), and PCR with specific primers for either MEFV(30 cycles) or β-actin (22 cycles) was performed. PCR fragments were blotted and hybridized with a gene-specific internal DIG-labeled oligonucleotide. (Sm. Intest. = small intestine; M = DNA length standard VI.)
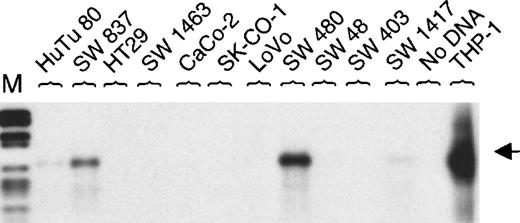
Initially, MEFV was reported to be expressed in the colon cancer cell line SW 480.6 We tested a panel of 11 colon cancer cell lines. Expression of MEFV in mRNA from SW 480 was confirmed. Additionally, MEFV mRNA was detected in the colon cancer cell line SW 837 and, at a very low level, it was detected in HuTu 80 and SW 1417 cells (Figure 4). Furthermore, we analyzed another 10 solid tumor–derived cell lines. Three prostate cancer cell lines (LNCaP, PC-3, and DU 145) expressed MEFV mRNA, whereas expression was not detected in 3 breast cancer cell lines (SK-BR2, MDA-MB-231, MCF7) tested (Table 1).
MEFV expression in colon cancer cell lines.
Total RNAs were isolated from 11 human colon cancer cell lines. Expression of MEFV was analyzed by RT-PCR (30 cycles) and hybridization using an MEFV-specific oligonucleotide. As a negative control, RT-PCR was performed without DNA and, as a positive control, cDNA from monocytoid THP-1 cells was amplified. (M = DNA length standard VI: 298, 394, 453, 517, 653, 1033, 1230, 1760, 2176 bp.)
MEFV expression in colon cancer cell lines.
Total RNAs were isolated from 11 human colon cancer cell lines. Expression of MEFV was analyzed by RT-PCR (30 cycles) and hybridization using an MEFV-specific oligonucleotide. As a negative control, RT-PCR was performed without DNA and, as a positive control, cDNA from monocytoid THP-1 cells was amplified. (M = DNA length standard VI: 298, 394, 453, 517, 653, 1033, 1230, 1760, 2176 bp.)
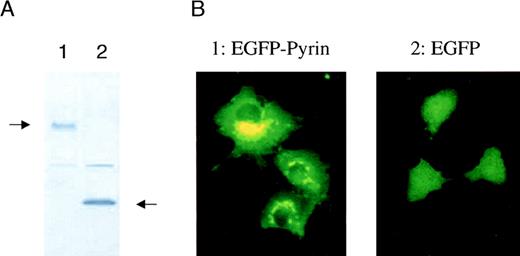
Intracellular localization of the pyrin protein
Determining the intracellular localization of a protein is important for understanding its function. Initial reports suggested that pyrin might act as a nuclear DNA binding factor based on analysis of its predicted amino acid sequence.3,4 12 To address the question of intracellular localization of pyrin, we fused MEFVcDNA to EGFP in a mammalian expression vector and analyzed expression of its corresponding protein, pyrin. COS-1 cells were transfected with an expression vector containing either EGFP or EGFP-pyrin. Western blot analysis of transfected COS-1 cells using a monoclonal anti-EGFP antibody showed that the cells expressed either the 27-kd EGFP protein or a 127-kd protein that corresponded to the calculated molecular mass of the EGFP-pyrin fusion protein (Figure5A). Fluorescent microscopy of transiently transfected cells demonstrated a diffuse expression in cells that were transfected with EGFP alone (Figure 5B). In contrast, COS-1 cells expressing the EGFP-pyrin fusion protein revealed a highly specific expression pattern consisting of distinct patches in the cytoplasm localized around the nucleus (Figure 5B). A similar expression pattern was found for EGFP-pyrin transiently expressed in U937 myeloid cells, which express low levels of endogenous pyrin (data not shown).
Intracellular localization of pyrin.
COS-1 cells were transiently transfected with a vector expressing either EGFP or EGFP-pyrin. (A) Western blot analysis of transfected cells (lane 1: EGFP-pyrin, lane 2: EGFP vector control) using a monoclonal anti-GFP antibody. (B) COS-1 cells either transfected with an EGFP-pyrin or an EGFP expression vector were grown on cover slips and analyzed by fluorescent microscopy (1000 × magnification).
Intracellular localization of pyrin.
COS-1 cells were transiently transfected with a vector expressing either EGFP or EGFP-pyrin. (A) Western blot analysis of transfected cells (lane 1: EGFP-pyrin, lane 2: EGFP vector control) using a monoclonal anti-GFP antibody. (B) COS-1 cells either transfected with an EGFP-pyrin or an EGFP expression vector were grown on cover slips and analyzed by fluorescent microscopy (1000 × magnification).
Expression of mutated pyrin does not alter its localization
In patients suffering from FMF, several point mutations have been reported.3-5 We introduced either of 2 of the most common point mutations (Met694Val and Val726Ala) into theMEFV cDNA by site directed mutagenesis. Mutated MEFVwas fused to EGFP and expressed in COS-1 cells. The intracellular localization of EGFP-pyrin was not affected by the presence of the point mutations. The mutants maintained the distinct perinuclear expression in the cytoplasm like the wild-type pyrin protein.
Discussion
FMF is an inherited disease that is characterized by recurrent attacks of fever. Positional cloning identified MEFV, the gene that is likely to cause FMF.3,4 So far, the function of the corresponding protein, named pyrin, remains unknown. Analysis of the predicted amino acid sequence of pyrin suggested that pyrin might be expressed as a nuclear factor that could regulate transcription.3,4,12 In contrast to this prediction, we demonstrate that the EGFP-pyrin fusion protein is expressed in the cytoplasm of COS-1 cells. To exclude that EGFP may interfere with nuclear localization, we expressed a known nuclear protein, cyclin A1, fused 3′ of EGFP, and found that it was clearly expressed in the nucleus. Our data suggest that pyrin is expressed in the cytoplasm and therefore may evoke functions other than a transcription factor. Alternatively, the intracellular localization of pyrin may change in response to appropriate stimuli, and pyrin might be translocated into the nucleus upon stimulation. For example, signal transducers and activators of transcription (STAT) proteins are expressed as monomeric cytoplasmic proteins and, in response to growth factors, they dimerize and translocate into the nucleus to act as transcription factors.13 Also, the transcription factor NFκB, when expressed as a GFP fusion protein in COS-7 cells, undergoes translocation from the cytoplasm to the nucleus upon stimulation with tumor necrosis factor.14 Although a variety of stimuli has been tested, none induced pyrin-EGFP to migrate into the nucleus of stably transfected U937 myeloid cells.
The distinct expression pattern of EGFP-pyrin in transfected COS cells might be correlated with the Golgi apparatus, as has been shown for another disease gene product.15 The Batten disease protein CLN3p is a Golgi integral membrane protein that displays a juxtanuclear asymmetric localization pattern when expressed as a GFP fusion protein in COS-7 cells.15 Our data suggest that pyrin might be expressed in association with the Golgi apparatus as well. This predicts a potential role for pyrin in Golgi-mediated protein assembly or transport.
Expression of MEFV mRNA was analyzed previously by Northern blot and detected in peripheral blood neutrophils and in 1 colon cancer cell line, SW 480.3 4 Hence, we tested both hematopoietic and nonhematopoietic normal human tissue and cell lines for expression of MEFV mRNA. Among the hematopoietic cell populations,MEFV mRNA was strongly expressed in neutrophils and was less pronounced in monocytes, CD19+ B cells, and CD3+ T cells. This low-level expression of MEFV may not be of physiologic relevance. So far, the expression levels of the corresponding protein, pyrin, have not been studied, and it remains an open question whether low levels of MEFV mRNA result in a significant amount of pyrin protein.
In nonhematopoietic tissues, MEFV mRNA was detected predominantly in spleen, lung, and muscle mRNA. This indicates that expression of MEFV is regulated in a tissue-specific manner with preferential expression in hematopoietic cells. Expression ofMEFV in lung and muscle might to some extent result from infiltrating macrophages or contaminating neutrophils.
To address a potential role for MEFV in tumorigenesis, we tested 43 human tumor cell lines for MEFV mRNA expression. Among the hematopoietic cell lines tested, we found expression ofMEFV predominantly in myeloid leukemic cell lines (8 of 11). Furthermore, we demonstrated prominent induction of MEFV mRNA during granulocytic differentiation of HL-60 cells. Expression ofMEFV in the colon cancer cell line SW 480 was confirmed, andMEFV expression was also found in 3 other colon cancer cell lines. In addition, MEFV transcripts were detectable in 3 prostate cancer cell lines. However, no dramatic differences inMEFV expression were observed between normal and matched cancerous colon or prostate samples from the same individuals (data not shown). At this time, we do not know if the relatively high expression of MEFV mRNA in several colon and prostate cancer cell lines has any clinical relevancy.
Missense mutations of the MEFV gene have been described in patients with FMF, but how these mutations affect the function of pyrin is unknown. Both activation and inactivation of pyrin as well as changes in either its localization or interaction with other proteins might result from the mutations. We expressed in COS-1 cells EGFP-pyrin bearing either of the 2 of the most frequent mutations in FMF patients. The perinuclear expression of EGFP-pyrin was not altered by the point muations. Interestingly, the mutations described so far primarily occur in the C-terminal B30.2 domain of pyrin.3-5 This domain is predicted to obtain a globular conformation when expressed as a protein and is present in different families of proteins (eg, nuclear factor Staf5016 or ret finger protein17) and might be involved in protein-protein interactions. The identification and characterization of interacting proteins might elucidate the functional role of pyrin.
Supported by grants from the National Institutes of Health, U.S. Army, and the C. and H. Koeffler and the Parker Hughes Funds. N. Tidow is recipient of a fellowship of the Deutsche Forschungsgemeinschaft. H. P. Koeffler holds the Mark Goodson Chair in Oncology Research and is a member of the Jonsson Cancer Center.
This work is in memory of Dr Marge Goldberg, an indefatigable assistant and a champion of individuals with familial Mediterranean fever.
Reprints:Nicola Tidow, Davis Bldg, RM5005, Division of Hematology/Oncology, Cedars-Sinai Medical Center/UCLA School of Medicine, 8700 Beverly Blvd, Los Angeles, CA 90048.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal