Abstract
The engraftment capacity of bone marrow–derived mesenchymal cells was investigated in 41 patients who had received a sex-mismatched, T-cell–depleted allograft from human leukocyte antigen (HLA)–matched or –mismatched family donors. Polymerase chain reaction (PCR) analysis of the human androgen receptor (HUMARA) or the amelogenin genes was used to detect donor-derived mesenchymal cells. Only 14 marrow samples (34%) from 41 consenting patients generated a marrow stromal layer adequate for PCR analysis. Monocyte-macrophage contamination of marrow stromal layers was reduced below the levels of sensitivity of HUMARA and amelogenin assays (5% and 3%, respectively) by repeated trypsinizations and treatment with the leucyl-leucine (leu-leu) methyl ester. Patients who received allografts from 12 female donors were analyzed by means of the HUMARA assay, and in 5 of 12 cases a partial female origin of stromal cells was demonstrated. Two patients who received allografts from male donors were analyzed by amplifying the amelogenin gene, and in both cases a partial male origin of stromal cells was shown. Fluorescent in situ hybridization analysis using a Y probe confirmed the results of PCR analysis and demonstrated in 2 cases the existence of a mixed chimerism at the stromal cell level. There was no statistical difference detected between the dose of fibroblast progenitors (colony-forming unit–F [CFU-F]) infused to patients with donor- or host-derived stromal cells (1.18 ± 0.13 × 104/kg vs 1.19 ± 0.19 × 104/kg; P ≥ .97). In conclusion, marrow stromal progenitors reinfused in patients receiving a T-cell–depleted allograft have a limited capacity of reconstituting marrow mesenchymal cells.
Introduction
Stromal cells of the marrow microenvironment and their associated biosynthetic products provide the physical framework within which hematopoiesis occurs, synthesize or present growth-stimulatory and growth-inhibitory factors, produce numerous extracellular matrix ligands, and express a broad repertoire of adhesion molecules that serve to mediate hematopoietic stem cell (HSC) homing and to transmit regulatory signals.1-4
Several in vitro and in vivo data provide circumstantial evidence of the existence of mesenchymal stem cells (MSCs) residing within the marrow microenvironment and having multilineage differentiation capacity.5-7 In nonhuman models, reinfused MSCs migrate to and become incorporated into several tissues of the recipient animals. In these animals the MSCs are capable of eliciting tissue-specific differentiation programs indicating that, similarly to HSCs, mesenchymal cells have a multi-organ homing capacity and an intrinsic degree of plasticity.8,9 Based on this peculiar functional activity, MSCs may be used either to replace the marrow microenvironment damaged by myeloablative chemotherapy; to correct acquired or inherited disorders of bone, muscle, or cartilage; or to serve as vehicles for gene therapy.10-13
Although demonstrated in several animal models,14-18 the transplantability of marrow stromal elements remains controversial in humans.19-26 Substantial modifications of stem cell transplantation (SCT) that have occurred during the last decade prompted us to re-evaluate the issue of MSC transplantability. Marrow stromal layers generated in vitro from patients who had undergone a T-cell–depleted, sex-mismatched hematopoietic transplantation were extensively depleted of monocyte-macrophages27 and analyzed for the presence of donor-derived stromal cells by polymerase chain reaction (PCR) amplification of the human androgen receptor alpha locus (HUMARA) or the amelogenin genes. To quantify donor-derived stromal cells, fluorescence in-situ hybridization (FISH) analysis with a Y probe was also performed. Our results demonstrate that marrow stromal progenitors reinfused in patients receiving a T-cell–depleted allograft have a limited capacity of reconstituting marrow microenvironment.
Patients, materials, and methods
Patients
This study included 41 consenting patients who had received a T-cell–depleted, sex-mismatched allogeneic transplantation from human leukocyte antigen (HLA)–matched or –mismatched family donors. Patients failing to achieve a quantitatively adequate stromal cell growth were, whenever possible, sequentially assayed at the time of subsequent marrow aspirations. A persistently defective stromal cell growth was observed in 27 of 41 patients, whereas 14 of 41 patients generated a confluent (defined as more than 75% of the flask surface covered by stromal cells) or subconfluent (defined as 50% to 60% of the flask surface covered by stromal cells) marrow layer at some time-points after allograft transplantation and could be evaluated (Table 1). Conditioning regimens, including either total body irradiation (TBI), cyclophosphamide (CTX), thiotepa (TT), and antithymocyte globulin (ATG) or TBI, TT, ATG, and fludarabine (FLU), have been reported in detail elsewhere.28-30 Sources of hematopoietic cells were either bone marrow (n = 5), mobilized peripheral blood (n = 1), or bone marrow plus mobilized blood (n = 8). Donor marrow was depleted of T lymphocytes by soybean agglutination and 2 rounds of E-rosetting, whereas mobilized blood underwent CD34+ cell enrichment. Post-transplantation immunosuppressive treatment was not given. Hematopoietic chimerism was assessed by PCR amplification of a panel of variable-number tandem-repeat (VNTR) regions with different DNA polymorphism patterns.31 Dilution experiments showed that by using this technique, the presence of host cells could be detected at a level approaching 1%. The diagnosis and degree of acute graft-versus-host disease (GVHD) were assessed according to consensus criteria.32 Normal values for bone marrow progenitor cell growth were provided from 57 healthy subjects with a median age of 42 years (range, 20 to 67 years).33
Characteristics of the patients
| Case no. . | Age/sex . | Diagnosis . | HLA typing . | Conditioning regimen29 30 . | Cell source . | Reinfused Cells . | Acute GVHD, grade . | Days to WBC, 1 000/μL . | Days to PLT, 50 000/μL . | Hematopoietic chimerism, % donor cells . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T-cell– depleted BM, × 106/kg . | CD34+ BM cells, × 106/kg . | CD34+ PB cells, × 106/kg . | ||||||||||
| 1 | 44/M | AML | matched | TBI/CTX/TT/ATG | BM + PB | 24 | 0.69 | 6.7 | 0 | 12 | 16 | ND |
| 2 | 38/M | CML | matched | TBI/CTX/TT/ATG | BM + PB | 55 | 1.05 | 5.4 | 0 | 10 | 15 | 100 |
| 3 | 31/M | AML | mismatched | TBI/CTX/TT/ATG | BM | 27 | ND | — | 0 | 28 | 40 | ND |
| 4 | 59/M | AML | matched | TBI/CTX/TT/ATG | BM + PB | 14 | 0.26 | 2.2 | 0 | 12 | 18 | 100 |
| 5 | 54/M | CML | matched | TBI/CTX/TT/ATG | BM | 23 | ND | — | 0 | 17 | 31 | 100 |
| 6 | 45/M | ALL | mismatched | TBI/CTX/TT/ATG | BM + PB | 11 | 0.54 | 4.9 | 0 | 14 | 28 | 100 |
| 7 | 26/M | RAEB-T | mismatched | TBI/FLU/TT/ATG | BM + PB | 13 | 0.26 | 6.9 | 0 | 11 | 48 | 100 |
| 8 | 45/M | CML | matched | TBI/CTX/TT/ATG | BM | 59 | ND | — | 0 | 52 | 72 | 100 |
| 9 | 50/M | CML | matched | TBI/CTX/TT/ATG | BM | 20 | 0.64 | — | 0 | 10 | 38 | 100 |
| 10 | 36/F | AML | matched | TBI/CTX/TT/ATG | BM + PB | 51 | 1.17 | 2.9 | 0 | 14 | 33 | 100 |
| 11 | 25/M | AML | mismatched | TBI/FLU/TT/ATG | PB | — | — | 7.7 | 0 | 12 | 17 | 100 |
| 12 | 51/M | CML | matched | TBI/CTX/TT/ATG | BM | 45 | ND | — | 0 | 22 | 26 | 100 |
| 13 | 47/M | AML | mismatched | TBI/FLU/TT/ATG | BM + PB | 12 | 0.19 | 4.6 | 0 | 11 | 27 | 100 |
| 14 | 29/F | CML | mismatched | TBI/CTX/TT/ATG | BM + PB | 12 | 0.48 | 9 | 0 | 13 | 16 | 100 |
| Case no. . | Age/sex . | Diagnosis . | HLA typing . | Conditioning regimen29 30 . | Cell source . | Reinfused Cells . | Acute GVHD, grade . | Days to WBC, 1 000/μL . | Days to PLT, 50 000/μL . | Hematopoietic chimerism, % donor cells . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T-cell– depleted BM, × 106/kg . | CD34+ BM cells, × 106/kg . | CD34+ PB cells, × 106/kg . | ||||||||||
| 1 | 44/M | AML | matched | TBI/CTX/TT/ATG | BM + PB | 24 | 0.69 | 6.7 | 0 | 12 | 16 | ND |
| 2 | 38/M | CML | matched | TBI/CTX/TT/ATG | BM + PB | 55 | 1.05 | 5.4 | 0 | 10 | 15 | 100 |
| 3 | 31/M | AML | mismatched | TBI/CTX/TT/ATG | BM | 27 | ND | — | 0 | 28 | 40 | ND |
| 4 | 59/M | AML | matched | TBI/CTX/TT/ATG | BM + PB | 14 | 0.26 | 2.2 | 0 | 12 | 18 | 100 |
| 5 | 54/M | CML | matched | TBI/CTX/TT/ATG | BM | 23 | ND | — | 0 | 17 | 31 | 100 |
| 6 | 45/M | ALL | mismatched | TBI/CTX/TT/ATG | BM + PB | 11 | 0.54 | 4.9 | 0 | 14 | 28 | 100 |
| 7 | 26/M | RAEB-T | mismatched | TBI/FLU/TT/ATG | BM + PB | 13 | 0.26 | 6.9 | 0 | 11 | 48 | 100 |
| 8 | 45/M | CML | matched | TBI/CTX/TT/ATG | BM | 59 | ND | — | 0 | 52 | 72 | 100 |
| 9 | 50/M | CML | matched | TBI/CTX/TT/ATG | BM | 20 | 0.64 | — | 0 | 10 | 38 | 100 |
| 10 | 36/F | AML | matched | TBI/CTX/TT/ATG | BM + PB | 51 | 1.17 | 2.9 | 0 | 14 | 33 | 100 |
| 11 | 25/M | AML | mismatched | TBI/FLU/TT/ATG | PB | — | — | 7.7 | 0 | 12 | 17 | 100 |
| 12 | 51/M | CML | matched | TBI/CTX/TT/ATG | BM | 45 | ND | — | 0 | 22 | 26 | 100 |
| 13 | 47/M | AML | mismatched | TBI/FLU/TT/ATG | BM + PB | 12 | 0.19 | 4.6 | 0 | 11 | 27 | 100 |
| 14 | 29/F | CML | mismatched | TBI/CTX/TT/ATG | BM + PB | 12 | 0.48 | 9 | 0 | 13 | 16 | 100 |
AML indicates acute myelogenous leukemia; CML, chronic myelogenous leukemia; ALL, acute lymphoblastic leukemia; RAEB-T, refractory anemia with excess of blasts in transformation; TBI, total body irradiation; CTX, cyclophosphamide; TT, thiotepa; ATG, antilymphocyte globulin; FLU, fludarabine; BM, bone marrow; PB, peripheral blood; GVHD, graft-versus-host disease; WBC, white blood cell counts; PLT, platelet counts; ND, not done.
CFU-F assay
The assay for colony-forming unit–fibroblast (CFU-F) was performed according to Castro-Malaspina et al.34 Briefly, 4 mL (1 × 105/mL) marrow mononuclear cells (MNCs) obtained by centrifugation on a Ficoll-Hypaque gradient (density, 1.077 g/mL) and resuspended in complete medium consisting of alpha-medium (Gibco BRL Life Technologies, Grand Island, NY) supplemented with 12.5% fetal bovine serum (Stem Cell Technologies, Vancouver, British Columbia, Canada), 12.5% horse serum (Stem Cell Technologies), 2 mmol/L L-glutamine, 10−4 mol/L 2-mercaptoethanol, 0.2 mmol/L inositol, 20 μmol/L folic acid, and 10−6 mol/L freshly dissolved hydrocortisone were plated in 60-mm Petri dishes, and fibroblast colony growth was evaluated after incubation (37°C, 5% carbon dioxide [CO2]) for 14 days in a humidified atmosphere. For scoring aggregates of at least 50 cells of CFU-F, the dishes were stained with crystal violet and examined under an inverted microscope.
Marrow stromal cells
To generate marrow stromal layers, 3-5 × 105/mL MNCs resuspended in complete medium were inoculated into 25 cm2 tissue culture flasks and incubated in a humidified atmosphere (37°C, 5% CO2). On a weekly basis the cultures were fed by complete replacement of the medium. Confluent or subconfluent stromal layers were trypsinized for at least 3 times to achieve a substantial depletion of stromal layers by monocyte-macrophages. Stromal layers were further depleted of monocyte-macrophages by incubation (22°C, 40 minutes) with the antilysosomal compound leucyl-leucine (leu-leu) methyl ester (5 mmol/L) (Sigma Chemical Co, St Louis, MO), which induces a selective depletion of monocyte-macrophages related to the characteristically rich endowment of lysosomes within these cells.27 At the end of the incubation period, the cells were washed twice and resuspended in phosphate-buffered saline (PBS). An aliquot of stromal cells was analyzed by flow cytometry to confirm the depletion of monocyte-macrophage cells.
Hematopoietic progenitor cell assays
The assay for multilineage CFU (CFU-Mix), erythroid burst (BFU-E), and CFU-GM (granulocyte-macrophage) was carried out as described elsewhere.35 Briefly, 5 × 104/mL MNCs were plated in 35-mm Petri dishes in 1-mL aliquots of Iscove's modified Dulbecco's medium (IMDM) (Seromed, Berlin, Germany) containing 30% FBS (Stem Cell Technologies), 10−4 mol/L 2-mercaptoethanol (Gibco), and 1.1% (wt/vol) methylcellulose. The cultures were stimulated with 50 ng/mL stem cell factor (SCF) (Amgen Inc, Thousand Oaks, CA), 10 ng/mL interleukin-3 (IL-3) (Sandoz, Basel, Switzerland), 10 ng/mL granulocyte colony-stimulating factor (G-CSF) (Amgen), 10 ng/mL GM-CSF (Sandoz), and 3 U/mL erythropoietin (Amgen). Colony growth was evaluated after incubation (37°C, 5% CO2) for 14-18 days in a humidified atmosphere.35 The long-term culture–initiating cell (LTC-IC) assay was performed according to Sutherland et al.36 Briefly, test cell suspensions (5 × 106 MNCs) resuspended in complete medium were seeded into cultures containing a feeder layer of irradiated (8000 cGy) 3 × 104/cm2 murine M2-10B4 cells (gift of Dr C. Eaves, Terry Fox Laboratory, Vancouver, British Columbia, Canada) engineered by retroviral gene transfer to produce human IL-3 and human G-CSF. After 5 weeks in LTC, nonadherent and adherent cells were harvested and assayed for clonogenic cells. Absolute LTC-IC values were calculated by dividing the total number of clonogenic cells by 4, which, according to limiting dilution analysis studies reported by others,36 is the average output of clonogenic cells per LTC-IC.
Flow cytometry
Marrow stromal cells (1 × 105/mL) were incubated for 30 minutes on ice with 10 μL CD45-FITC (fluorescein isothiocyanate)/CD14-PE (phycoerythrin). Each fluorescence analysis included a double-negative isotype control immunoglobulin G1 [IgG1]FITC/IgG1PE), and antibodies and isotype controls were purchased (Becton Dickinson, San Jose, CA). The percentage of positive cells was determined by subtracting the percentage of fluorescent cells in the control from the percentage of cells positively stained with the appropriate antibody. The cells were analyzed on a fluorescent-activated cell sorter (FACSort) laser flow cytometry system (Becton Dickinson) equipped with a Macintosh Quadra 650 computer (Apple Computer Inc, Cupertino, CA) using Cell Quest software (Becton Dickinson).
HUMARA assay
The HUMARA gene was analyzed to identify the sex origin of stromal cells. The highly polymorphic trinucleotide CAG repeat in the first exon allowed us to distinguish the maternal and paternal alleles in heterozygotic females; the alleles can be identified after DNA amplification as 2 fragments of different length. In male cells only one fragment corresponding to the maternal allele can be detected. The PCR assay was performed according to Allen et al.37Briefly, 250 ng DNA was amplified with 25 pmol primer A, 5′-GCTGTGAAGGTTGCTGTTCCTCAT-3′, and primer B, 5′- TCCAGAATCTGTTCCAGAGCGTGC-3′.
The PCR reaction was performed in a final volume of 60 μL containing 1.5 U Taq polymerase and 5 μL 10 times PCR buffer comprising 500 mmol/L potassium chloride (KCl), 0.1 mol/L Tris HCl (tris[hydroxymethyl] aminomethane hydrochloride) (pH 8.8), 0.025 mol/L magnesium dichloride (MgCl2), 2 mg/mL bovine serum albumin (BSA), and 0.002 mol/L of each dNTP (dinucleoside 5′-triphosphate). The samples were amplified for 32 cycles (each cycle consisted of 60 seconds at 95°C, 50 seconds at 59°C, and 60 seconds at 72°C), with an initial denaturation at 95°C for 5 minutes. The PCR product was loaded on 60 cm 8% polyacrylamide gel in TAE buffer and subjected to electrophoresis for 20 hours at 300 V. The gel was then visualized using ethidium bromide.
Amelogenin assay
Amelogenin gene amplification was allowed to simultaneously detect both male and female cells by a single-step PCR reaction as described by Pugatsch et al.38 The female cells were detected by a single amplification product of 977 base pair (bp) (AMGX), and male cells gave rise to 2 products of 977 bp (AMGX) and 788 bp (AMGY), respectively. DNA was isolated from stromal cells by phenylchloroform extraction using standard techniques.39For each DNA sample, a PCR was performed using primer A, 5′-CTGATGGTTGGCCTCAAGCCTGTG-3′; primer B, 5′-TAAAGAGATTCATTAACTTGACTG-3′; and primer C, 5′-GCCCAAAGTTAGT- AATTTTAC-3′.
The PCR assay was performed in a final volume of 30 μL containing 1 μg DNA, 100 pmol of each amplification primer, 1.5 U Taqpolymerase, and 3 μL 10 times PCR buffer comprising 500 mmol/L KCl, 0.1 mol/L Tris HCl (pH 8.8), 0.025 mol/L MgCl2, 0.002 mol/L of each dNTP, and 2 mg/mL BSA. PCR was carried out for 30 cycles (each cycle consisted of 1 minute at 94°C, 2 minutes at 60°C, and 2 minutes at 72°C), with an initial denaturation at 95°C for 5 minutes. Amplification products (10 μL) were analyzed on 1.6% agarose gel in TBE buffer with 0.5 μg/mL ethidium bromide.
FISH analysis
FISH analysis using a digoxygenin-labeled probe for chromosome Y (Ingen Laboratories, Inc) was performed on trypsinized stromal cells according to the manufacturer's instructions. Briefly, methanol/acetic acid–fixed cell samples were placed on glass slides and air dried for 10-15 minutes at room temperature. Following the addition of 10 μL DNA probe to the sample area, the probe and target DNA were denaturated simultaneously (9 minutes, 90°C). The slides were then transferred to a Hyb Box, where hybridization occurred spontaneously. The slides were subsequently washed in 50% formamide/1 times side scatter criteria (SSC) (5 minutes, 45°C) and then in a 1 times SSC (2-5 minutes, 45°C). For digoxygenin detection, 15 μL antidigoxygenin Fab fragments labeled with fluorescein (FITC) were placed over the sample area. The slides were incubated (5 minutes, 37°C), washed in 2 times SSC (2 minutes, room temperature), and counterstained with propidium iodide (PI) in 10 μL antifade solution before being examined under an Axiophot fluorescent microscope (Zeiss). A dual wavelength filter was used to view both FITC and PI simultaneously.
Statistical analysis
Four plates were scored for each data point per experiment, and the results were expressed as the mean plus or minus 1 SEM. Statistical analysis was performed with the statistical package Statview (BrainPower Inc, Calabazas, CA) run on a Macintosh 6300 personal computer (Apple Computer). The Student t test for unpaired data (2-tailed) was used to test the probability of significant differences between samples.
Results
A total of 68 samples from 41 consenting patients were plated under long-term culture conditions in order to generate marrow stromal layers to be analyzed for the presence of donor-derived mesenchymal cells. A persistently defective stromal cell growth that prevented molecular analysis was observed in 27 of 41 patients, whereas a confluent or subconfluent stromal cell layer was generated by 14 of 41 patients (34%) at some time-points after transplantation. The median interval between allografting and marrow aspiration was not different in the 2 groups of patients who generated or failed to generate a stromal cell layer (6 vs 5 months,P ≥ .05).
At the time of the study all patients were in complete hematological remission and displayed a full donor hematopoiesis, as demonstrated by DNA polymorphism analysis (Table 1). In vitro culture of hematopoietic cells revealed significant damage of hematopoietic progenitor cell compartments (Table 2). Compared to healthy controls, allografted patients showed a statistically significant reduction of the mean (mean ± SEM) number of multipotent CFU-Mix (5 ± 1 vs 0.3 ± 0.1; P ≤ .05), erythroid BFU-E (69 ± 5 vs 8 ± 1; P ≤ .05), and CFU-GM (200 ± 11 vs 27 ± 5; P ≤ .05) as well as the more primitive LTC-IC progenitors (266 ± 31 vs 17 ± 4;P ≤ .001) (Table 2).
In vitro growth of marrow hematopoietic progenitors
| Case no. . | Assay time, months post-SCT . | CFU-mix . | BFU-E . | CFU-GM . | LTC-IC . |
|---|---|---|---|---|---|
| 1 | 2 | 0.5 | 4 | 40 | 22 |
| 6 | 0 | 9 | 93 | 11 | |
| 2 | 2 | 0 | 11 | 39 | ND |
| 3 | 50 | 0 | 2 | 7 | cont |
| 62 | ND | ND | ND | ND | |
| 4 | 3 | 0 | 0 | 6 | ND |
| 5 | 44 | 1 | 9 | 41 | 5 |
| 59 | 0 | 14 | 38 | ND | |
| 6 | 2 | 0 | 3 | 14 | 0 |
| 7 | 3 | 0 | 0 | 1 | cont |
| 6 | 0 | 9 | 9 | 3 | |
| 9 | 0 | 16 | 37 | 42 | |
| 8 | 48 | 0.25 | 12 | 8 | 5 |
| 79 | 0 | 9 | 7 | ND | |
| 9 | 2 | 0 | 2 | 3 | ND |
| 10 | 3 | 0.5 | 9 | 25 | 6 |
| 6 | 0.3 | 1 | 9 | ND | |
| 10 | 1 | 3 | 6 | ND | |
| 18 | 1 | 6 | 12 | 3 | |
| 31 | 0 | 1 | 5 | ND | |
| 11 | 1 | 0 | 1 | 21 | ND |
| 12 | 82 | 0.5 | 5 | 31 | ND |
| 13 | 3 | 0 | 7 | 61 | cont |
| 6 | 1 | 22 | 76 | cont | |
| 17 | 2 | 32 | 64 | 64 | |
| 14 | 17 | 0 | 8 | 21 | 35 |
| Normal controls (n = 55) | |||||
| Mean ± SEM | 5 ± 1 | 69 ± 5 | 200 ± 11 | 266 ± 31 | |
| Case no. . | Assay time, months post-SCT . | CFU-mix . | BFU-E . | CFU-GM . | LTC-IC . |
|---|---|---|---|---|---|
| 1 | 2 | 0.5 | 4 | 40 | 22 |
| 6 | 0 | 9 | 93 | 11 | |
| 2 | 2 | 0 | 11 | 39 | ND |
| 3 | 50 | 0 | 2 | 7 | cont |
| 62 | ND | ND | ND | ND | |
| 4 | 3 | 0 | 0 | 6 | ND |
| 5 | 44 | 1 | 9 | 41 | 5 |
| 59 | 0 | 14 | 38 | ND | |
| 6 | 2 | 0 | 3 | 14 | 0 |
| 7 | 3 | 0 | 0 | 1 | cont |
| 6 | 0 | 9 | 9 | 3 | |
| 9 | 0 | 16 | 37 | 42 | |
| 8 | 48 | 0.25 | 12 | 8 | 5 |
| 79 | 0 | 9 | 7 | ND | |
| 9 | 2 | 0 | 2 | 3 | ND |
| 10 | 3 | 0.5 | 9 | 25 | 6 |
| 6 | 0.3 | 1 | 9 | ND | |
| 10 | 1 | 3 | 6 | ND | |
| 18 | 1 | 6 | 12 | 3 | |
| 31 | 0 | 1 | 5 | ND | |
| 11 | 1 | 0 | 1 | 21 | ND |
| 12 | 82 | 0.5 | 5 | 31 | ND |
| 13 | 3 | 0 | 7 | 61 | cont |
| 6 | 1 | 22 | 76 | cont | |
| 17 | 2 | 32 | 64 | 64 | |
| 14 | 17 | 0 | 8 | 21 | 35 |
| Normal controls (n = 55) | |||||
| Mean ± SEM | 5 ± 1 | 69 ± 5 | 200 ± 11 | 266 ± 31 | |
The mean values of CFU-Mix, BFU-E, and CFU-GM are given per 5 × 104 cells, and the mean values of LTC-IC are given per 2 × 106 cells.
ND indicates not done; cont, fungal contamination.
As shown in Table 3, a stromal progenitor cell growth allowing molecular analysis could be detected 1-3 months after transplantation in 6 patients (case Nos. 1, 2, 4, 6, 9, and 11), whereas a substantially delayed stromal growth was observed in 3 patients (case Nos. 7, 10, and 13). The remaining patients (case Nos. 3, 5, 8, 12, and 14) were not studied early after allografting, thus these cases were not informative regarding the time course of stromal cell regeneration. Marrow stromal layers reached confluence or subconfluence after 4-5 weeks of culture (Table 3). Compared to healthy controls, a significant reduction of the mean incidence of CFU-F progenitors was evident (56 ± 6 vs 9 ± 2;P ≤ .05) (Table 3).
In vitro growth of marrow mesenchymal progenitors
| Case no. . | Assay time, months post-SCT . | CFU-F, per 1 × 106 MNCs . | Marrow stromal layers . | ||
|---|---|---|---|---|---|
| Type of growth . | Confluence, week . | Adipogenesis, week . | |||
| 1 | 2 | ND | confluent3-150 | 5 | 2 |
| 6 | 30 | confluent | 5 | 2 | |
| 2 | 2 | 21 | subconfluent3-151 | 5 | 4 |
| 3 | 50 | 7 | confluent | 5 | 3 |
| 62 | ND | confluent | 4 | 3 | |
| 4 | 3 | 10 | subconfluent | 5 | 4 |
| 5 | 44 | 0 | no growth | — | — |
| 59 | 7 | confluent | 4 | 3 | |
| 6 | 2 | ND | subconfluent | 4 | 3 |
| 7 | 3 | 0 | no growth | — | — |
| 6 | 3 | defective3-152 | NR | — | |
| 9 | 20 | confluent | 4 | 2 | |
| 8 | 48 | 0 | no growth | — | — |
| 79 | 10 | confluent | 5 | 4 | |
| 9 | 2 | 20 | confluent | 5 | 4 |
| 10 | 3 | 0 | no growth | — | — |
| 6 | 0 | no growth | — | — | |
| 10 | 0 | no growth | — | — | |
| 18 | 3 | defective | NR | — | |
| 31 | 16 | subconfluent | 4 | 3 | |
| 11 | 1 | ND | subconfluent | 5 | 4 |
| 12 | 82 | ND | subconfluent | 5 | 4 |
| 13 | 3 | 0 | no growth | — | — |
| 6 | 2 | defective | NR | — | |
| 17 | 12 | subconfluent | 4 | 3 | |
| 14 | 17 | 18 | confluent | 4 | 3 |
| Normal controls (n = 55) | |||||
| Mean ± SEM | 56 ± 6 | confluent | 4 ± 1 | 3 ± 0.4 | |
| Case no. . | Assay time, months post-SCT . | CFU-F, per 1 × 106 MNCs . | Marrow stromal layers . | ||
|---|---|---|---|---|---|
| Type of growth . | Confluence, week . | Adipogenesis, week . | |||
| 1 | 2 | ND | confluent3-150 | 5 | 2 |
| 6 | 30 | confluent | 5 | 2 | |
| 2 | 2 | 21 | subconfluent3-151 | 5 | 4 |
| 3 | 50 | 7 | confluent | 5 | 3 |
| 62 | ND | confluent | 4 | 3 | |
| 4 | 3 | 10 | subconfluent | 5 | 4 |
| 5 | 44 | 0 | no growth | — | — |
| 59 | 7 | confluent | 4 | 3 | |
| 6 | 2 | ND | subconfluent | 4 | 3 |
| 7 | 3 | 0 | no growth | — | — |
| 6 | 3 | defective3-152 | NR | — | |
| 9 | 20 | confluent | 4 | 2 | |
| 8 | 48 | 0 | no growth | — | — |
| 79 | 10 | confluent | 5 | 4 | |
| 9 | 2 | 20 | confluent | 5 | 4 |
| 10 | 3 | 0 | no growth | — | — |
| 6 | 0 | no growth | — | — | |
| 10 | 0 | no growth | — | — | |
| 18 | 3 | defective | NR | — | |
| 31 | 16 | subconfluent | 4 | 3 | |
| 11 | 1 | ND | subconfluent | 5 | 4 |
| 12 | 82 | ND | subconfluent | 5 | 4 |
| 13 | 3 | 0 | no growth | — | — |
| 6 | 2 | defective | NR | — | |
| 17 | 12 | subconfluent | 4 | 3 | |
| 14 | 17 | 18 | confluent | 4 | 3 |
| Normal controls (n = 55) | |||||
| Mean ± SEM | 56 ± 6 | confluent | 4 ± 1 | 3 ± 0.4 | |
NR indicates not reached.
Flask surface covered by stromal cells ≥75%.
Flask surface covered by stromal cells from 50% to 60%.
Flask surface covered by stromal cells ≤20%.
Because marrow stroma contamination with donor-derived monocyte-macrophages is the major cause of false-positive results,40 depletion of monocyte-macrophages represents a critical issue in studies aimed at investigating the origin of marrow mesenchymal cells following SCT. In the present study the strategy used to eliminate monocyte-macrophage cells contaminating marrow stroma was based on repeated trypsinizations of stromal layers and treatment of cells before analysis with the leu-leu methyl ester.27Flow cytometry analysis of stromal cells with anti-CD45 and anti-CD14 monoclonal antibodies demonstrated that the depletion procedure consistently resulted in cell populations which were CD45−(on average, at least 98%) and contained percentages of CD14+ cells (on average, less than or equal to 2%) (Table4) that were below the detection limits of our PCR assays. In fact, preliminary experiments performed to standardize the PCR assays demonstrated that the levels of sensitivity for HUMARA and amelogenin analysis were 5% and 3%, respectively. Therefore, these levels of sensitivity prevented the detection of donor-derived CD14+ cells and avoided false-positive results.
Analysis of marrow stromal cell origin
| Case no. . | Assay time, months post-SCT . | BM CFU-F, × 104/kg . | CD14+cells within marrow stroma, % . | MSC chimerism . | |
|---|---|---|---|---|---|
| PCR analysis4-150 . | FISH analysis, % pos cells4-151 . | ||||
| 1 | 2 | 1.10 | <1 | donor | ND |
| 6 | — | <1 | donor | ND | |
| 2 | 2 | 0.77 | 2 | donor | ND |
| 3 | 50 | ND | <1 | donor | ND |
| 62 | — | 2 | donor | ND | |
| 4 | 3 | 0.81 | <1 | host | ND |
| 5 | 59 | ND | 2 | donor | ND |
| 6 | 2 | 0.93 | 2 | host | ND |
| 7 | 9 | 1.40 | <1 | host | ND |
| 8 | 79 | ND | <1 | host | ND |
| 9 | 2 | 1.54 | <1 | donor | ND |
| 10 | 31 | 1.35 | <1 | donor | 50 |
| 11 | 1 | NA | 2 | host | ND |
| 12 | 82 | ND | 2 | host | 99 |
| 13 | 17 | 1.62 | 2 | host | 97 |
| 14 | 17 | 1.15 | <1 | donor | 38 |
| Case no. . | Assay time, months post-SCT . | BM CFU-F, × 104/kg . | CD14+cells within marrow stroma, % . | MSC chimerism . | |
|---|---|---|---|---|---|
| PCR analysis4-150 . | FISH analysis, % pos cells4-151 . | ||||
| 1 | 2 | 1.10 | <1 | donor | ND |
| 6 | — | <1 | donor | ND | |
| 2 | 2 | 0.77 | 2 | donor | ND |
| 3 | 50 | ND | <1 | donor | ND |
| 62 | — | 2 | donor | ND | |
| 4 | 3 | 0.81 | <1 | host | ND |
| 5 | 59 | ND | 2 | donor | ND |
| 6 | 2 | 0.93 | 2 | host | ND |
| 7 | 9 | 1.40 | <1 | host | ND |
| 8 | 79 | ND | <1 | host | ND |
| 9 | 2 | 1.54 | <1 | donor | ND |
| 10 | 31 | 1.35 | <1 | donor | 50 |
| 11 | 1 | NA | 2 | host | ND |
| 12 | 82 | ND | 2 | host | 99 |
| 13 | 17 | 1.62 | 2 | host | 97 |
| 14 | 17 | 1.15 | <1 | donor | 38 |
NA indicates not applicable.
Indicates that the HUMARA assay was performed in all but 2 patients (case nos. 10 and 14) who were analyzed by the amelogenin assay.
Indicates that the FISH analysis was performed for the Y chromosome of 500 cells.
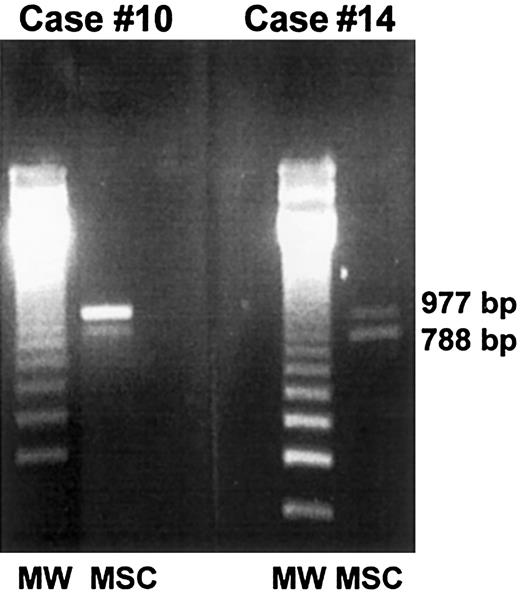
Twelve male patients who received allografts from female donors were analyzed by means of the HUMARA assay. In 5 of 12 cases, 2 alleles were detected after PCR amplification, thus demonstrating the partial female origin of stromal cells (Table 4, Figure1). Female patients who received allografts from male donors were analyzed by amplifying the amelogenin gene (case Nos. 10 and 14). In both cases, a 788 fragment located on the Y chromosome and a 977 fragment located on the X chromosome were detected, thus demonstrating the partial male origin of stromal cells (Table 4, Figure 2). In cases Nos. 1 and 3, the patients were studied on 2 separate occasions, and donor-derived stromal cells were detected in both patients at both time-points (Table4).
Representative results of stromal cell analysis by PCR-based HUMARA analysis.
Marrow stromal cells from 3 male patients (case nos. 2, 4, and 5) allografted into female donors were generated in vitro, extensively depleted of monocyte-macrophages, and analyzed by PCR for the presence of donor-derived mesenchymal cells. In case nos. 2 and 5, 2 HUMARA alleles were detected by PCR amplification, thus demonstrating the partial female origin of stromal cells, whereas in case no. 4 only one allele was detected, thus demonstrating the host origin of stromal cells. MW indicates molecular weight marker; MSC, marrow stromal cells.
Representative results of stromal cell analysis by PCR-based HUMARA analysis.
Marrow stromal cells from 3 male patients (case nos. 2, 4, and 5) allografted into female donors were generated in vitro, extensively depleted of monocyte-macrophages, and analyzed by PCR for the presence of donor-derived mesenchymal cells. In case nos. 2 and 5, 2 HUMARA alleles were detected by PCR amplification, thus demonstrating the partial female origin of stromal cells, whereas in case no. 4 only one allele was detected, thus demonstrating the host origin of stromal cells. MW indicates molecular weight marker; MSC, marrow stromal cells.
PCR-based amelogenin gene amplification.
Marrow stromal cells from 2 female patients who received allografts from male donors (case nos. 10 and 14) were generated in vitro, extensively depleted of monocyte-macrophages, and analyzed by PCR for the presence of donor-derived mesenchymal cells. In both cases, a 788 fragment located on the Y chromosome and a 977 fragment located on the X chromosome were detected, thus demonstrating the partial male origin of stromal cells.
PCR-based amelogenin gene amplification.
Marrow stromal cells from 2 female patients who received allografts from male donors (case nos. 10 and 14) were generated in vitro, extensively depleted of monocyte-macrophages, and analyzed by PCR for the presence of donor-derived mesenchymal cells. In both cases, a 788 fragment located on the Y chromosome and a 977 fragment located on the X chromosome were detected, thus demonstrating the partial male origin of stromal cells.
To investigate whether stromal cells were totally or partially donor-derived (ie, having the existence of either a mixed or complete chimerism), FISH analysis using a Y probe was performed in 4 patients (cases Nos. 10, 12, 13, and 14). As shown in Table 4, the percentages of Y-positive cells ranged from 38% to 99%. In these 4 patients, the results of FISH analysis were in keeping with the results of PCR analysis, and in 2 patients (case Nos. 10 and 14) the existence of a mixed chimerism at the stromal cell level was demonstrated.
In 9 of 14 patients the stromal cell content of the graft could be analyzed by assaying the growth of fibroblast CFU-F (Table 4). Overall, the mean dose of reinfused CFU-F was 1.2 ± 0.1 × 104/kg (range, 0.77-1.62 × 104/kg) body weight, with no statistical difference detected between the dose of CFU-F reinfused to patients with donor- or host-derived stromal cells (1.18 ± 0.13 × 104/kg vs 1.19 ± 0.19 × 104/kg; P ≥ .97). All 7 patients in whom donor-derived stromal cells were detected had received allografts with T-cell–depleted bone marrow, and 4 of 7 patients had also received mobilized CD34+ cells. Again, there was no statistical difference found between the dose of reinfused hematopoietic cells or the time to hematopoietic reconstitution and the detection of donor- versus host-derived stromal cells (Tables 1 and 4). We reinfused 1 of 14 patients (case No. 11) with mobilized blood only, and no evidence of donor-derived stroma could be demonstrated.
Discussion
In the present work we demonstrate that following T-cell–depleted SCT using either HLA-matched or -mismatched family donors, stromal progenitors reinfused with HSCs induce a mixed chimerism at the stromal cell level in a limited number of patients, ie, 7 of 41 patients (17%).
Marrow stromal cells generated ex vivo from patients with hematological malignancies who had undergone autologous or allogeneic transplantation show a significant impairment of their proliferative and differentiative potential as well as impairment of their ability to support the growth of allogeneic CD34+cells.41 In keeping with these data, only 34% of the patients reported herein generated a confluent or subconfluent stromal layer, which further confirms the substantial microenvironment damage induced by chemoradiotherapy. No distinct clinical or biological feature could identify patients who either generated (14 of 41 patients) or failed to generate (27 of 41 patients) a stromal cell layer. In particular, the interval between allografting and the time of the study was not predictive for the capacity of in vitro stromal cell generation.
Substantial changes of SCT methodology that have occurred during the last decade, including the introduction of highly immunosuppressive and myeloablative conditioning regimens or the use of blood-derived stem cells, might play a role in stromal cell transplantability. Indeed, in our patients the detection of donor-derived stromal cells could not be predicted by any specific clinical feature or graft-related parameter, such as the dose of reinfused cells or the source of allografted stem cells, noted prior to transplantation. However, all patients with chimeric marrow stroma were characterized by the following: (1) an allograft with extensively T-cell–depleted marrow (7 of 7 patients) and mobilized blood (4 of 7 patients) stem cells; (2) no evidence of acute GVHD; and (3) a TBI-based regimen conditioning. Whether or not these features could have conditioned an in vivo peculiar cytokine environment, which in turn might have been responsible for stromal progenitor engraftment, remains a matter of hypothesis. Incidence and severity of acute GVHD, as well as differences inherent to graft cell composition, may play a relevant role in preventing stromal cell engraftment. Acute GVHD, which is characterized by the production of cytokines that might negatively regulate marrow homing and/or proliferation of stromal cells, was not observed in our patients who received allografts with extensively T-cell–depleted hematopoietic cells.42 Additionally, we can hypothesize that T-cell depletion performed in our patients might have eliminated a cell population suppressing stromal progenitors, thus facilitating stromal cell engraftment.
In addition to “qualitative” changes in cell composition, the reinfusion of a T-cell–depleted graft is associated with the “quantitative” requirement of a large cell inoculum of CD34+ cells. Recent data of the nonobese diabetic/severe combined immunodeficiency (NOD/SCID) model suggest that transplantation of mobilized blood progenitors is associated with the engraftment of mesenchymal cells within a marrow microenvironment, which is probably due to the existence of circulating stromal progenitors.43Therefore, in our patients the detection of donor-derived stromal cells may reflect the number of infused cells, including hematopoietic and mesenchymal progenitors, and also the reinfusion of still unidentified cells of stromal origin. In the present study the mesenchymal cell content of the graft was analyzed in 9 of 14 cases by assaying the clonogenic growth of CFU-F progenitors. Indeed, there was no evident correlation between the number of reinfused CFU-F progenitors and the post-transplantation detection of donor-derived stromal cells. However, this finding may reflect intrinsic limitations of the CFU-F assay in detecting a population of primitive stromal cells with reconstitutive capacity. Characterization of the stromal cell content of the graft will require more extensive studies, including immunophenotyping and functional assays, on a larger number of patients.
Several studies in animal models have clearly demonstrated that stromal cells not only seed the bone marrow but also enhance hematopoietic recovery. However, the transplantability of marrow stromal elements still remains an open issue in humans.45 Controversial results so far reported by different groups can be explained by either methodological differences in detecting donor-derived mesenchymal cells or differences in the doses of reinfused stromal cells. In addition, the heterogeneity of conditioning regimens, immunosuppression after transplantation, graft manipulation, and patient-related biological differences might account for such conflicting results. Indeed, the low frequency of stromal progenitors within adult bone marrow and the small number of mesenchymal progenitors infused during a conventional allogeneic transplantation might represent the major cause explaining the lack of stromal cell transplantability in humans.
Regeneration of stromal cells soon after transplantation was associated with donor-derived stroma in 3 of 6 patients (case Nos. 1, 2, and 9). Among patients who had a substantial delay in stromal cell regeneration (case Nos. 7, 10, and 13), engraftment of donor-derived stromal cells was detected only in one patient (case No. 10) at more than 31 months after allografting. These data demonstrate the existence of different patterns of CFU-F regeneration after T-cell–depleted allografting, including “early” and “late” regeneration, and suggest that engraftment of donor-derived stromal progenitors is an early event after transplantation, which can eventually be underestimated by sampling error or methodological limitations inherent to the culture systems we have used.
The fundamental role of marrow microenvironment in hematopoietic regulation has been demonstrated by several in vitro and in vivo studies which show that marrow stroma components can regulate and influence the production of early and mature hematopoietic cells.3,46-48 The importance of the marrow microenvironment has recently been highlighted by the demonstration that the developmental potential of hematopoietic stem cells could be reprogrammed by changing their microenvironment.49 In addition, the existence of unique niches that promote either short-term or long-term hematopoiesis has been affirmed by the isolation of murine stromal cell lines which differ markedly in their ability to maintain enriched populations of repopulating stem cells in vitro.50 Overall, these findings strongly support a crucial role of MSCs in improving the rate and quality of myeloid as well as lymphoid engraftment after lymphomyeloablative and stroma-damaging treatment. Despite the recent demonstration of the feasibility and safety of infusing culture-expanded autologous MSCs in advanced breast cancer patients undergoing peripheral blood SCT,51 the homing properties and the long-term reconstitutive capacity of these culture-expanded MSCs remain largely unknown.
In conclusion, we have demonstrated that marrow stromal progenitors reinfused with HSCs in patients receiving a T-cell–depleted allograft have a limited capacity of reconstituting bone marrow microenvironment. Such a limited functional capacity strongly suggests that conventional hematopoietic grafts should be supplemented with ex vivo generated mesenchymal cells that are capable of long-term functional capacity in order to improve marrow stromal reconstitution following SCT. However, several MSC-related issues, including marrow homing, generation of a differentiated progeny in the recipient marrow, capacity of infused stromal cells to elicit the desired biological activity, and the life-span of infused cells, still need to be adequately explored and dissected in preclinical models.
Supported in part by a grant from the Ministero dell'Università e della Ricerca Scientifica e Tecnologica, Rome, Italy; the Associazione Italiana per la Ricerca sul Cancro, Milan, Italy.
D.C. and C.C.-S. contributed equally to this work.
Submitted February 28, 2000; accepted July 19, 2000.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Carmelo Carlo-Stella, Bone Marrow Transplantation Unit, Istituto Nazionale Tumori, Via Venezian 1, 20133 Milano, Italy; e-mail: carmelo.carlostella@unimi.it.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal