Abstract
The IL-3 family of cytokines transduces signals through Stat5 and regulates myeloid development. Previous studies have determined that a carboxy terminally truncated isoform of Stat5 is activated in immature myeloid cells. This isoform, which lacks a transcriptional activation domain, is generated by a protein-processing event. To determine whether Stat5 cleavage plays an important role in the growth and maturation of myeloid progenitors, the FdCP1 model of myeloid maturation was evaluated. FdCP1 cells are IL-3–dependent myeloid progenitors that differentiate into monocytes when cultured in granulocyte macrophage–colony-stimulating factor (GM-CSF). Consistent with their immature phenotype, when FdCP1 cells are cultured in IL-3 they exhibit robust protease activity and signal through truncated Stat5 isoforms. In contrast, maturation leads to a loss of protease activity and a switch to the expression to full-length Stat5 isoforms. Introduction of a noncleavable, full-length Stat5 mutant into undifferentiated FdCP1 cells leads to a partially differentiated phenotype and prevents further differentiation in response to GM-CSF. These results support our hypothesis that Stat5 processing is important for myeloid maturation.
Introduction
The IL-3 family of cytokines (IL-3, IL-5, and granulocyte macrophage–colony-stimulating factor [GM-CSF]) plays an important role in the growth, differentiation, and activation of cells from the myeloid lineage.1 On binding their receptors, these ligands induce the activation of several signaling pathways, including the MAP kinase, PI-3 kinase, and the JAK-STAT pathways (reviewed in deGroot et al2). Of these, the JAK-STAT pathway is the most specific and has been shown to regulate many events during leukocyte maturation and activation (reviewed in Darnell3 and Schindler and Strehlow4). STATs (signal transducers and activators of transcription) are latent cytoplasmic transcription factors that become activated by receptor-associated tyrosine kinases from the JAK (Janus kinase) family. Activated STATs dimerize, translocate to the nucleus, and bind to members of the GAS (gamma activation site) family of enhancers to promote transcription.3,5,6 IL-3, IL-5, and GM-CSF induce the activation of Stat5a and Stat5b, which are encoded for by 2 distinct genes.7 8
An increasing number of studies had implicated Stat5 in hematopoiesis. For example, Stat5 null mice (Stat5a−/− and Stat5b−/−) exhibit important defects in erythropoiesis, granulopoiesis, and lymphopoiesis.2,9-12 In vitro studies, using either dominant negative or constitutively activated mutants of Stat5, have also implicated Stat5 in IL-3–dependent proliferation.2,10,12-14 In addition, Stat5 activation has been linked to Bcr/Abl-induced transformation and myelogenous leukemia.14 15
Recent studies have determined that IL-3 stimulates the activation of carboxy terminally truncated Stat5a (77 kd) and Stat5b (80 kd) isoforms in myeloid progenitors.7,16-20 These truncated isoforms lack a transcriptional activation domain. Consistent with this, genes that are activated by full-length Stat5 (ie, in mature cells) fail to be induced by IL-3 in immature myelocytes.17 The truncated Stat5 isoforms may, however, contribute to the activation of a distinct set of genes important for the immature phenotype.
Truncated Stat5 isoforms are generated through a unique protein-processing event.17,20,21 The responsible protease, which has been characterized extensively, is found exclusively in myeloid progenitors. This protease is a member of the serine family of proteases and has a molecular weight of approximately 25 kd. It cleaves Stat5a between tyrosine 721 (Y721) and methionine 722 (M722). Stat5b is cleaved between the corresponding amino acids, Y724 and M725 (Y724↓M725). Mutation of these 2 residues in Stat5b (Stat5bm/m) renders the protein resistant to cleavage.21
Numerous cell lines have been successfully exploited to characterize events that occur during early myeloid development. This includes FdCP1 cells, which are IL-3–dependent myeloid progenitors that transduce signals through truncated Stat5 isoforms.21 The c.19 subclone of these cells differentiates into monocytes after several days of culture in GM-CSF.22 Hence, these cells provide a unique opportunity to study the role of the Stat5 protease and its substrate in myeloid development. Introduction of a noncleavable Stat5b into FdCP1 leads to a significant defect in differentiation. This provides strong support for our model that Stat5 cleavage plays a crucial role in myeloid development.
Materials and methods
Cell culture
FdCP1 clone 19 (c.19) cells were a generous gift from L. Rohrschneider and are a clonal derivative of FdCP1 cells.22 FdCP1(c.19) cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum, penicillin/streptomycin, and 10% WEHI3b conditioned medium.22 Ba/F3 and FdTrk cells were grown as described previously, in RPMI supplemented with 10% heat-inactivated fetal calf serum, penicillin/streptomycin, and conditioned media.7 23Cells from the 293 cell line were obtained from American Type Culture Collection (Rockville, MD) and grown in DMEM supplemented with 10% heat-inactivated fetal calf serum and penicillin/streptomycin. IL-3–dependent cell lines were starved of growth factors for 5 hours before treatment with either mIL-3 (10 ng/mL; Peprotech, Rocky Hill, NJ) or mGM-CSF (10 ng/mL; Peprotech).
Cloning
The murine Stat5b wild-type gene and a Stat5bY724S↓M725A mutant21 were cloned into a vector in which the myeloid-specific MRP8 promoter drives expression.24 A FLAG epitope was added to the amino terminus of Stat5b through site-directed mutagenesis (Quick-Change; Stratagene, La Jolla, CA) and was verified by sequencing. Ten micrograms of each construct was transfected with 1 μg pSV2-neo plasmid25 into FdCP1(c.19) cells by electroporation (ECM 600; 260 V, 1050 F, infinite resistance; Gentronics). Neomycin (neo)-resistant clones were selected in 0.5 mg/mL G418 (Geneticin; GIBCO/BRL, Baltimore, MD) by limited dilution.26
Differentiation assays
FdCP1 clones, grown in 10% WEHI-conditioned media (WCM), were washed in factor-free medium and then incubated in mIL-3 (10 ng/mL or 10% WCM) or mGM-CSF (10 ng/mL) for 3 to 10 days.22 27 At days 3 and 10, aliquots were removed and evaluated by flow cytometry (FACSstar Plus), histology, or biochemical studies (below). Flow cytometric analysis entailed staining 5 × 106 cells with a biotinylated rat antimouse F4/80 monoclonal antibody or a biotinylated rat IgG2b isotype control antibody (generous gifts of J. Siu, San Diego, CA). Binding was revealed by a Texas Red-conjugated streptavidin (PharMingen, San Diego, CA). Samples were also stained with 1 μg/mL propidium iodine (Sigma, St Louis, MO) to identify viable cells and were then evaluated on a FACSstar Plus with CellQuest software. Histologic analysis entailed collecting cells by cytospin and staining with Wright-Giemsa (Accustain; Sigma). They were then evaluated under a Nikon Eclipse TE300 with a 100× objective (Melville, NY).
Western blotting
Nuclear, cytoplasmic, or whole-cell extracts were prepared as previously described.28-30 Protein concentration was determined through a Bradford assay (Bio-Rad, Hercules, CA). These samples were then fractionated by 8% SDS-PAGE and transferred to nitrocellulose membrane (Schleicher & Schuell, Keene, NH) as previously described.7 17 Filters were probed with several antibodies including anti-FLAG M5 (Sigma), anti-phospho-Stat5a/b (Upstate Biotechnology, Lake Placid, NY), and pan-Stat5 (Transduction Laboratories, Lexington, KY), as recommended by the manufacturers.
Protease assay
Stat5 protease activity was evaluated as previously described.17,21 Briefly, 1 μL recombinant murine Stat5b was incubated with 5 μL of a CHAPS (3[(3-cholamidopropyl) dimethylammonio]-1-propane sulfonate; Sigma) extract for 4 hours at 37°C. These extracts were prepared by lysing cells in CHAPS buffer (0.1% CHAPS, 10 mmol/L HEPES [pH 7.4], 2 mmol/L EDTA, 1 mmol/L dithiothreitol, and 10% glycerol17 31). Lysates were cleared by centrifugation (12 000g for 5 minutes at 4°C) and then assayed on the rStat5b substrate. Cleavage products were evaluated by immunoblotting with a pan-Stat5–specific monoclonal antibody (Transduction Laboratories).
Northern blotting
FdCP1 cells were serum starved for 5 hours and then stimulated with mIL-3 (10 ng/mL) for 30 minutes. Total cellular RNA was isolated from 2 × 107 cells with Trizol Reagent (Life Technologies) as specified by the manufacturer. Thirty-five micrograms total RNA was fractionated on a 0.8% agarose–formaldehyde gel and transferred to a nylon membrane (Schleicher & Schuell) by capillary transfer.17 Membranes were hybridized with preparations of purified (Qiagen, Valencia, CA), randomly labeled (Boehringer Mannheim, Indianapolis, IN) cDNA fragments (specific activity, approximately 1 × 108 cpm/μg). CIS-1 (0.1 kb) and OSM (0.3 kb) probes were prepared from EcoRI fragments as previously described.17 The β-actin probe was prepared as anXbaI–KpnI cDNA fragment (1244 bp; Ambion). Relative band intensity was determined by Phosphor-imaging (GS-525/GS-505; Bio-Rad) and evaluated with Molecular Analyst software.
Results
Stat5 expression in FdCP1(c.19) cells
The murine nonleukemic cell line FdCP1 represents a myeloid progenitor that is IL-3 dependent for growth (ie, self-renewal32). A subclone, c.19, has been shown to differentiate into monocytes when stimulated with GM-CSF.22,27 Previous studies from our laboratory have determined that carboxy terminally truncated isoforms of Stat5a (p77) and Stat5b (p80) are activated in response to IL-3 in FdCP1 cells.16,17,21 This contrasts the full-length isoforms shown to be activated in other, more mature cells. To determine whether FdCP1(c.19) cells exhibit a change in the Stat5 isoforms they express and activate on differentiation, nuclear extracts were prepared from cells cultured either in WCM, a source of IL-3, or GM-CSF. As previously reported,17,21 only truncated Stat5 was recovered in nuclear extracts of IL-3–stimulated immature cells (Figure 1A, lane 1). However, after differentiation there was a marked decrease in the level of truncated Stat5 and a corresponding increase in the level of full-length Stat5 recovered in IL-3–stimulated nuclear extracts (lane 3). Immunoblotting with a Stat5 phosphotyrosine-specific antibody (upper panel) or a pan-Stat5–specific antibody (lower panel) yielded similar results. Of note, Stat5 activation was more modest in the GM-CSF–differentiated cells. This may in part reflect the relatively shorter half-life of full-length Stat5.33 In addition, some Stat5 was recovered in extracts prepared from unstimulated, mature (GM-CSF–differentiated) cells, most likely because these cells were more resistant to starvation (lane 4).
Change in Stat5 isoform activation during GM-CSF–induced maturation of FdCP1 cells.
(A) FdCP1(c.19) cells were either cultured in WCM or differentiated in GM-CSF for 3 days. Cells were then starved (−) and stimulated with IL-3 (+; 10 ng/mL, 15 minutes). Nuclear extracts were evaluated by sequential immunoblotting with Stat5 phospho-specific (upper panel) and pan–Stat5-specific antibodies (lower panel). Recombinant inactive Stat5b (rStat5b) was included as a control (lane 5). The mobility of p96 (Stat5a), p94 (Stat5b), p80 (truncated Stat5b), and p77 (truncated Stat5a) are indicated in the left margin of each panel. (B) FdCP1(c.19) cells were cultured as in A. Differentiated cells, with increased forward and side scatter, were collected by flow cytometry to increase their purity to more than 95% (compare with Figure 3). CHAPS extracts were prepared and assayed on a recombinant FLAG-tagged Stat5b (rFlagSt5b) substrate prepared by overexpression in 293 cells.21 The mobility of rFlagSt5b (p94) and the cleaved product (p80) are indicated in the left margin of each panel.
Change in Stat5 isoform activation during GM-CSF–induced maturation of FdCP1 cells.
(A) FdCP1(c.19) cells were either cultured in WCM or differentiated in GM-CSF for 3 days. Cells were then starved (−) and stimulated with IL-3 (+; 10 ng/mL, 15 minutes). Nuclear extracts were evaluated by sequential immunoblotting with Stat5 phospho-specific (upper panel) and pan–Stat5-specific antibodies (lower panel). Recombinant inactive Stat5b (rStat5b) was included as a control (lane 5). The mobility of p96 (Stat5a), p94 (Stat5b), p80 (truncated Stat5b), and p77 (truncated Stat5a) are indicated in the left margin of each panel. (B) FdCP1(c.19) cells were cultured as in A. Differentiated cells, with increased forward and side scatter, were collected by flow cytometry to increase their purity to more than 95% (compare with Figure 3). CHAPS extracts were prepared and assayed on a recombinant FLAG-tagged Stat5b (rFlagSt5b) substrate prepared by overexpression in 293 cells.21 The mobility of rFlagSt5b (p94) and the cleaved product (p80) are indicated in the left margin of each panel.
The truncated isoforms of Stat5 are generated through the activity of a lineage-specific protease.21 The observed GM-CSF–dependent switch to full-length isoforms (Figure 1A) implied a corresponding loss of Stat5 protease activity. To test this directly, extracts were prepared from 2 pools of GM-CSF–differentiated FdCP1(c.19) cells (FdCP1-GM1 and FdCP1-GM2) and assayed for protease activity. This entailed incubating the extracts with a recombinant FLAG-tagged Stat5b substrate (rFStat5b).17,21 As previously reported, the protease activity recovered from FdTrk (an FdCP1 derivative) and undifferentiated FdCP1 cells was able to cleave some of the rFStat5b substrate into the appropriate 80-kd truncated isoform (Figure 1B, lanes 1, 2). In contrast, extracts prepared from differentiated cells did not exhibit Stat5 cleaving activity (lanes 3, 4). These data are consistent with previous studies comparing immature and mature cells, and they support our hypothesis that a switch toward the full-length Stat5 isoforms occurs during murine myeloid differentiation.7,17 20 They also highlight the potential usefulness of FdCP1(c.19) cells for studies on protease function.
Expression of noncleavable Stat5b (Y724S↓M725A) in FdCP1 cells
The Stat5 protease cleaves Stat5b between amino acids 724 and 725. Mutation of these 2 amino acids (Y724S↓M725A) yields a noncleavable Stat5 isoform.21 To determine whether expression of this mutant might affect myeloid differentiation, FdCP1 clones expressing a FLAG epitope-tagged version of Stat5bY724S↓M725A (FLAG-Stat5bm/m) were established. Of the many positive clones isolated, 2 independent clones that expressed near physiological levels of FLAG-Stat5bm/m(clones 19-M1 and 19-M2) were selected for further evaluation. A neomycin-resistant clone (19-neo) and a clone expressing a FLAG-epitope–tagged wild-type Stat5b (FLAG-Stat5bwt; 19-WT) were included as controls. Nuclear extracts were prepared from these cells before and after stimulation with IL-3 and were evaluated by immunoblotting with a FLAG epitope-specific antibody. Although a set of nonspecific bands was detected in all extracts, specific bands were detected in nuclear extracts prepared from IL-3–stimulated 19-WT, 19-M1, and 19-M2 cells (Figure 2A). As anticipated, truncated FLAG-Stat5b was recovered from the 19-WT cells (Figure 2, lane 3). In contrast, full-length FLAG-Stat5b was recovered from 19-M1 and 19-M2 cells (Figure 2, lanes 5, 7), albeit at considerably different levels. Similar results were obtained when the filter was reprobed with Stat5 phospho-specific and pan-specific antibodies (Figures 2B,C, 4). Additionally, the truncated Stat5 recovered from IL-3–stimulated 19-neo extracts could now be visualized (lane 1). Of note, these latter 2 experiments also identified a small amount of endogenous (ie, not epitope tagged) truncated Stat5 that had accumulated in the nucleus of IL-3–stimulated 19-M1 cells. An analogous, but less robust, signal was detected in the 19-M2 cells. Similar results were obtained when cytoplasmic extracts were evaluated with the Stat5 phospho-specific antibody (Figure 2D). These results demonstrate that a specific mutation in the Stat5b cleavage site is sufficient to render Stat5 resistance to cleavage in vivo. They also demonstrate that this mutation does not interfere either with Stat5 phosphorylation or nuclear translocation. However, expression of the mutant Stat5 does appear to lead to a substantial reduction in the level of endogenous Stat5 processing.
Expression of Stat5 in transfected FdCP1 cells.
Nuclear extracts were prepared as outlined in Figure 1 and sequentially immunoblotted with a FLAG epitope-tagged specific antibody (A), a Stat5 phospho-specific antibody (B), and a pan–Stat5-specific antibody (C). Recombinant inactive FLAG-Stat5b (rFLAG-Stat5b) was included as a control (lane 11). Matched cytoplasmic extracts are evaluated in D. The mobility of full length (ie, p94/p96) and truncated (ie, p77/p80) Stat5a and Stat5b are indicated in the left margin of each panel. 19-neo (lanes 1, 2) and 19-WT (lanes 3, 4) are clones of cells stably transfected with pSV2-neo and FLAG-Stat5bWT, respectively. 19-M1 (lanes 5, 6) and 19-M2 (lanes 7, 8) are 2 independent clones of cells transfected with FLAG-Stat5bm/m.
Expression of Stat5 in transfected FdCP1 cells.
Nuclear extracts were prepared as outlined in Figure 1 and sequentially immunoblotted with a FLAG epitope-tagged specific antibody (A), a Stat5 phospho-specific antibody (B), and a pan–Stat5-specific antibody (C). Recombinant inactive FLAG-Stat5b (rFLAG-Stat5b) was included as a control (lane 11). Matched cytoplasmic extracts are evaluated in D. The mobility of full length (ie, p94/p96) and truncated (ie, p77/p80) Stat5a and Stat5b are indicated in the left margin of each panel. 19-neo (lanes 1, 2) and 19-WT (lanes 3, 4) are clones of cells stably transfected with pSV2-neo and FLAG-Stat5bWT, respectively. 19-M1 (lanes 5, 6) and 19-M2 (lanes 7, 8) are 2 independent clones of cells transfected with FLAG-Stat5bm/m.
19-M1 and 19-M2 cells exhibit a defective differentiation phenotype
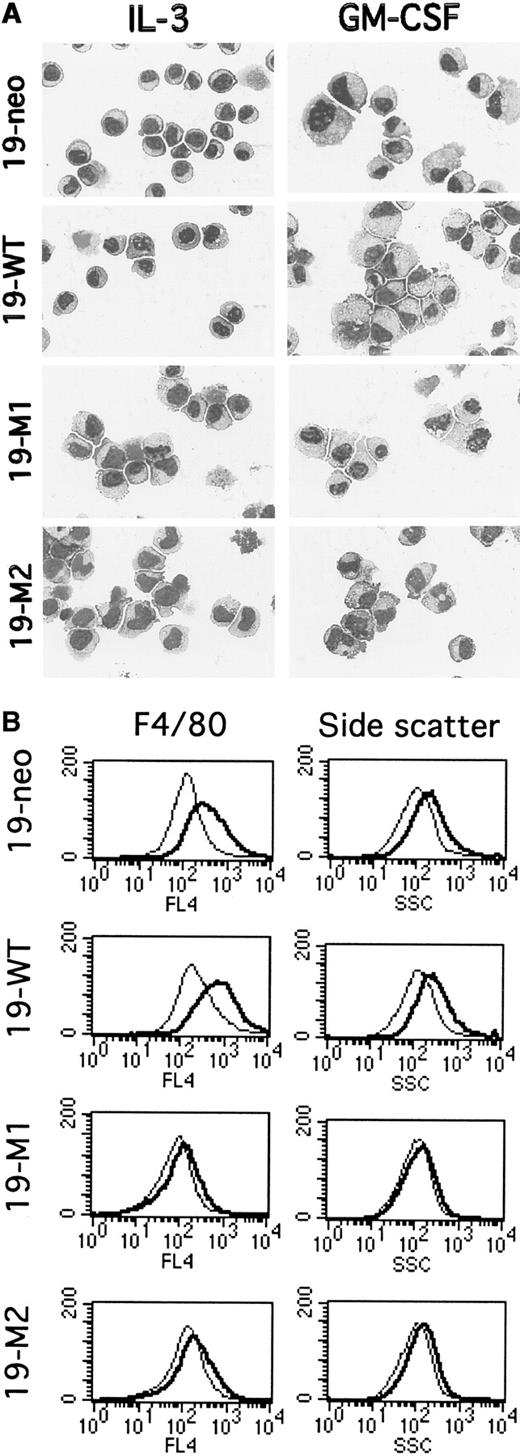
The ability of FdCP1(c.19) cells to differentiate has been demonstrated by changes in morphology and in the expression of cell-surface markers.22,27 Undifferentiated cells exhibit a characteristic myeloblastic morphology, with a generous nucleus and a thin, basophilic cytoplasm (Figure 3A, panel A). On differentiation they acquire a monocytic morphology, with increased size, enlarged vacuolated cytoplasm, and indented nuclei (Figure 3A, panel B). As anticipated, 19-neo and 19-WT exhibited the characteristic parental (ie, myeloid progenitor) morphology when grown in IL-3 (Figure 3A, panels A, C). After 3 days of culture in GM-CSF approximately 70% to 75% of the cells exhibited a characteristic monocytic (ie, differentiated) morphology (Figure 3A, panels B, D). In contrast, when cells expressing FLAG-Stat5bm/m 19-M1 and 19-M2 were cultured in IL-3, they exhibited a partially differentiated phenotype (Figure 3A, panels E, G) and failed to differentiate further in the presence of GM-CSF (Figure 3A, panels F, H). Similar observations have been made with several additional clones expressing a nonepitope–tagged version of the Stat5bm/m.21Additional time in GM-CSF (up to 10 days) failed to yield any further differentiation in these clones (data not shown).
Evaluation of myeloid differentiation in mutant FdCP1 cells.
(A) Histologic analysis. FdCP1-derived cell lines expressing pSV2-neo alone (19-neo; A, B), FLAG-Stat5b WT (19-WT; C, D), FLAG-Stat5bm/m (19-M1 and 19-M2; E-H) were stained with Wright–Giemsa before or after differentiation in GM-CSF for 3 days. These photomicrographs are representative of the results obtained from 2 independent experiments. (B) Flow cytometric analysis. 19-neo, 19-WT, 19-M1, and 19-M2 cells were evaluated for expression of F4/80 (left column) or side scatter (SSC; right column) before (light tracing) or after differentiation (dark tracing) in GM-CSF for 3 days. These studies were carried out on the same set of cells evaluated by Wright–Giemsa staining. These histograms are representative of several independent experiments.
Evaluation of myeloid differentiation in mutant FdCP1 cells.
(A) Histologic analysis. FdCP1-derived cell lines expressing pSV2-neo alone (19-neo; A, B), FLAG-Stat5b WT (19-WT; C, D), FLAG-Stat5bm/m (19-M1 and 19-M2; E-H) were stained with Wright–Giemsa before or after differentiation in GM-CSF for 3 days. These photomicrographs are representative of the results obtained from 2 independent experiments. (B) Flow cytometric analysis. 19-neo, 19-WT, 19-M1, and 19-M2 cells were evaluated for expression of F4/80 (left column) or side scatter (SSC; right column) before (light tracing) or after differentiation (dark tracing) in GM-CSF for 3 days. These studies were carried out on the same set of cells evaluated by Wright–Giemsa staining. These histograms are representative of several independent experiments.
Next, the ability of these 4 cell lines to differentiate was evaluated by flow cytometry. Previous studies had determined that FdCP1(c.19) differentiation correlates with an up-regulation in the expression of F4/80 (a monocyte marker)34 and an increase in granularity, as determined by side scatter.22,27 When cultured in GM-CSF for 3 days, 19-neo and 19-WT cells exhibited the previously reported increase in both F4/80 and side scatter22 (Figure 3B). In contrast, the 19-M1 and 19-M2 cells failed to change in either of these parameters when stimulated with GM-CSF for 3 days. Moreover, the level of F4/80 expression in mutant cells (before or after GM-CSF) correlated with that of unstimulated wild-type cells, indicating a block in the expression of this gene.
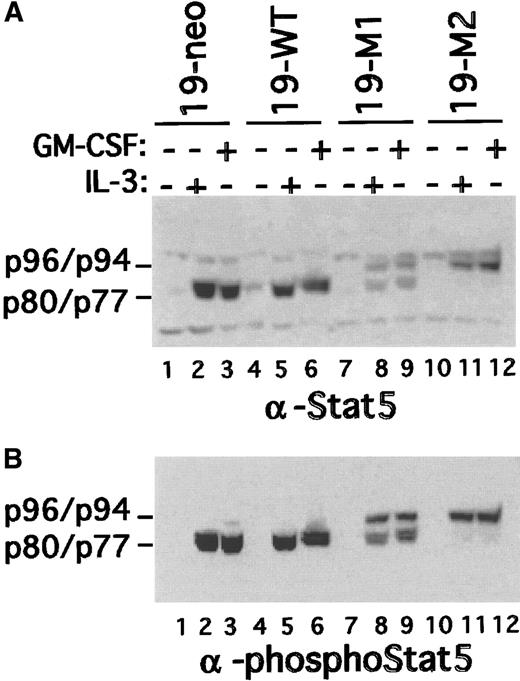
To exclude the possibility that the block in differentiation was caused by a defect in the response to GM-CSF, Stat5 activation was evaluated. Nuclear extracts were prepared from all 4 cell types before and after a short stimulation with IL-3 or GM-CSF. The extracts were then evaluated by immunoblotting both with phospho-Stat5– and pan-Stat5–specific antibodies. Consistent with the results shown in Figure 2, IL-3 stimulated the nuclear accumulation of truncated Stat5a in 19-neo and 19-WT cells (Figure 4). Again, predominantly full-length Stat5 was recovered in 19-M1 and 19-M2 nuclear extracts. However, a modest amount of truncated (ie, endogenous) Stat5 accumulated in the nucleus of 19-M1 cells. Identical results were obtained with GM-CSF–stimulated cells, indicating an intact response to this ligand. Moreover, GM-CSF and IL-3 were equivalent in their ability to promote survival in both wild-type and mutant clones (data not shown). These studies indicated that there is no significant defect in the ability of mutant clones to respond to GM-CSF.
Stat5 activation in mutant FdCP1 clones.
Nuclear extracts were prepared from cells (19-neo, 19-WT, 19-M1, and 19-M2 cells) treated with either GM-CSF or IL-3 as outlined in Figure1. The extracts were sequentially immunoblotted with a pan–Stat5-specific antibody (A) and Stat5 phospho-specific antibody (B). Mobility of p96 (Stat5a), p94 (Stat5b), p80 (truncated Stat5b), and p77 (truncated Stat5a) is indicated on the left margin of each panel.
Stat5 activation in mutant FdCP1 clones.
Nuclear extracts were prepared from cells (19-neo, 19-WT, 19-M1, and 19-M2 cells) treated with either GM-CSF or IL-3 as outlined in Figure1. The extracts were sequentially immunoblotted with a pan–Stat5-specific antibody (A) and Stat5 phospho-specific antibody (B). Mobility of p96 (Stat5a), p94 (Stat5b), p80 (truncated Stat5b), and p77 (truncated Stat5a) is indicated on the left margin of each panel.
Expression of Stat5 target genes in 19-M1 and 19-M2 cells
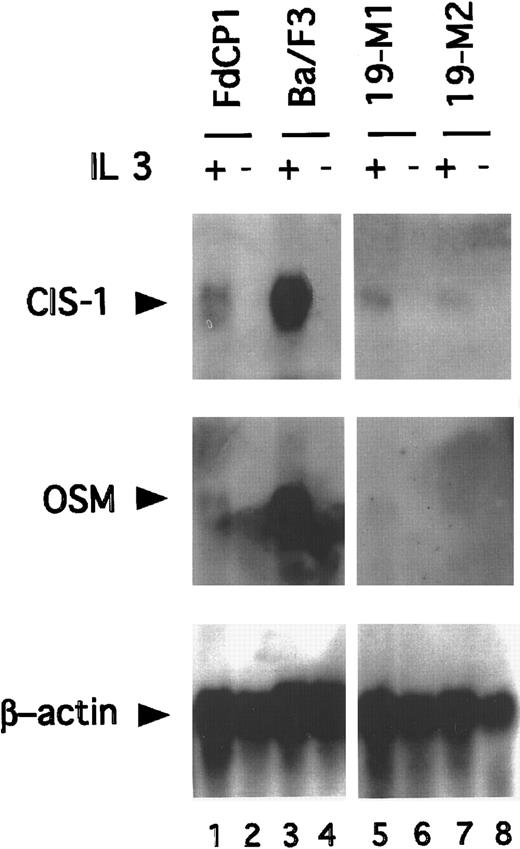
The truncated isoforms of Stat5 lack a transcriptional activation domain. Consistent with this, cells signaling through these isoforms have been found to be defective in their IL-3–dependent induction of several Stat5 target genes (eg, CIS, OSM, and pim1).17,35-37 To determine whether the IL-3–dependent transcription of these genes would be restored in immature cells expressing Stat5bm/m, Northern blotting studies were carried out. RNA was prepared from 19-M1 and 19-M2 cells before and after stimulation with IL-3 and then sequentially hybridized with CIS, OSM, and β-actin–specific probes. Ba/F3 and FdCP1(c.19) cells were included as controls. As anticipated, IL-3 stimulated a strong induction (approximately 8 fold) of CIS and OSM in Ba/F3 cells, but not FdCP1 cells (Figure 5). However, expression remained low in the 2 mutant cell lines. Analogous results were obtained with the pim-1 probe (data not shown). These observations demonstrated that the expression of the noncleavable isoform of Stat5 does not restore the IL-3–dependent expression of “mature” Stat5 target genes. Although this mutation is proximal to the well defined transcriptional activation domain of Stat5,33,38 39 we could not exclude the possibility that this mutant cripples transcriptional activity. We do not, however, favor this model because of the significant phenotypic changes found in cells expressing Stat5bm/m. Rather, we suspect that the failure of these genes to be expressed reflected the absence of an additional differentiation-specific transcription factor(s), required to fully activate the corresponding promoters.
Northern blot analysis demonstrating expression of Stat5 target genes.
RNA was prepared from FdCP1 (lanes 1, 2), Ba/F3 (lanes 3, 4), 19-M1 (lanes 5, 6), and 19-M2 (lanes 7, 8) cells before (−) or after (+) stimulation with IL-3 for 30 minutes. Thirty-five micrograms total RNA was fractionated on a formaldehyde gel and sequentially hybridized with a CIS-1 (top panel), OSM 1 (middle panel), and β-Actin (bottom panel) probe. Quantitation of signal intensity was measured on a Phosphor-imager.
Northern blot analysis demonstrating expression of Stat5 target genes.
RNA was prepared from FdCP1 (lanes 1, 2), Ba/F3 (lanes 3, 4), 19-M1 (lanes 5, 6), and 19-M2 (lanes 7, 8) cells before (−) or after (+) stimulation with IL-3 for 30 minutes. Thirty-five micrograms total RNA was fractionated on a formaldehyde gel and sequentially hybridized with a CIS-1 (top panel), OSM 1 (middle panel), and β-Actin (bottom panel) probe. Quantitation of signal intensity was measured on a Phosphor-imager.
Stat5 protease activity in 19-M1 and 19-M2 cells
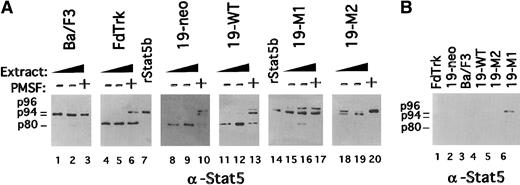
Stat5 protease activity is lost as myelocytes mature (Figure1).17,20,21 These observations indicate that Stat5 protease activity is down-regulated during myeloid maturation. Corresponding with a decrease in the expression of endogenous truncated Stat5, 19-M1 and 19-M2 cells exhibited a partially differentiated phenotype (Figures 2B,C, 4). To evaluate more directly the Stat5 protease activity in these cells, an in vitro cleaving assay was performed. Increasing amounts of CHAPS extracts from these cells were incubated with a recombinant Stat5b (rStat5b) substrate (Figure6A). Although the substrate in these studies was not tagged, immunoblotting studies demonstrated that these CHAPS extracts were not significantly contaminated with endogenous Stat5b (Figure 6B). There was, however, a modest contamination with endogenous Stat5a (p96) in some extracts (eg, 19-M1). Again, extracts from FdTrk and Ba/F3 cells served as positive and negative controls for protease activity.7,21 As anticipated, 19-neo and 19-WT exhibited near normal levels of protease activity, and this activity was inhibited by the addition of phenylmethylsulfonyl fluoride (Figure6).17 20 In marked contrast, the 19-M1 and 19-M2 cells exhibited a substantially reduced level of protease activity, suggesting a more mature phenotype. Consistent with the immunoblotting studies (Figures 2, 4), 19-M1 exhibited a slightly increased level of protease activity.
Evaluation Stat5 protease activity in mutant FdCP1 cell lines.
(A) Recombinant Stat5b (rStat5b) was incubated for 4 hours at 37°C with either 35 μg (lanes 1, 8, 11, 15, 18) or 70 μg (lanes 2, 9, 12, 16, 19) CHAPS extract from Ba/F3 (lanes 1-3), FdTrk (lanes 4-6), 19-neo (lanes 8-10), 19-WT (lanes 11-13), 19-M1 (lanes 15-17), and 19-M2 (lanes 18-20) cells. Protease activity was blocked in some samples by the addition of 0.5 mmol/L phenylmethylsulfonyl fluoride (+). Control samples of undigested rStat5b are found in lanes 7 and 14. Digestion was evaluated by immunoblotting with pan-Stat5 antibody. Mobility of intact endogenous Stat5a (p96), rStat5b (p94) and cleaved Stat5b (p80) is indicated in the left margin. (B) Equivalent amounts of CHAPS extracts are evaluated for contamination with Stat5b by immunoblotting with pan-Stat5 antibody. Mobility of intact endogenous Stat5a (p96), Stat5b (p94), and truncated Stat5b (p80) is indicated in the left margin.
Evaluation Stat5 protease activity in mutant FdCP1 cell lines.
(A) Recombinant Stat5b (rStat5b) was incubated for 4 hours at 37°C with either 35 μg (lanes 1, 8, 11, 15, 18) or 70 μg (lanes 2, 9, 12, 16, 19) CHAPS extract from Ba/F3 (lanes 1-3), FdTrk (lanes 4-6), 19-neo (lanes 8-10), 19-WT (lanes 11-13), 19-M1 (lanes 15-17), and 19-M2 (lanes 18-20) cells. Protease activity was blocked in some samples by the addition of 0.5 mmol/L phenylmethylsulfonyl fluoride (+). Control samples of undigested rStat5b are found in lanes 7 and 14. Digestion was evaluated by immunoblotting with pan-Stat5 antibody. Mobility of intact endogenous Stat5a (p96), rStat5b (p94) and cleaved Stat5b (p80) is indicated in the left margin. (B) Equivalent amounts of CHAPS extracts are evaluated for contamination with Stat5b by immunoblotting with pan-Stat5 antibody. Mobility of intact endogenous Stat5a (p96), Stat5b (p94), and truncated Stat5b (p80) is indicated in the left margin.
The reduction of protease activity found in mutant clones could be attributed to an intrinsic loss in the expression of protease or a potentially dominant inhibitory property of the FLAG-Stat5bm/m transgene. To determine whether mutant Stat5b can block protease activity, increasing amounts of rStat5bm/m were added to a protease assay in which FLAG-Stat5bWT was the substrate (Figure7, lanes 4-6). In a parallel experiment (Figure 7, lanes 7-9), increasing amounts of wild-type rStat5b were added as a specificity control. Cleavage was determined by probing with a substrate-specific FLAG-epitope antibody. rStat5bm/m and rStat5bWT both significantly reduced protease activity when they were added at a significant molar excess (eg, 8-fold excess; Figure 7, panel B, lanes 6, 9). These results indicated that large quantities of rStat5bm/m and rStat5bWT can antagonize protease activity, but this was not the case when they were present at more physiological levels. For example, protease activity was normal in 19-WT cells, in which FLAG-Stat5bWTexpression was at near physiological levels (Figure 6). Similarly, in 19-M1 and 19-M2 cells, FLAG-Stat5bm/m was expressed at physiological levels. Hence, the reduction of protease activity in these cells likely reflected an intrinsic decrease in activity (model 1). In other words, we do not believe that this reduction was caused by a competitive inhibition by Stat5bm/m in vivo.
Evaluation of potential inhibitory activity of Stat5bm/m on protease activity.
Recombinant FLAG-Stat5b was incubated for 4 hours at 37°C with 35 μg CHAPS extract from FdCP1 cells, either with (lane 3) or without (lane 2) 0.5 mmol/L phenylmethylsulfonyl fluoride. Increasing amounts of either recombinant Stat5bm/m (lanes 4-6) or recombinant Stat5bWT (lanes 7-9) were added to the reaction in ratios of 1:1 (lanes 4, 7), 2:1 (lanes 5, 8), or 8:1 (lanes 6, 9) relative to the substrate. Samples were analyzed by sequential immunoblotting with a anti-FLAG antibody (A) or a pan-Stat5 antibody (B). Mobility of intact Stat5b (p94) and cleaved Stat5b (p80) is indicated in the left margin.
Evaluation of potential inhibitory activity of Stat5bm/m on protease activity.
Recombinant FLAG-Stat5b was incubated for 4 hours at 37°C with 35 μg CHAPS extract from FdCP1 cells, either with (lane 3) or without (lane 2) 0.5 mmol/L phenylmethylsulfonyl fluoride. Increasing amounts of either recombinant Stat5bm/m (lanes 4-6) or recombinant Stat5bWT (lanes 7-9) were added to the reaction in ratios of 1:1 (lanes 4, 7), 2:1 (lanes 5, 8), or 8:1 (lanes 6, 9) relative to the substrate. Samples were analyzed by sequential immunoblotting with a anti-FLAG antibody (A) or a pan-Stat5 antibody (B). Mobility of intact Stat5b (p94) and cleaved Stat5b (p80) is indicated in the left margin.
Discussion
Cytokines regulate multiple aspects of lineage selection and hematopoietic differentiation. Cell-specific responses to extracellular signals are integrated into a genetic program of differentiation through the activation of key transcription factors.40 The IL-3 cytokine family, which transduces its signals through Stat5, plays an important role in myeloid development.2,9-12 Moreover, myeloid maturation appears to entail a switch in Stat5 isoforms, from the truncated (in immature cells) to the full-length (in mature cells).16,18 Additionally, the identification of constitutively active Stat5 in several types of leukemia provides further evidence that Stat5 plays an important role in hematopoietic development.15
The expression/activation of functionally distinct isoforms of transcription factors is not unique to Stat5 and myeloid development. A functionally distinct and developmentally regulated truncated isoform of Stat3 has also been observed during neutrophil–granulocyte differentiation.41 This isoform appears to mediate the induction of a distinct set of genes by associating with additional transcription factors.41,42 Other transcription factors are also known to be regulated by protein processing, including NFκB43 and SREBP (sterol regulatory element-binding proteins),44 suggesting that proteases can regulate signaling cascades.
The identification of a functionally distinct set of Stat5 isoforms (ie, p70 and p80) in myeloid progenitors led to the hypothesis that they are important in the “immature” response to IL-3 (and related ligands). Because these isoforms lack a transcriptional activation domain, they are unable to induce Stat5 target genes.16,17In contrast, more mature myeloid cell lines express full-length Stat5. This correlates with a loss in protease activity and an induction of Stat5-dependent, “mature” genes.10,12,17,35-37 In the current study we took advantage of an FdCP1 cell line that can be induced to differentiate in vitro, and we explored this model of Stat5 isoform-dependent maturation. Consistent with findings from previous studies, when FdCP1(c.19) cells are in their basal immature state (ie, cultured in IL-3), they respond to IL-3 with the activation of truncated Stat5 isoforms (Figure 1). As anticipated, these cells exhibited a robust level of Stat5 protease activity (Figures 1, 6). Consistent with studies on more mature myelocytes,8,16,20 21 when FdCP1(c.19) cells are induced to differentiate (ie, cultured in GM-CSF), there is a switch to the expression of full-length Stat5 (Figure 1). This correlates with a loss in protease activity (Figure 1), providing convincing evidence that maturation is linked to a switch in Stat5 isoform expression.
To test more rigorously the role of Stat5 in myeloid maturation, a noncleavable isoform of Stat5 was expressed in immature FdCP1(c.19) cells. We anticipated that this would lead to the expression mature genes and would perturb the phenotype of these cells and their ability to differentiate. Consistent with our prediction, FdCP1(c.19) cells expressing an epitope-tagged noncleavable Stat5 (ie, 19-M1 and 19-M2 cells) exhibited a number of important changes, most significant of which were morphologic changes characteristic of a mature phenotype. Similar observations have been made for cells expressing a different version of noncleavable Stat5.21 Although these changes correlated directly with the expression of the noncleavable Stat5 (FLAG-Stat5bm/m), a reduction in the expression of endogenous Stat5 was also noted. This, however, appeared to be caused by a decrease of Stat5 protease activity rather than a potential dominant inhibitory effect of the noncleavable mutant (Figure 7). Hence, there appears to be a direct correlation with maturation and loss in protease activity. Although these studies do not determine how protease activity is down-regulated, it is intriguing to speculate that this may be achieved through the up-regulation of a serpin (serine protease inhibitor), at least one of which is known to be regulated by Stat5.45Alternatively, the expression of the protease may be directly down-regulated during myeloid maturation.46
It was surprising to find that the partially differentiated phenotype in 19-M1 and 19-M2 cells did not correlate with an up-regulation in the IL-3–dependent expression of several target genes (Figure 5). This suggests that other lineage-specific factors may contribute to the regulation of these genes. Consistent with this possibility, studies on the common GM-CSF receptor chain4 have identified distinct domains that transduce important Stat5-independent signals.47 48 Several additional potential explanations for the inability to express target genes should be considered. For example, noncleavable Stat5 may be transcriptionally inactive, but this is more difficult to reconcile with the observed phenotypic changes. Similarly, loss in protease activity may affect an additional pathway that is important in maturation.
In addition to acquiring a partially differentiated phenotype, 19-M1 and 19-M2 cells are also resistant to further differentiation in GM-CSF. This suggests that the mis-expression of full-length Stat5 sufficiently perturbs FdCP1 cells, perhaps through the expression of inappropriate genes, to prevent further differentiation. This is not surprising considering the important roles gene targeting and overexpression studies have attributed to Stat5 in lymphoid, erythroid, and myeloid development.9-12,14 49 This compelling body of data supports our conclusion that Stat5 processing and isoform switching play a crucial role in myeloid maturation. Future studies will test the ability of FLAG-Stat5m/m to perturb myeloid development in vivo through the generation of transgenic mice.
Acknowledgments
We thank Carolyn Lee and Vincent Sahi for excellent technical support; Cristina Angelin-Duclos and Chris Park for helpful discussions; and Riccardo Dalla Favera and Kursad Turgay for critical reading of the manuscript.
Supported by National Institutes of Health grant RO1-HL55413, the Burroughs-Wellcome Foundation, and the Leukemia Society of America (C. S.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Christian Schindler, Departments of Microbiology and Medicine, Columbia University, HHSC-1212, 701 West 168th Street, New York, NY 10032; e-mail: cws4@columbia.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal