Abstract
Human hematopoietic progenitor cells express L-selectin and also express PSGL-1, a ligand for all selectins. Using a shear-based adhesion assay, a hematopoietic cell L-selectin ligand (HCLL) that is expressed on the hematopoietic cell line KG1a and on normal human hematopoietic progenitors was previously identified. To characterize the structural biology of HCLL and to define its relationship to PSGL-1, the effects of chemical and enzymatic treatments on HCLL activity of KG1a cells and membrane preparations were analyzed. Protease digestions and chemical treatments of KG1a cells and membranes indicated that HCLL is an integral membrane glycoprotein. Glycosidase digestions of membrane protein preparations and metabolic treatments of KG1a cells with glycosylation processing modifiers revealed that L-selectin binding determinants on HCLL are sialofucosylated structures presented on complex-type N-glycans. Adhesion assays and biochemical studies showed that this glycoprotein is also expressed on circulating blasts in native acute leukemias. HCLL is distinguishable from PSGL-1: (1) KG1a cells sorted for PSGL-1 expression had equivalent HCLL activity; (2) anti–PSGL-1 blocking antibodies and proteases known to eliminate L-selectin binding to PSGL-1 had no effect on HCLL binding activity of KG1a cells; (3) blasts from native leukemias with low expression of PSGL-1 and CD34 display high HCLL activity; and (4) despite high level expression of PSGL-1, HCLL activity was absent on HL60 cells. These data provide first evidence of a naturally expressed membrane L-selectin ligand expressing binding determinant(s) on an N-linked glycoconjugate. This novel ligand may help mediate L-selectin–dependent cell-cell adhesive interactions within the cytoarchitecture of the bone marrow microenvironment.
Introduction
The selectins (L-, E- and P-selectin) are a family of membrane proteins that behave as Ca++-dependent lectins and mediate adhesive interactions under shear with their respective ligands. Although they are best known for mediating “rolling” adhesive interactions between leukocytes and endothelium, which are critical to leukocyte migration from vascular to extravascular compartments,1 the selectins—and their ligands—are also expressed among cells within the marrow2,3 where they help create the specialized architecture of the marrow microenvironment.4,5 Cell migration experiments, clonogenic assays, and clinical studies indicate that selectins may play important roles in the trafficking of hematopoietic cells into the marrow and in hematopoiesis.3 6-11
While E- and P-selectin are found on endothelial cells and platelets (P-selectin only), L-selectin is expressed on mature leukocytes and, notably, on early hematopoietic progenitor cells.2 Expression of L-selectin on human stem cells is associated with higher clonogenic potential of progenitors,7 and recovery of platelet and neutrophil counts after clinical stem cell transplantation correlates with the number of L-selectin+/CD34+ cells infused.8,9 Moreover, cross-linking of L-selectin on stem cell membranes before culture increases the clonogenic capacity of the cells in in vitro assays,12 whereas chronic addition of anti–L-selectin antibody suppresses colony formation.13Collectively, these data suggest that L-selectin–mediated receptor/ligand interactions may promote hematopoiesis, and heighten interest in defining the nature of L-selectin ligand(s) in the marrow microenvironment.
Although several L-selectin ligands have been identified, our understanding of the structure and function of L-selectin ligands is incomplete. For all naturally expressed glycoprotein L-selectin ligands described to date, the binding determinants are present on sialylated, fucosylated lactosamines displayed on O-linked carbohydrates.14,15 On lymph node high endothelial venule (HEV) cells, multiple L-selectin ligands have been described,16-19 all of which are glycoproteins reactive with the monoclonal antibody MECA 79.20 One of these ligands is a glycoform of CD34.21 Although present on endothelial cells in most tissues, CD34 is an integral membrane glycoprotein best known as a marker of the multilineage colony-forming hematopoietic “stem cell.”22 This marker was originally identified on KG1a cells, a human leukemia line that phenotypically resembles the native stem cell.22-24
In early studies, we investigated whether CD34 on hematopoietic cells functions as an L-selectin ligand. By using the shear-based L-selectin–specific Stamper-Woodruff assay,25 we found that the CD34 glycoform present on hematopoietic cells is not an L-selectin ligand. However, our studies showed that an L-selectin ligand is expressed on KG1a cells and on normal human bone marrow CD34+ hematopoietic progenitor cells.2,26 Like endothelial L-selectin ligands, this hematopoietic cell L-selectin ligand (HCLL) requires sialylation for activity. However, HCLL differs in several fundamental ways from previously described endothelial L-selectin ligands: It does not contain epitopes recognized by MECA 79, it is resistant to digestion with O-sialoglycoprotease (OSGP), a peptidase that cleaves at sites of O-linked sialylated carbohydrate modifications, and its activity is sulfation independent.27
Subsequent to our description of HCLL activity on hematopoietic progenitors, several groups reported that P-selectin glycoprotein ligand-1 (PSGL-1), a cell surface mucin-like glycoprotein that serves as a P-selectin and E-selectin ligand,28,29 can also serve as an L-selectin ligand.30-33 The binding determinants for both L- and P-selectins are localized to the N-terminal region of mature PSGL-1 and are sensitive to cleavage by mocarhagin, a protease that cleaves a 10 residue fragment from the N-terminus.32PSGL-1 is expressed on neutrophils and on a variety of human myeloid cell lines (including HL60 and KG1a cells34-36), and is also expressed on normal hematopoietic progenitors.36-38However, contrary to L-selectin, engagement of PSGL-1 on progenitors appears to inhibit hematopoiesis.6 This finding suggests that the increased clonogenic activity associated with L-selectin binding may be mediated by interactions with ligands other than PSGL-1. In this report we show, by biochemical analysis of KG1a cells and native circulating blasts from patients with acute leukemias, that HCLL is a naturally expressed integral membrane glycoprotein L-selectin ligand distinct from PSGL-1. The data also show that HCLL is uniquely distinguished from all other L-selectin ligands in that L-selectin binding determinant(s) are presented on N-linked complex carbohydrates.
Materials and methods
Cells, antibodies, and enzymes
KG1a and HL60 cells (from ATCC) were cultured in RPMI 1640/10% fetal bovine serum (FBS). Citrated peripheral blood was obtained from patients with acute myelocytic leukemia (AML) and acute lymphocytic leukemia (ALL) with peripheral blasts representing more than 80% of differential, and blasts were isolated by Ficoll-Hypaque (Sigma, St Louis, MO) density centrifugation.Antibodies: Anti–PSGL-1 antibody PL-1 (which blocks P- and L-selectin binding to PSGL-130-32,39) was from Pharmingen (San Diego, CA). Functional P-selectin–Ig fusion protein, murine moAbs PSL-275, 2G3, 4F9, and 4D8 (and isotype control moAbs), and rabbit polyclonal antiserum (Rb3443) directed against PSGL-1 were a gift of Dr R. Camphausen (Genetics Institute, Cambridge, MA); with the exception of PSL-275, these reagents block the binding of P-selectin to PSGL-1,28,40,41 yet the binding epitope of PSL-275 is cleaved by mocarhagin.42 Phycoerythrin (PE)-conjugated goat antimouse IgG antibodies were from Jackson ImmunoResearch (West Grove, PA). Unconjugated anti-CD43 moAb L60 (which recognizes an O-linked sialic acid epitope27) and isotype control was from Becton-Dickinson (San Jose, CA). Hamster antirat L-selectin moAb HRL-1 (a function-blocking antibody) and isotype control were from Pharmingen. Murine antihuman L-selectin moAb LAM 1-3, anti-CD34 moAb QBEND10, and isotype controls were from Coulter-Immunotech (Miami, FL). Enzymes: O-sialoglycoprotease was from Accurate Chemical (Westbury, NY); bromelain, chymotrypsin, pronase, human neutrophil elastase, and porcine pancreas elastase were from Sigma; mocarhagin was a gift of Dr R. Camphausen; recombinant phospholipase C (PI-PLC) was from Oxford Glycosystems (Oxfordshire, United Kingdom); Vibrio cholera neuraminidase, Newcastle neuraminidase, α-L-fucosidase, N-glycosidase F, endoglycosidase H, and recombinant endoglycosidase F2 were from Roche Molecular Biochemicals (Indianapolis, IN).
Immunofluorescence studies
Measurement of PSGL-1 expression and PSGL-1 activity (binding of P-selectin–Ig) on cell suspensions was performed by flow cytometry. Cell suspensions (1 × 106 cells in 100 μL phosphate-buffered saline (PBS)/2% FBS on ice) were incubated with anti–PSGL-1 moAbs (3 μg each) or with equivalent amounts of isotype control antibody for 30 minutes. After washing in PBS/2% FBS, the cells were treated with 5 μg PE-conjugated goat antimouse IgG secondary antibody, followed by additional washes. Flow cytometric analysis of PSGL-1 activity was performed by incubating cells with P-selectin–Ig or human IgG1 isotype control (each at 4 μg/106 cells) precomplexed with protein A-FITC (Zymed, San Francisco, CA) as described.41 For cell sorting, KG1a were stained with a cocktail of anti–PSGL-1 moAbs or with P-selectin–Ig and populations were separated by fluorescence intensity into cells with highest (top 5%) and lowest (bottom 5%) PSGL-1 levels. Flow cytometry and cell sorting were performed on a FACStar apparatus (Becton Dickinson).
Lymphocyte adherence assay
The Stamper-Woodruff lymphocyte adherence assay was performed on glutaraldehyde-fixed cytocentrifuged (cytospin) preparations of cells as previously described,26 or on glutaraldehyde-fixed slides containing cell membrane protein preparations (see below). For all adherence assays, lymphocyte suspensions (107 per milliliter rat thoracic duct lymphocytes or human peripheral blood lymphocytes isolated by density centrifugation of citrated blood) in RPMI 1640 medium were overlaid onto glass slides containing cytospin preparations of cells, containing membrane protein preparations, or containing glutaraldehyde-fixed 8-μm–thick frozen sections of murine lymph node (which served as control to assess L-selectin–mediated lymphocyte binding to HEV). Slides were placed on a rotating platform for incubation under shear (80 rpm) at 4°C for 30 minutes. Slides were then rinsed in PBS to remove nonadherent lymphocytes, fixed in 3% glutaraldehyde, and stained with methyl green-thionin. Slides were examined for lymphocyte adherence to cells or to spotted membrane protein material by light microscopy. As previously described,26 L-selectin–specific lymphocyte adherence was verified in every assay by performing parallel incubations with phorbol myristate acetate (PMA)-treated lymphocytes (PMA induces shedding of L-selectin), by preincubation of lymphocytes with blocking anti–L-selectin moAb (either HRL-1 for rat or LAM1-3 for human lymphocytes, or with respective control isotype moAb), and by performing assays in the presence of 10 mmol/L EDTA.
For cytospin preparations, L-selectin ligand activity was measured by counting the number of lymphocytes adherent to a confluent area of cytocentrifuged cells, using an ocular grid under 100 × magnification. In experiments in which cells were treated with enzymes, metabolic agents or antibodies, untreated (control) and treated cells were assayed in parallel; ligand activity was measured by counting respective adherent lymphocytes on a minimum of 3 separate grid fields of cytospins, 3 slides per experiment, 3 separate experiments. The mean lymphocyte binding for each treatment and control slide was quantified, and data were expressed as the percentage of lymphocyte binding of treated compared with untreated cells, as previously described.26 27 To assess blocking activity of anti–PSGL-1 antibodies, KG1a cytospins were preincubated in RPMI 1640 containing anti–PSGL-1 antibodies or isotype-matched antibody (controls) at concentrations as high as 50 μg/mL for 30 minutes at 4°C; slides were then rinsed and cell suspensions containing the respective antibodies at concentrations equivalent to those used in the preincubation were overlaid onto the slides. For experiments using cytospin mixtures of HL60 and KG1a cells, immunohistochemistry for CD34 antigen was performed as per manufacturer's (Dako, Carpinteria, CA) recommendations, after the adherence assay to identify KG1a cells (alkaline phosphatase, New Fuchsin chromagen; stained cells are pink).
For Stamper-Woodruff adherence assays on preparations of cell membrane proteins, 1 μg membrane protein (prepared as described below) was spotted onto glass slides, allowed to dry at room temperature, and then fixed in 3% glutaraldehyde. Adherence assays were performed on these slides exactly as described for cytospin preparations of cells. After enzyme treatments of membrane protein, lymphocytes adhering to enzyme-treated and buffer-treated preparations were quantified as described above, with data expressed as a percentage of control lymphocyte adherence (enzyme-treated samples compared with corresponding buffer-treated membrane proteins).
Enzyme digestions
Cell suspensions (20 × 106 cells/mL RPMI) were treated with proteases or with buffer alone (control) before cytocentrifugation. Protease concentrations for digestions were as follows: 100 U/mL chymotrypsin; 0.1% bromelain; 50 mg/mL human neutrophil elastase or porcine pancreas elastase; O-sialoglycoprotease at 60 μg /mL; mocarhagin at 10 μg/mL. All digestions were 1 hour at 37°C in RPMI 1640 medium, except for mocarhagin digestion for which cells were suspended in 0.15 mol/L NaCl, 2 mmol/L CaCl2, 1 mg/mL BSA, 0.01 mol/L Tris, pH 7.4, for 20 minutes at 37°C. To verify the efficiency of O-sialoglycoprotease and of mocarhagin digestions, flow cytometry was performed to measure QBEND10 (an OSGP-sensitive epitope of CD34) and the PSL-275 epitope of PSGL-1, respectively. After digestions, cells were washed in RPMI 1640 medium, then cytocentrifuged onto slides for adherence assays. For digestion with Newcastle virus neuraminidase, glutaraldehyde-fixed cytospins of KG1a cells were rinsed twice with enzyme buffer (50 mmol/L NaAcetate, 154 mol/L NaCl, 9 mmol/L CaCl2, pH 5.5), and incubated at 37°C for 1 hour with 50 μL buffer (control) or 10 mU/mL neuraminidase in buffer; the slides were then washed in RPMI 1640/1% FBS, and the lymphocyte adherence assay was performed on the cytospins.
For α-L-fucosidase, N-glycosidase F, endoglycosidase F2, or endoglycosidase H digestions, KG1a membrane preparations (as described below) were denatured in 1% SDS and boiled 5 minutes. Denatured membrane preparations were then diluted in buffers (as noted in parentheses) specific for each glycosidase and incubated for 24 hours at 37°C in either 0.8 U/mL α-L-fucosidase (0.1 mol/L sodium acetate, pH 5.5), 8 U/mL N-glycosidase F (50 mmol/L sodium phosphate,12.5 mmol/L EDTA,1% 2-mercaptoethanol, pH 7.5), 4 mU/mL endoglycosidase F2 (0.5 mol/L sodium acetate, 1% NP-40, pH 5.5), or 50 mU/mL endoglycosidase H (0.1 mol/L sodium acetate, 1% 2-mercaptoethanol, 0.5 mmol/L PMSF, pH 5.5). Confirmation of enzyme activity was made by analysis of shifts in SDS-PAGE protein profiles (Coomassie staining) of enzyme-treated versus buffer-treated preparations. After buffer treatments or enzyme digestions, 1 μg total protein of each suspension was spotted onto slides for adherence assays as described above.
Analysis of ligand reexpression after neuraminidase or bromelain treatment of KG1a cells
KG1a (20 × 106 cells/mL) were treated with 0.1 U/mL neuraminidase (V cholerae neuraminidase) in RPMI 1640 without bicarbonate (pH 7.4) for 1 hour at 37°C; for controls, an equivalent volume of enzyme buffer (one-tenth volume of 50 mmol/L sodium acetate, 154 mmol/L NaCl, 9 mmol/L CaCl2, pH 5.5) was added to KG1a in RPMI 1640 without bicarbonate (pH 7.4) under identical conditions (final pH 6.8). Bromelain digestion was performed as described above. After neuraminidase or bromelain digestion, cells were washed 3 times in RPMI 1640 without bicarbonate and an aliquot, designated as t = 0, was tested in the adherence assay to verify complete loss of ligand activity. The remaining cells were cultured in RPMI 1640/10% FBS and an aliquot of these cells was removed at serial time points over 24 hours for analysis of the reappearance of ligand activity in the adherence assay.
Effects of metabolic inhibition of N-linked glycosylation or protein synthesis
Tunicamycin (15 μg/mL) or diluent control (DMSO, final concentration 0.3%) was added to cell cultures (107cells/mL in RPMI 1640 with 10% FBS) after either V choleraneuraminidase treatment (0.1 U/mL) or respective enzyme buffer treatment of the KG1a cells for 1 hour as described above. For metabolic studies utilizing either swainsonine or deoxymannojirimycin, cells were treated with neuraminidase and then washed and suspended in RPMI 1640/10% FBS containing either 0.4 mg/mL deoxymannojirimycin or 40 μg/mL swainsonine. Cells were incubated with metabolic inhibitors of glycosylation for 24 hours, then washed and cytocentrifuged onto glass slides for analysis of L-selectin–dependent lymphocyte adherence.
For inhibition of protein synthesis, cycloheximide (Sigma, endotoxin-cleared27) was added to cultures of neuraminidase-digested, bromelain-digested, or buffer-treated KG1a (107 cells/mL in RPMI 1640/10% FBS) at 1.25 μg/mL final concentration. In each case, cells were cultured for 20 hours and the L-selectin ligand activity was measured by adherence assay at the end of the culture period. For all studies utilizing metabolic inhibitors, parallel cultures were established in the presence of35S-methionine/cysteine, and protein synthesis was measured by scintillation counting of trichloroacetic acid (TCA)-precipitable protein as previously described.27
Salt and pH treatments of KG1a cells
KG1a cells (50 × 106 cells) were incubated for 5 minutes at 37°C with vigorous agitation in 2 mL high salt solution (to 1 mol/L NaCl) under a variety of pH conditions: 0.1 mol/L Tris, pH 9; 0.1 mol/L sodium phosphate, pH 7; 0.1 mol/L sodium acetate, pH 5; and 0.1 mol/L acetic acid, pH 3. After these treatments, cells were centrifuged and supernatants collected. Cells were then washed in RPMI 1640 × 2, and cells and supernatants were assessed for ligand activity. Supernatants were spotted onto glass slides and air-dried, then glutaraldehyde fixed for Stamper-Woodruff assays. L-selectin ligand activity of these cells and supernatants was compared with cells similarly treated with agitation in RPMI 1640 medium alone.
Isolation of KG1a membranes and organic solvent extraction of membranes
KG1a membranes were isolated by nitrogen cavitation, followed by differential centrifugation to isolate membrane fractions.43 KG1a were suspended (4 × 108cells/mL) in PI buffer (150 mmol/L NaCl, 50 mmol/L Tris-HCl pH 7.4, 1 mmol/L EDTA, 0.02% sodium azide, containing protease inhibitors [20 μg/mL PMSF, 0.5 μg/mL aprotinin, 0.5 μg/mL pepstatin A, 0.5 μg/mL leupeptin, 20 μg/mL trypsin inhibitor]) and subjected to nitrogen cavitation (400 psi). Ruptured cells were then centrifuged (3600g) to pellet nuclear and mitochondrial debris; the supernatant was saved, and the pellet was washed with PI buffer and recentrifuged. The 2 supernatants were then pooled and centrifuged (22 000g) to pellet membrane material that was then suspended in PI buffer.
For lipid extractions, membrane preparations (0.2 mg/mL protein) were mixed (1:5) with organic solvents: water-saturated butanol or chloroform: methanol (2:1) for 3 minutes. Appropriate solvent to PBS mixtures (1:1) were then added at 10% of the above final volume and phases were separated by centrifugation. Aqueous and organic phases were evaporated, and resulting material was resuspended in original volume of PBS and spotted on glass slides, air-dried, then glutaraldehyde fixed for analysis in the Stamper-Woodruff assay.
Results
PSGL-1 is not the principal L-selectin ligand on KG1a cells
With the Stamper-Woodruff assay, lymphocyte adherence to cytospin preparations of cells or to membrane material spotted on glass slides was mediated only by L-selectin, as determined by complete elimination of lymphocyte adherence after PMA-treatment of lymphocytes, preincubation of lymphocytes with anti–L-selectin moAb, and by performing the assay in the presence of EDTA. Incubation of KG1a cytospin monolayers with anti–PSGL-1 monoclonal antibodies singly and in combination, including PL-1, 4F9, 4D8, and 2G3 (each of which block L- and P-selectin binding to PSGL-1), did not inhibit lymphocyte attachment in the adherence assay (Table1). These incubations were performed both before and after glutaraldehyde fixation of KG1a onto glass slides, with antibody concentrations as high as 100 μg/mL, both singly and in combination. Moreover, incubation of KG1a with undiluted anti–PSGL-1 rabbit antisera Rb3443 did not interfere with L-selectin ligand activity in the adherence assay. In each case where antibodies were used, KG1a cells were stained with the respective antibody and isotype (or preimmune sera) control and analyzed by flow cytometry to confirm attachment to PSGL-1. HCLL activity was also unaffected by preincubation of KG1a with P-selectin–Ig chimera (Table 1).
Effects of anti-PSGL-1 antibodies, P-selectin-IgG chimera, and anti-L-selectin antibody on lymphocyte adherence to KG1a cells
| Blocking treatments . | Percentage control binding* (SEM) . |
|---|---|
| Anti–PSGL-1 monoclonal antibodies (20 μg/mL of each in a cocktail) | 103.0 (7.0) |
| P-selectin–IgG chimera, 50 μg/mL | 97.6 (7.4) |
| Anti–L-selectin antibody | 1.9 (0.6)† |
| Blocking treatments . | Percentage control binding* (SEM) . |
|---|---|
| Anti–PSGL-1 monoclonal antibodies (20 μg/mL of each in a cocktail) | 103.0 (7.0) |
| P-selectin–IgG chimera, 50 μg/mL | 97.6 (7.4) |
| Anti–L-selectin antibody | 1.9 (0.6)† |
Number of lymphocytes adherent to confluent area of KG1a were counted by light microscopy using an ocular grid under 100× magnification (quantified a minimum of 3 fields per slide, 3 slides per experiment, 3 separate experiments). Results are presented as percentage binding compared with corresponding isotype MoAb-treated control KG1a cytospin preparations.
Similar abrogation of binding was observed in assays performed in the presence of EDTA and utilizing PMA-treated lymphocytes (see text for details).
Mocarhagin digestion abrogates both P-selectin and L-selectin binding activity of PSGL-1.32 KG1a cells incubated with this protease were analyzed by flow cytometry for P-selectin binding utilizing P-selectin–Ig chimera. As shown in Figure1, mocarhagin digestion completely eliminated P-selectin binding to PSGL-1. Staining of KG1a cells with N-terminal–specific moAb PSL-275 was similarly eliminated after mocarhagin treatment, confirming the high efficiency of digestion of the N-terminal region of PSGL-1. However, treatment of KG1a with mocarhagin or enzyme buffer (control) did not affect L-selectin–specific lymphocyte adherence in the Stamper-Woodruff assay (Table 2). OSGP digestion of KG1a also eliminated P-selectin–Ig binding to KG1a cells (data not shown), without affecting HCLL activity (Table 2).
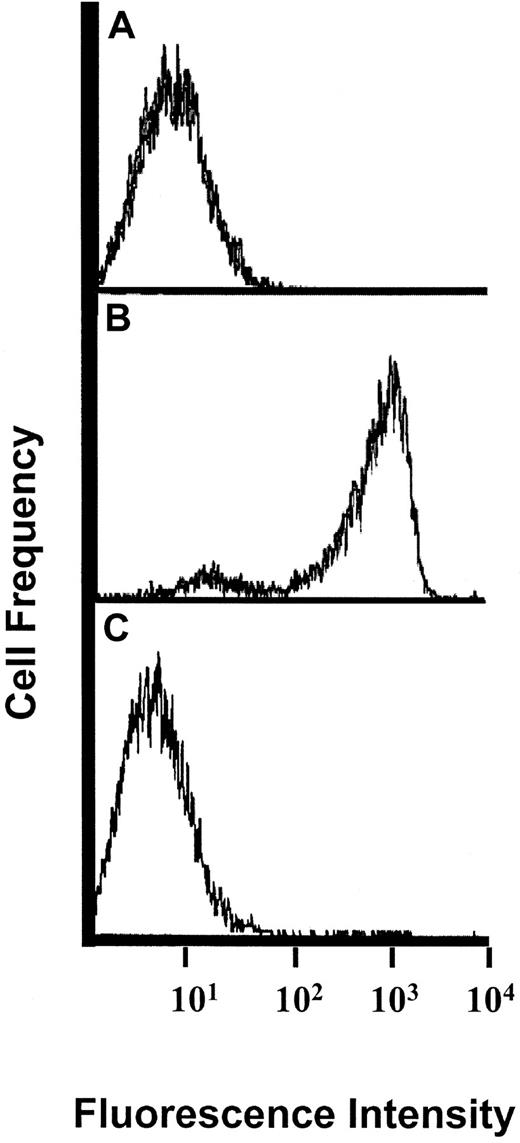
Flow cytometric analysis of P-selectin–Ig binding to PSGL-1 on KG1a cells after mocarhagin digestion.
(A) staining pattern of isotype control (human IgG1) antibody, followed by protein A-FITC; (B) buffer-treated cells stained with P-selectin–Ig chimera, followed by protein A-FITC; (C) mocarhagin-treated cells stained with P-selectin–Ig, followed by protein A-FITC. Note shift in fluorescence intensity of P-selectin–Ig binding to PSGL-1 after mocarhagin digestion, compared with buffer-treated cells.
Flow cytometric analysis of P-selectin–Ig binding to PSGL-1 on KG1a cells after mocarhagin digestion.
(A) staining pattern of isotype control (human IgG1) antibody, followed by protein A-FITC; (B) buffer-treated cells stained with P-selectin–Ig chimera, followed by protein A-FITC; (C) mocarhagin-treated cells stained with P-selectin–Ig, followed by protein A-FITC. Note shift in fluorescence intensity of P-selectin–Ig binding to PSGL-1 after mocarhagin digestion, compared with buffer-treated cells.
L-selectin ligand activity of KG1a cells after protease or phospholipase C treatment
| Enzymatic treatments . | Percentage control binding* (SEM) . |
|---|---|
| O-sialoglycoprotease | 98.4 (2.3) |
| Mocarhagin | 98.9 (6.3) |
| Bromelain | 3.8 (0.4)† |
| Chymotrypsin | 6.7 (0.7)† |
| Pronase | 9.4 (1.3)† |
| Porcine pancreas elastase | 47.3 (17.6)† |
| Human neutrophil elastase | 25.4 (13.5)† |
| PI-PLC | 98.1 (2.0) |
| Enzymatic treatments . | Percentage control binding* (SEM) . |
|---|---|
| O-sialoglycoprotease | 98.4 (2.3) |
| Mocarhagin | 98.9 (6.3) |
| Bromelain | 3.8 (0.4)† |
| Chymotrypsin | 6.7 (0.7)† |
| Pronase | 9.4 (1.3)† |
| Porcine pancreas elastase | 47.3 (17.6)† |
| Human neutrophil elastase | 25.4 (13.5)† |
| PI-PLC | 98.1 (2.0) |
Number of lymphocytes adherent to confluent area of KG1a were counted by light microscopy using an ocular grid under 100× magnification (quantified a minimum of 3 fields per slide, 3 slides per experiment, 3 separate experiments). Results are presented as percentage binding compared with corresponding buffer-treated control KG1a cytospin preparations.
Statistically significant (paired t test;P < .01) difference compared with enzyme buffer alone group.
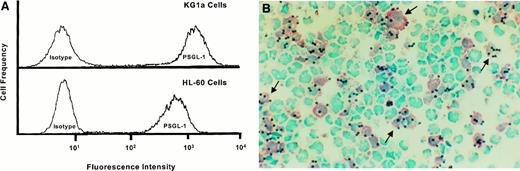
The hematopoietic cell line HL60 has been utilized extensively for studies of PSGL-1 structure and function,28,29,34,37,40,44and PSGL-1 on HL60 has been reported to be an L-selectin ligand.30,31 As measured by binding of P-selectin–Ig chimera, KG1a express functional levels of PSGL-1 comparable to HL60 cells (Figure 2A). However, in Stamper-Woodruff assays of cytospin mixtures of KG1a and HL60 cells, lymphocytes bind only to KG1a cells (pink staining cells, Figure 2B). The selectivity and specificity of binding to KG1a in mixing studies indicate that the absence of lymphocyte adherence to HL60 cells under Stamper-Woodruff assay conditions is not due to an indirect biologic effect of HL60 on lymphocytes rendering them incapable of attaching to L-selectin ligands (such as activation-induced shedding of L-selection45 46), as any such effect would concomitantly prevent binding to KG1a in these cytospin mixtures.
HCLL activity is independent of PSGL-1.
(A) Flow cytometric analysis of PSGL-1 levels on KG1a cells and HL60 cells. Data shown are results obtained using P-selectin–Ig to stain cells, and therefore are reflective of PSGL-1 functional levels. Note that PSGL-1 expression is characteristic of both HL60 and KG1a cells. (B) L-selectin–mediated adherence of lymphocytes occurs on KG1a cells but not HL60 cells. Representative result of lymphocyte adherence assay performed on cytospin mixture of KG1a cells and HL60 cells. Immunohistochemical staining for CD34 antigen (pink stain) identifies KG1a cells. As demonstrated by arrows, note that lymphocytes (solid blue dots) adhere only to pink (KG1a) cells (background stain is methyl green-thionin, 250 × magnification).
HCLL activity is independent of PSGL-1.
(A) Flow cytometric analysis of PSGL-1 levels on KG1a cells and HL60 cells. Data shown are results obtained using P-selectin–Ig to stain cells, and therefore are reflective of PSGL-1 functional levels. Note that PSGL-1 expression is characteristic of both HL60 and KG1a cells. (B) L-selectin–mediated adherence of lymphocytes occurs on KG1a cells but not HL60 cells. Representative result of lymphocyte adherence assay performed on cytospin mixture of KG1a cells and HL60 cells. Immunohistochemical staining for CD34 antigen (pink stain) identifies KG1a cells. As demonstrated by arrows, note that lymphocytes (solid blue dots) adhere only to pink (KG1a) cells (background stain is methyl green-thionin, 250 × magnification).
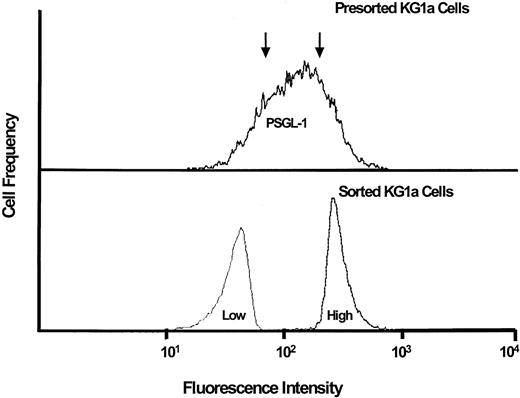
To further examine the role of PSGL-1 in the L-selectin ligand activity of KG1a cells, we separated KG1a cells into subpopulations with high- and low-level expression of PSGL-1 by fluorescence-activated cell sorting utilizing a cocktail of anti–PSGL-1 antibodies. Cytospin preparations of the separated populations were made and cells were then analyzed for L-selectin ligand activity in the Stamper-Woodruff assay. The adherence of lymphocytes was identical in the 2 populations (compared with presorted KG1a cells, mean [SEM] percentage binding was 100.6 [9.5] and 98.6 [11.1]) in the “low” and “high” PSGL-1 groups, respectively), despite high-efficiency separation of PSGL-1 expressing cells as verified by repeat staining using anti–PSGL-1 antisera (Figure 3).
L-selectin–mediated lymphocyte adherence to PSGL-1–deficient KG1a cells.
KG1a cells were sorted by FACS into populations (separated at arrows, upper panel) representing high-level PSGL-1 expression (“high”) and low-level PSGL-1 expression (“low”), as shown in lower panel. Adherence assays were performed on cells from both populations. L-selectin ligand activity was equivalently high in both cell populations (see text for details).
L-selectin–mediated lymphocyte adherence to PSGL-1–deficient KG1a cells.
KG1a cells were sorted by FACS into populations (separated at arrows, upper panel) representing high-level PSGL-1 expression (“high”) and low-level PSGL-1 expression (“low”), as shown in lower panel. Adherence assays were performed on cells from both populations. L-selectin ligand activity was equivalently high in both cell populations (see text for details).
Hematopoietic cell L-selectin ligand is an integral membrane protein
As shown in Table 2, treatment of KG1a cells with bromelain, chymotrypsin, and pronase completely abrogates L-selectin ligand activity, whereas digestion with pancreas and neutrophil elastase partially affects activity and digestion with mocarhagin, and O-sialoglycoprotease has no effect on ligand activity. Ligand activity is also resistant to digestion with PI-PLC (Table 2). Similar to previous data from our laboratory after neuraminidase treatment of KG1a,27 cycloheximide treatment of cells for as long as 20 hours after protease (bromelain) digestion completely prevents ligand reexpression without significantly affecting cell numbers or viability (trypan blue exclusion more than 95%) (Table3).
Reexpression of L-selectin ligand activity after neuraminidase or protease treatment of KG1a cells is dependent on de novo protein synthesis
| KG1a treatment3-150 . | Mean (SEM) % control binding3-151 . |
|---|---|
| Neuraminidase (t = 0, no culture) | 0.2 (0.1)+ |
| Neuraminidase buffer control (no culture) | 98.5 (7.5) |
| Neuraminidase, 24-h culture | 101.3 (6.5) |
| Bromelain (t = 0, no culture) | 3.8 (0.4)++ |
| Bromelain, 20-h culture | 95.2 (7.1) |
| Neuraminidase, 20-h culture with cycloheximide (1.25 μg/mL) | 0.5 (0.2)+ |
| Bromelain, 20-h culture with cycloheximide (1.25 μg/mL) | 0.6 (0.3)++ |
| 20-h culture with cycloheximide (1.25 μg/mL) (no neuraminidase or bromelain digestion) | 91.8 (5.8) |
| KG1a treatment3-150 . | Mean (SEM) % control binding3-151 . |
|---|---|
| Neuraminidase (t = 0, no culture) | 0.2 (0.1)+ |
| Neuraminidase buffer control (no culture) | 98.5 (7.5) |
| Neuraminidase, 24-h culture | 101.3 (6.5) |
| Bromelain (t = 0, no culture) | 3.8 (0.4)++ |
| Bromelain, 20-h culture | 95.2 (7.1) |
| Neuraminidase, 20-h culture with cycloheximide (1.25 μg/mL) | 0.5 (0.2)+ |
| Bromelain, 20-h culture with cycloheximide (1.25 μg/mL) | 0.6 (0.3)++ |
| 20-h culture with cycloheximide (1.25 μg/mL) (no neuraminidase or bromelain digestion) | 91.8 (5.8) |
After neuraminidase or bromelain treatment, all cells were washed before culturing as indicated.
Number of lymphocytes adherent to confluent area of KG1a were counted by light microscopy using an ocular grid under 100× magnification (quantified a minimum of 3 fields per slide, 3 slides per experiment, 3 separate experiments). Results are presented as percentage binding compared with corresponding untreated control KG1a cytospin preparations.
+,++Statistically significant (P < .01, by paired t test) difference compared with undigested, neuraminidase buffer-incubated (+) or RPMI 1640-incubated (++) cells. All other values are not statistically different (P > .05).
To determine whether HCLL is an integral membrane structure, KG1a cells were treated with high salt conditions (to 1 mol/L NaCl) over a range of pH levels (pH 3-9), and supernatant (elution) and residual membrane fractions were tested in the Stamper-Woodruff assay. HCLL activity was detected only on membranes after all treatments (data not shown). To examine whether membrane lipid contributed to HCLL activity, organic solvent extractions of cells were performed, and respective organic and aqueous phases were then tested in the Stamper-Woodruff assay. In each case, ligand activity partitioned in the aqueous phase, and there was no activity within the lipid (solvent) compartment (data not shown).
Hematopoietic cell L-selectin ligand L-selectin binding determinant(s) are sialylated, fucosylated structures displayed on complex N-linked glycans
We previously showed that V cholera neuraminidase treatment completely abrogates HCLL activity on KG1a.24This enzyme has broad specificity for cleavage of terminal sialic acids in either α2-3, α2-6, or α2-8 linkage to relevant oligosaccharides. To further characterize the sialic acid linkage, we used the Newcastle disease virus neuraminidase that cleaves terminal sialic acids in α2-3 and α2-8 linkages, but not in α2-6 linkage.47 48 As with results of V choleraneuraminidase digestions, treatment of KG1a cells with Newcastle neuraminidase before the Stamper-Woodruff assay abrogated HCLL binding activity (mean binding of 9.8% [SEM of 2.2%] compared with respective buffer treatment alone, P < .001 by paired t test). These data, specifically, indicate that L-selectin binding determinant(s) on HCLL consist of oligosaccharides principally bearing α2-3-and/or α2-8-linked sialic acids. To examine whether fucosylation is required for ligand activity, we digested denatured KG1a membrane preparations with α-L-fucosidase, which cleaves terminal fucose residues in α1,2-, α1,3-, and α1,4-linkages. Treatment with this enzyme markedly diminished HCLL binding activity (mean binding of 11.5% [SEM of 2.9%] compared with respective buffer treatment alone, P < .001 by paired t test). Collectively, the results of sialidase and fucosidase digestions indicate that the relevant binding determinant of HCLL is a sialylated, fucosylated glycoconjugate.
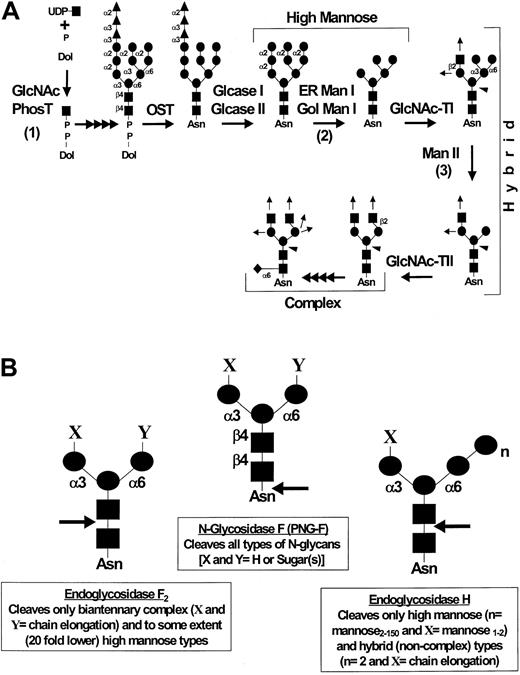
Inhibition of protein synthesis with cycloheximide after either neuraminidase27 or protease treatment (Table 3) prevents ligand reexpression. These data indicate that ligand reexpression after such treatments results from de novo glycoprotein synthesis and processing; thus, incubation of the cells with metabolic inhibitors of glycosylation during the period of ligand reexpression would result in replacement of original membrane ligand glycoprotein(s) with ligand molecules affected by the relevant metabolic treatments. Accordingly, to investigate the effect of modifications in N-linked oligosaccharide processing on the activity of HCLL, we treated KG1a cells with V cholera neuraminidase, and cultured these treated cells for 24 hours in the presence of glycosylation inhibitors, tunicamycin, swainsonine, and deoxymannojirimycin (Figure4A shows schema depicting the effects of these inhibitors on the different stages of carbohydrate processing). Before culturing the cells, the cleavage of sensitive sialic acid epitopes was confirmed in every case by testing for loss of binding activity in the Stamper-Woodruff assay and by measuring the expression of the sialic acid–dependent L60 epitope of CD43 by flow cytometry.27 For each inhibitor, effects on glycosylation of membrane proteins were verified by analysis of protein band shifts by SDS-PAGE of membrane lysates (data not shown); however, treatment of cells with the glycosylation inhibitors did not affect cell viability (trypan blue exclusion more than 98% for each inhibitor). As shown in Table 4, treatment with glycosylation inhibitors without prior neuraminidase digestion did not affect L-selectin ligand activity of KG1a cells, suggesting that the membrane turnover of fully processed HCLL is relatively slow and there is no direct toxic effect of the inhibitors on the activity of HCLL already expressed on the membrane. However, after neuraminidase digestion, tunicamycin treatment completely prevented HCLL reexpression and both swainsonine and deoxymannojirimycin markedly diminished expression of HCLL activity. Return of the L60 epitope of CD43 was not inhibited by treatments with either tunicamycin, swainsonine, or deoxymannojirimycin (data not shown), indicating that these agents did not affect sialylation directly and, in particular, did not affect synthesis of O-linked sialylated structures. Moreover, similar to our findings in previous studies,27 these metabolic agents had no effect on de novo protein synthesis as TCA-precipitable35S-methionine/cysteine radiolabeled protein counts were similar to that of cells not treated with the inhibitors (data not shown).
N-glycan processing and endoglycosidase sensitivity.
(A) Schema of N-glycan biosynthesis: assembly and processing. Note the synthesis of high mannose, hybrid, and complex-type N-glycans and the sites of inhibition of N-glycan synthesis and processing by tunicamycin (1), deoxymannojirimycin (2), and swainsonine (3). GlcNAc-PhosT indicates N-acetylglucosamine-1-phosphotransferase; OST, oligosaccharyl transferase; Glcase I, α1,2 glucosidase I; Glcase II, α1,3 glucosidase II; ER Man I, ER-localized α1,2 mannosidase I; Gol Man I, Golgi-localized α1,2 mannosidase I; GlcNAc-TI, β1,2 N-acetylglucosaminyltransferase; Man II, α1,3/6 mannosidase II; GlcNAc-TII, β1,2 N-acetylglucosaminyltransferase. ● indicates mannose (man); ▴, glucose (Glc); ▪, N-acetylglucosamine (GlcNAc); ♦, fucose; ↑, potential sites of chain elongation; ◂, potential addition of β4 GlcNAc. (B) N-glycan branch specificity of endo-N-glycosidases. The specificities of N-glycan cleavage for N-glycosidase F (N-glycanase), endoglycosidase F2, and endoglycosidase H are as shown by arrows. ●, mannose; ▪, N-acetylglucosamine.
N-glycan processing and endoglycosidase sensitivity.
(A) Schema of N-glycan biosynthesis: assembly and processing. Note the synthesis of high mannose, hybrid, and complex-type N-glycans and the sites of inhibition of N-glycan synthesis and processing by tunicamycin (1), deoxymannojirimycin (2), and swainsonine (3). GlcNAc-PhosT indicates N-acetylglucosamine-1-phosphotransferase; OST, oligosaccharyl transferase; Glcase I, α1,2 glucosidase I; Glcase II, α1,3 glucosidase II; ER Man I, ER-localized α1,2 mannosidase I; Gol Man I, Golgi-localized α1,2 mannosidase I; GlcNAc-TI, β1,2 N-acetylglucosaminyltransferase; Man II, α1,3/6 mannosidase II; GlcNAc-TII, β1,2 N-acetylglucosaminyltransferase. ● indicates mannose (man); ▴, glucose (Glc); ▪, N-acetylglucosamine (GlcNAc); ♦, fucose; ↑, potential sites of chain elongation; ◂, potential addition of β4 GlcNAc. (B) N-glycan branch specificity of endo-N-glycosidases. The specificities of N-glycan cleavage for N-glycosidase F (N-glycanase), endoglycosidase F2, and endoglycosidase H are as shown by arrows. ●, mannose; ▪, N-acetylglucosamine.
L-selectin ligand activity of KG1a cells treated with inhibitors of N-linked glycosylation
| KG1a treatment4-150 . | Percentage control adhesion (SEM)4-151 . |
|---|---|
| Neuraminidase (1 h) | 1.0 (0.7)‡ |
| Neuraminidase (1-h then 24-h recovery) | 99.3 (1.3) |
| Tunicamycin alone | 96.5 (7.4) |
| Deoxymannojirimycin alone | 107.5 (6.7) |
| Swainsonine alone | 95.5 (10.3) |
| Neuraminidase + tunicamycin | 1.0 (0.7)‡ |
| Neuraminidase + deoxymannojirimycin | 22.5 (0.4)‡ |
| Neuraminidase + swainsonine | 26.6 (2.5)‡ |
| KG1a treatment4-150 . | Percentage control adhesion (SEM)4-151 . |
|---|---|
| Neuraminidase (1 h) | 1.0 (0.7)‡ |
| Neuraminidase (1-h then 24-h recovery) | 99.3 (1.3) |
| Tunicamycin alone | 96.5 (7.4) |
| Deoxymannojirimycin alone | 107.5 (6.7) |
| Swainsonine alone | 95.5 (10.3) |
| Neuraminidase + tunicamycin | 1.0 (0.7)‡ |
| Neuraminidase + deoxymannojirimycin | 22.5 (0.4)‡ |
| Neuraminidase + swainsonine | 26.6 (2.5)‡ |
KG1a cells were cultured in the presence of tunicamycin (15 μg/mL), deoxymannojirimycin (0.4 mg/mL), or swainsonine (40 μg/mL) for 24 hours after a 1-hour (preculture) treatment with Vibrio cholera neuraminidase (0.1 U/mL). Cytospin preparations of these cells were then subjected to Stamper-Woodruff assay as detailed in the text. L-selectin–specific lymphocyte binding to cytospins was quantified by light microscopy.
Values represent mean percentage lymphocyte adherence (SE of the mean) compared with untreated control group from 3 independent experiments. Adherent lymphocytes were counted from 4 fields on each cytospin in triplicate slides using an optical grid under 100× magnification.
Statistical significance (paired t test;P < .01) compared with neuraminidase (1-hour then 24-hour recovery) group.
To further analyze the structural biology of the critical glycosylations conferring ligand activity, Stamper-Woodruff assays were performed on N-glycosidase and buffer (control) treated membrane preparations of KG1a cells. Interestingly, HCLL binding activity was maintained after SDS solubilization and 2-mercaptoethanol treatment of membrane proteins, indicating that the protein scaffold can be denatured significantly without disturbing ligand activity. Figure 4B displays the relevant specificity for each endo-N-glycosidase. As shown in Table 5, N-glycosidase F digestion completely abrogates HCLL activity, whereas endoglycosidase F2 digestion moderately affects activity and endoglycosidase H digestion has no effect.
L-selectin ligand activity of KG1a cells treated with N-glycosidases
| N-glycosidase treatment of KG1a membrane preparation5-150 . | Percentage control adhesion (SEM)5-151 . |
|---|---|
| Endoglycosidase H | 105.5 (2.5) |
| Endoglycosidase F2 | 66.9 (0.1)5-152 |
| N-glycosidase F | 16.0 (0.7)5-153 |
| N-glycosidase treatment of KG1a membrane preparation5-150 . | Percentage control adhesion (SEM)5-151 . |
|---|---|
| Endoglycosidase H | 105.5 (2.5) |
| Endoglycosidase F2 | 66.9 (0.1)5-152 |
| N-glycosidase F | 16.0 (0.7)5-153 |
KG1a membrane preparations (0.2 mg/mL) were incubated with either Endoglycosidase H (50 mU/mL), Endoglycosidase F2 (4 mU/mL), N-glycosidase F (8 U/mL), or respective buffers (controls) for 24 hours at 37°C, then spotted onto glass slides (1 μg per spot) and subjected to Stamper-Woodruff assays as detailed in the text. L-selectin-specific lymphocyte binding to spotted samples was quantified by light microscopy.
Values represent mean percentage adherence (SE of the mean) compared with buffer control groups from 3 independent experiments. Adherent lymphocytes were counted from 4 fields on each spot in triplicate slides using an optical grid under 100× magnification.
Statistical significance (Paired t test;P = .03) compared with buffer control group.
Statistical significance (Paired t test; P< .01) compared with buffer control group.
Hematopoietic cell L-selectin ligand is present on blasts from native acute leukemias
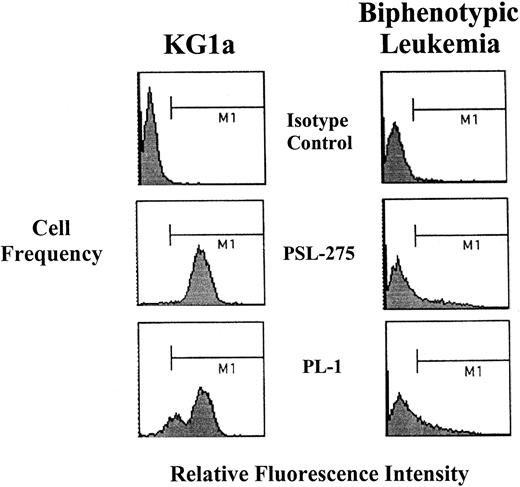
Peripheral blood mononuclear cells (PBMCs) were isolated from blood of 9 patients with AML or ALL, in which blasts represented more than 80% of differential counts (by morphology and flow cytometry, blasts comprised more than 95% of isolated PBMCs). These specimens represent all samples available to date. CD34 and PSGL-1 levels were measured by flow cytometry, and OSGP- and mocarhagin-treated blasts were evaluated for HCLL activity by Stamper-Woodruff assay. All surveyed blasts showed OSGP- and mocarhagin-resistant HCLL activity at levels comparable to KG1a cells (Table6). Blasts from a biphenotypic leukemia (Figure 5) and from an M2 were deficient in PSGL-1, and blasts from 2 patients (one M4 and one M5) lacked CD34, yet HCLL activity was high in all the blasts. As with KG1a cells, ligand activity on all blasts was sensitive to neuraminidase, bromelain, and chymotrypsin digestion, and N-glycosidase F digestion of membrane fractions prepared from 3 specimens (M2, M4, and M5) abrogated binding activity (binding less than 10% of untreated control). Of note, morphologically normal PBMCs (and granulocytes) from patients lacked HCLL activity.
HCLL activity of peripheral blood blasts from patients with acute leukemias
| Cell types6-150 . | Lymphocyte adherence6-151 . | |||
|---|---|---|---|---|
| Untreated . | OSGP . | Mocarhagin . | Neuraminidase . | |
| KG1a cells (positive control) | ++++ | ++++ | ++++ | − |
| Biphenotypic leukemia6-153 | ++++ | ++++ | nd | − |
| AML (M1)6-152 | ++++ | ++++ | ++++ | − |
| AML (M2)6-155 | +++ | +++ | +++ | − |
| AML (M4)6-152,6-154 | ++++ | ++++ | ++++ | − |
| AML (M5)6-152,6-154 | ++++ | ++++ | ++++ | − |
| ALL (L2, pre-B) | ++++ | ++++ | ++++ | − |
| Cell types6-150 . | Lymphocyte adherence6-151 . | |||
|---|---|---|---|---|
| Untreated . | OSGP . | Mocarhagin . | Neuraminidase . | |
| KG1a cells (positive control) | ++++ | ++++ | ++++ | − |
| Biphenotypic leukemia6-153 | ++++ | ++++ | nd | − |
| AML (M1)6-152 | ++++ | ++++ | ++++ | − |
| AML (M2)6-155 | +++ | +++ | +++ | − |
| AML (M4)6-152,6-154 | ++++ | ++++ | ++++ | − |
| AML (M5)6-152,6-154 | ++++ | ++++ | ++++ | − |
| ALL (L2, pre-B) | ++++ | ++++ | ++++ | − |
For all leukemic specimens analyzed, KG1a cell HCLL activity was assessed in parallel to serve as a comparative control. Cytospins of buffer-, OSGP-, mocarhagin- or neuraminidase (Newcastle)-treated blasts or KG1a cells were prepared and underwent Stamper-Woodruff assay as described in “Materials and methods.” Lymphocytes adherent to leukemic cell monolayers were counted by light microscopy using an ocular grid under 100 × magnification (3 fields per slide, 3 slides per experiment, 3 separate experiments). L-selectin dependence lymphocyte adhesion was confirmed in every experiment as described in “Materials and methods.” Note that L-selectin ligand activity in all treated groups was resistant to OSGP and mocarhagin, and sensitive to neuraminidase.
All leukemia specimens showed >80% expression of CD34 and of PSGL-1 except as where indicated by footnote symbols
Results from Stamper-Woodruff assay are presented as percentage of cytocentrifuged cells with adherent lymphocytes: −, 0 to 10%, +, 11 to 35%, ++, 36 to 60%, +++, 61 to 85%, and ++++, 86 to 100%.
Two different specimens of leukemias with these subtypes were evaluated.
This leukemia was negative for all histochemical markers and displayed both myeloid (CD13+/CD33+) and B-lymphoid (CD10+/CD19+/TdT+) cell markers; PSGL-1 expression was low (18% PSGL-1+, see below for staining profiles).
PSGL-1 expression was low (14% PSGL-1+ blasts).
CD34 expression was absent (<5%) on leukemic cells from one of the 2 patients with M4 and one of the 2 patients with M5 as determined by flow cytometry.
nd, not determined
Flow cytometric analysis of PSGL-1 expression (PSL-275 and PL-1) on blasts from biphenotypic leukemia.
Note that PSGL-1 expression was markedly lower than that of KG1a, though HCLL activity was identical (Table 6).
Flow cytometric analysis of PSGL-1 expression (PSL-275 and PL-1) on blasts from biphenotypic leukemia.
Note that PSGL-1 expression was markedly lower than that of KG1a, though HCLL activity was identical (Table 6).
Discussion
Previous studies from our laboratory have identified an L-selectin ligand expressed on the human hematopoietic progenitor cell line KG1a and on normal human CD34+ hematopoietic progenitor cells.2,26 This ligand, designated HCLL, is distinguished from its endothelial counterparts by: (1) sulfation-independent binding activity; (2) resistance to inactivation by OSGP digestion; and (3) absence of MECA79 antigen.26 27 This study was undertaken to characterize the structural biology of HCLL. The data presented here indicate that HCLL is an integral membrane glycoprotein that is biochemically distinct from PSGL-1 and is unique among naturally expressed L-selectin ligands in that the L-selectin binding determinant(s) are presented on N-linked complex glycans.
The earliest hematopoietic progenitor cells express PSGL-1, which is a ligand for all 3 selectins.49,50 The L-selectin binding determinant of PSGL-1 overlaps that of P-selectin in an N-terminal region, which is sensitive to OSGP and mocarhagin digestion.31-33,51 P-selectin and L-selectin binding to PSGL-1 is dependent on sulfation of tyrosines within this region,52,53 whereas critical sulfation occurs on carbohydrates on endothelial L-selectin ligands.18,54-56KG1a express abundant levels of PSGL-1 and our previous studies did not directly address the relationship of HCLL to PSGL-1. Although the sulfation-independence and OSGP-resistance of HCLL contrasts with the L-selectin binding activity of PSGL-1, there is precedent for structural modifications in PSGL-1 conferring selective selectin binding: skin-homing T cells express PSGL-1 specifically decorated with the CLA epitope (a posttranslational carbohydrate modification recognized by the moAb HECA-452), which confers E-selectin binding.57 We therefore sought more direct evidence to establish whether HCLL activity was attributable to PSGL-1. The data reported here clearly demonstrate that the L-selectin binding activity of HCLL is independent of PSGL-1: (1) Mocarhagin and OSGP digestion were each completely effective in cleaving the L-selectin/P-selectin binding domain of the PSGL-1 molecule, but had no effect on HCLL activity; (2) experiments utilizing mixtures of KG1a cells and HL60 cells (which express equivalent levels of PSGL-1) reveal that only KG1a cells support L-selectin–dependent lymphocyte attachment in the Stamper-Woodruff assay; (3) antibody blocking studies utilizing both monoclonal and polyclonal reagents directed against L/P-selectin binding epitopes of PSGL-1 did not affect L-selectin–mediated lymphocyte binding of KG1a; (4) no difference in L-selectin–mediated lymphocyte adherence was observed in KG1a cells sorted for high and low levels of PSGL-1 expression; and (5) blasts from patients with acute leukemias deficient in PSGL-1 displayed high HCLL activity.
Although apparently in contrast to results of previous studies,30-33 our finding that HCLL binding activity is not attributable to PSGL-1 does not negate a role for PSGL-1 as an L-selectin ligand. Rather, these data indicate that the biophysical requirements for PSGL-1–specific L-selectin binding are not met under the operative conditions of the conventional Stamper-Woodruff assay. Threshold biophysical shear forces are necessary to both promote and maintain L-selectin adherence to its ligands.58 Many studies directed at identifying L-selectin ligands have utilized static assay conditions and have relied on probes such as L-selectin–Ig chimeras or isolated molecules presented in nonphysiologic conditions (such as in high concentrations in solution or as displayed on artificial substrates). Compared with these approaches, the Stamper-Woodruff assay offers an important advantage in that it measures functional adhesive interactions under shear conditions of native L-selectin, as expressed naturally on the membrane of a physiologic cell type (the lymphocyte), with its relevant ligand(s) expressed in their native state in natural lipid bilayers of cell membranes.
A variety of selectin ligands have been described, including membrane glycoproteins and glycolipids, secreted glycoproteins, and even extracellular matrix elements.15 49 The results of glycosidase and protease digestions and of metabolic inhibition of both protein and glycoconjugate synthesis indicate that the relevant HCLL carbohydrate binding determinants are sialofucosylated glycans expressed on a protein core. The partitioning of the activity in the aqueous phase of organic solvent extractions argues strongly against a significant component of glycolipid to HCLL activity. Moreover, resistance to PI-PLC digestion and to elution after treatment with high salt buffers over various pH levels indicates that HCLL is an integral membrane glycoprotein.
All other glycoprotein L-selectin ligands (including PSGL-1) are sialomucins sensitive to OSGP, which express relevant carbohydrate L-selectin binding determinant(s) on O-linked glycans.31-33,51,59 60 The results presented here provide 2 independent lines of evidence, obtained from separate but complementary experimental approaches (N-glycosidase digestions and treatment of cells with metabolic inhibitors of glycosylation), which show that critical L-selectin binding determinants on HCLL are presented on N-linked glycans. The diminished reexpression of ligand activity in the presence of glycosylation inhibitors after desialylation was not due to a general inhibition of sialylation because the L60 epitope of CD43 was reexpressed completely in the presence of these agents. Moreover, it was not secondary to inhibition of protein synthesis of the cells, as35S-methionine/cysteine incorporation into nascent protein (assessed by measurement of TCA-precipitable radioactive protein) was not significantly changed in the presence of any of the N-linked glycosylation inhibitors.
The relevant N-linked glycan modification of HCLL that contains the L-selectin binding determinant(s) is, at minimum, a biantennary complex (ie, modified at both Man-1,6 and Man-1,3 arms) structure (Figure 4A). N-glycosidase F hydrolyzes all types of N-glycan structures in mammalian glycoproteins,61 whereas the substrate specificity of endoglycosidase H is high mannose and hybrid structures61,62 and endoglycosidase F2 hydrolyzes only complex biantennary and high mannose structures62 63 (Figure 4B). The observation that endoglycosidase F2 and N-glycosidase F digestions significantly affect HCLL binding activity, whereas endoglycosidase H digestion has no effect on HCLL activity, excludes unmodified high mannose and hybrid structures from consideration and, in particular, indicates that biantennary complex and multiantennary complex structures display the relevant L-selectin binding determinant(s). Moreover, metabolic treatments with either swainsonine or deoxymannojirimycin inhibit recovery of HCLL binding activity after neuraminidase treatment of the cells, which is also consistent with a minimum requirement for a biantennary complex structure (Figure 4A). Specifically, the inhibition by swainsonine suggests that modification of the Man-1,6 arm (in addition to the Man-1,3 arm) is crucial for full expression of ligand activity (Figure 4A). Of note, HCLL activity withstands protein denaturation in 1% SDS and 1% 2-mercaptoethanol (as present in the glycosidase buffers), suggesting that the core protein functions predominantly as a scaffold for the relevant ligand-specific carbohydrate modifications and that these N-linked carbohydrates direct the conformational/structural specificity for binding activity.
The results of this study offer important new insights into the complexity and breadth of L-selectin ligands. The data presented here provide first evidence of a naturally expressed L-selectin ligand with N-glycan–dependent binding activity. We have previously described that this novel L-selectin ligand is expressed among normal CD34+ hematopoietic progenitor cells but is absent in CD34−/lin+ progenitors and in mature granulocytes and lymphocytes.2,64 We now present evidence that native blasts in both AML and ALL express HCLL, a finding that warrants further investigation of the role of HCLL in the hematopoietic process. Studies in vitro indicate that L-selectin expression is associated with higher clonogenic activity of human hematopoietic progenitors,7,12,13 suggesting that L-selectin–dependent receptor-ligand interactions enhance proliferation and/or differentiation of progenitors. In contrast, engagement of PSGL-1 appears to inhibit hematopoiesis,6 and increased myelopoiesis is observed both in fucosyltransferase VII-deficient mice (which have defective PSGL-165) and in E- and P-selectin double-deficient mice.66 Of note, L-selectin knockout mice appear to have normal peripheral blood and marrow granulocyte counts,67 but, to our knowledge, no direct investigations of hematopoiesis in L-selectin–deficient progenitors, by clonogenic assays or by competitive repopulation studies, have been performed.
L-selectin/ligand interactions confer shear-resistance, a property critical to initial seeding of progenitors within the hematopoietic microenvironment after stem cell transplantation. On bone marrow cells, L-selectin expression is absent on stromal elements, but mature leukocytes and the earliest hematopoietic progenitors are typically L-selectin+, whereas intermediate stages of leukocyte development and erythrocyte and megakaryocytic lineage cells are L-selectin−.13,68,69 A recent study showed that normal CD34+ human bone marrow and fetal liver hematopoietic cells roll on immobilized L-selectin under shear, with the highest rolling efficiency in the more immature (CD38−) subpopulation of cells.38 This study also showed that anti–PSGL-1 moAb PL1 has no significant effect on progenitor cell rolling on L-selectin over a wide range of shear forces, but the ligand(s) that conferred this shear-resistant adherence were unidentified.38 We have evidence from parallel plate flow chamber experiments that HCLL displays high shear resistance,70 making HCLL a prime candidate for the L-selectin ligand activity observed on primitive hematopoietic cells. Additional exploration of these observations awaits the isolation of HCLL. At present, our knowledge of the role of selectins in hematopoiesis is incomplete, and further characterization of HCLL should provide important insights into the biology of selectin-dependent adhesive interactions in progenitor growth, in stem cell trafficking and in marrow recovery after stem cell transplantation.
Acknowledgments
We are grateful to Dr Xhizhuang Shu, Jack Y. Lee, Geoffrey E. Grove, and Bozena Antoniu for excellent technical assistance. We wish to thank Dr Richard M. Stone, Dr Jeffrey Kutok, Dr Daniel Deangelo, and Ms Ilene A. Galinsky for assistance in procuring leukemia specimens, and Drs Dale A. Cumming, Anthony L. Tarantino, Robert C. Fuhlbrigge, and Thomas S. Kupper for helpful discussions and for critical review of the manuscript. This report is dedicated to the memory of Ms Ann M. (“Annie”) Ferguson.
Supported by National Institutes of Health grant HL60528 (R.S.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Robert Sackstein, Harvard Institutes of Medicine, 77 Ave Louis Pasteur, Rm 671, Boston, MA 02115; e-mail:rsackstein@rics.bwh.harvard.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal