Abstract
The red blood cell membrane (RBCM) is a primary model for animal cell plasma membranes. One of its major organizing centers is the cytoplasmic domain of band 3 (cdb3), which links multiple proteins to the membrane. Included among its peripheral protein ligands are ankyrin (the major bridge to the spectrin-actin skeleton), protein 4.1, protein 4.2, aldolase, glyceraldehyde-3-phosphate dehydrogenase, phosphofructokinase, deoxyhemoglobin, p72syk protein tyrosine kinase, and hemichromes. The crystal structure of cdb3 is reported at 0.26 nm (2.6 Å) resolution. A tight symmetric dimer is formed by cdb3; it is stabilized by interlocked dimerization arms contributed by both monomers. Each subunit also includes a larger peripheral protein binding domain with an α+ β-fold. The binding sites of several peripheral proteins are localized in the structure, and the nature of the major conformational change that regulates membrane-skeletal interactions is evaluated. An improved structural definition of the protein network at the inner surface of the RBCM is now possible.
Introduction
The red blood cell membrane (RBCM) is constructed primarily of an anastomosing spectrin-actin protein skeleton connected at regular intervals to the integral membrane proteins, band 3 and glycophorin C, via the bridging proteins, ankyrin, and protein 4.1.1 Because the membrane is easily isolated, architecturally simple, and compositionally related to many other membranes, it is commonly presented as a model of animal cell plasma membranes in biochemistry textbooks.2,3 However, although each of the major membrane proteins has been scrutinized functionally, high resolution crystallographic data are available only for actin and a single domain of spectrin.4-6
Band 3 constitutes the most abundant polypeptide in the RBCM, comprising 25% of the total membrane protein. Band 3, also termed the anion exchanger (AE1), can be cleaved into 2 independent structural domains that retain the functional characteristics of their counterparts in the intact protein. The 55-kd membrane-spanning domain is thought to traverse the bilayer 12 times and serves to catalyze the exchange of anions (mainly Cl− for HCO3−) across the membrane during gas transport in the blood.7 The membrane-spanning domain may also mediate removal of senescent RBCs from circulation,8-10 and it has been shown to carry a number of common blood group antigens.11 A low (2.0 nm [20 Å]) resolution structure of the membrane-spanning domain has been reported.12
The cytoplasmic domain of band 3 (cdb3) functions primarily as an anchoring site for other membrane-associated proteins. Included among the protein ligands of cdb3 are ankyrin,13 protein 4.2,14,15 protein 4.1,16,17glyceraldehyde-3-phosphate dehydrogenase (GAPDH),18phosphofructokinase,19 aldolase,20hemoglobin,21,22 hemichromes,23 and the protein tyrosine kinase (p72syk).24Importantly, each of these band 3 interactions has nontrivial consequences for the structure and function of the cell, ranging from control of cell flexibility and shape25 to regulation of glucose metabolism,26 ion transport,27 and cell lifespan.10 Not surprisingly, mutations mapped to band 3 have been reported to alter each of the above characteristics including cell shape.27-33
Recognizing that cdb3 constitutes a major organizing center of the RBCM and that structural information on cdb3 might allow a more accurate model of the architecture and function of the erythrocyte, we undertook to solve the crystal structure of cdb3 and to re-evaluate the organization of the membrane in its vicinity. We report here the structure of cdb3 at 0.26 nm (2.6Å) resolution. Using previous information on the sites of cdb3 association with its peripheral protein ligands, we also map these sites on cdb3 and reconstruct a model of the membrane at this important locus.
Materials and methods
Experimental procedures
Residues 1-379 of the human erythrocyte band 3, encompassing the entire sequence of cdb3 released by chymotryptic cleavage of erythrocyte membranes, was expressed in Escherichia coli and purified to homogeneity, as described elsewhere.34 X-ray diffraction patterns were measured from 1 selenomethionyl (SeMet)-substituted cdb3 crystal at 3 wavelengths near the selenopotassium (SeK) absorption edge by the use of facilities associated with BioCars beamline BM-14D (Advanced Photon Source, Argonne National Laboratories, ). Diffraction patterns were measured by the rotation/oscillation method over a total range of 360° at each wavelength. The diffraction patterns were analyzed with the HKL programs35 and were consistent with the space group and unit cell of the native crystal form previously described.34 The crystal was flash frozen immediately prior to exposure to x-rays and was maintained at a nominal temperature of 108 K throughout the diffraction experiments. It had dimensions of 0.2 × 0.25 × 0.3 mm and diffracted to 0.26 nm (2.6 Å). Representative statistics describing the diffraction data are presented in Table 1.
Summary of diffraction data and selected refinement statistics
| Diffraction measurements | |||
| Wavelength, nm (Å) | 0.09789 (0.9789) | 0.09796 (0.9796) | 0.09537 (0.9537) |
| Resolution, nm (Å) | 3.5-0.26 (35.0-2.6) | 3.5-0.26 (35.0-2.6) | 3.5-0.26 (35.0-2.6) |
| Rsym, % (last shell, %)* | 0.057 (0.208) | 0.049 (0.192) | 0.052 (0.175) |
| Completeness, % (last shell, %) | 99.1 (94.5) | 98.2 (92.9) | 97.6 (56.1) |
| Current model and refinement statistics | |||
| R, Rfree (reflections)† | 0.216 (41 136) | 0.290 (3 612) | |
| Atoms, no. | |||
| Nonhydrogen | 9 687 | ||
| Protein | 9 509 | ||
| Water | 178 | ||
| R.m.s. deviations‡ | |||
| Bonds, nm (Å) | 0.00080 (0.0080) | ||
| Angles, deg | 1.67 | ||
| Diffraction measurements | |||
| Wavelength, nm (Å) | 0.09789 (0.9789) | 0.09796 (0.9796) | 0.09537 (0.9537) |
| Resolution, nm (Å) | 3.5-0.26 (35.0-2.6) | 3.5-0.26 (35.0-2.6) | 3.5-0.26 (35.0-2.6) |
| Rsym, % (last shell, %)* | 0.057 (0.208) | 0.049 (0.192) | 0.052 (0.175) |
| Completeness, % (last shell, %) | 99.1 (94.5) | 98.2 (92.9) | 97.6 (56.1) |
| Current model and refinement statistics | |||
| R, Rfree (reflections)† | 0.216 (41 136) | 0.290 (3 612) | |
| Atoms, no. | |||
| Nonhydrogen | 9 687 | ||
| Protein | 9 509 | ||
| Water | 178 | ||
| R.m.s. deviations‡ | |||
| Bonds, nm (Å) | 0.00080 (0.0080) | ||
| Angles, deg | 1.67 | ||
Deg indicates degrees.
Rsym indicates ςIHKL − < IHKL > ςIHKL, where IHKL is the intensity for one observation of a reflection and < > is the mean for all observations of that reflection.
R indicates ςFobs − Fcal/ςFobs, where Fobs and Fcal are the observed and calculated structure factor amplitudes, respectively. Rfree was calculated using the same formula as Rcrystal, but with only 8% of the total amplitudes chosen randomly.
R.m.s. deviation is the root-mean-square deviation from the expected bond lengths or angles.
Initial phase estimates were obtained by the multiple-wavelength anomalous diffraction method. The program SOLVE36 was used to establish positional and scattering parameters for 16 Se sites within the asymmetric unit and to calculate initial phase estimates. The CCP4 package37 was used for most of the subsequent crystallographic calculations. Prior to model building, the phases and electron density maps were improved through density modification without averaging by use of the CCP4 program DM.38 The secondary structure was easily recognized in the maps. The backbone structures of the 4 monomers were independently traced and then compared to establish initial polyalanine models for each monomer. These models were then independently elaborated to include side chains prior to refinement.
The structure was refined by the use of XPLOR39 and O.40 Following initial refinement by simulated annealing in the presence of non-crystallographic symmetry (NCS) restraints, the model was further refined by restrained positional and temperature factor adjustments as well as manual model rebuilding. The NCS restraints were not used in the latter stages of refinement. The properties of the refined model are also provided in Table 1.
Evaluation of the locations of the C-termini was problematic because the 4 monomers in the asymmetric unit exhibited variably ordered structures beyond residue 348. Although the P and S monomers are not well defined beyond this point, the Q and R monomers have very similar, well-defined backbone structures through residue 356, and the C-terminal residues of monomer Q are not directly affected by crystal contacts. Therefore, to compare the locations of the C-termini among the 3 possible tetrameric complexes, monomer Q was superimposed on all monomers, and the transformed locations of Cα:356Q were used as markers for the C-termini. Similarly, the transformed location of Cα55Q was used as the last resolvable N-terminal residue for each subunit.
Results
Crystallization and data collection
The cdb3 was crystallized at pH 4.8 by the sitting drop vapor diffusion method followed by streak-seeding of the harvested crystals into solutions of higher protein content, as described previously.34 Crystals of the longest dimension (approximately 0.3 mm) that diffracted x-rays to resolutions up to approximately 0.21 nm (2.1Å) were obtained. Unfortunately, isomorphous heavy atom derivatives using various reagents could not be prepared either due to poor heavy atom substitution or because of an induced change in crystal structure. Therefore, SeMet-substituted cdb3 was expressed in an E coli host that was inhibited in endogenous methionine biosynthesis,41 and the crystals were grown as described above. X-ray diffraction analysis revealed a C2 space group with the following unit cell dimensions: a, 18.159 nm (181.59 Å); b, 9.258 nm (92.58 Å); c, 12.333 nm (123.33 Å); and β, 131.86°. The asymmetric unit includes the equivalent of 4 cdb3 monomers. The current structure consists of 9509 nonhydrogen protein atoms and 178 water molecules and was refined to 0.26 nm (2.6 Å) resolution and an R factor of 22% (Rfree = 29%). Properties of the refined model are provided in Table 1.
Structure of cdb3 monomer
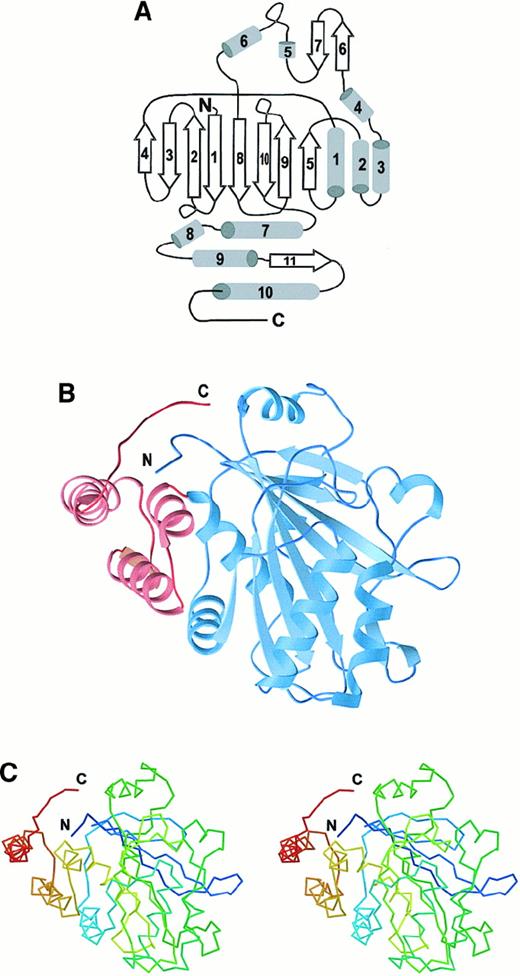
The cdb3 monomer includes 11 β-strands and 10 helical segments and belongs to the α + β–fold class (Figure1A). Eight of the β-strands are assembled into a central β-sheet of mixed parallel and antiparallel strands (Figure 1B). Two of the remaining strands (β6 and β7, residues 176-185) form a β-hairpin that likely constitutes the core of the ankyrin binding site, as described below. Along with the first 6 helices, these elements assemble into a globular domain spanning residues 55-290. A short helix and loop segment, residues 291-303, connects this large domain to a dimerization arm, which comprises residues 304-357. This arm is largely helical, but it includes a single β-strand, which plays a critical role in the formation of an interlocked dimer (Figure 2A). The monomer is compact, having dimensions of approximately 4.3 × 4.5 × 5.3 nm (43 × 45 × 53 Å). There are no major structural differences among the 4 monomers in the asymmetric unit, although there are variations in the regions of intermolecular contact and in the extent of order in the N- and C-terminal segments.
Structure of cdb3 monomer.
(A) Diagram of cdb3 secondary structure with the following sequence assignments: β157-66, β273-82, β384-88, β492–96, α1104–116, β5117–122, α2128–141, α3146–158, α4166–170, β6176–180, β7182–185, α5196–201, α6212–220, β8224–234, β9239–247, β10263–271, α7278–290, α8292–300, α9304–316, β11318–323, and α10328–347. (B) Ribbon diagram of cdb3 monomer, with blue corresponding to the peripheral protein binding domain and red to the dimerization arm. (C) Stereo drawing of the α-carbon trace for the cdb3 monomer in approximately the same orientation as in panel B. Colors change from blue at the N-terminus to red at the C-terminus.
Structure of cdb3 monomer.
(A) Diagram of cdb3 secondary structure with the following sequence assignments: β157-66, β273-82, β384-88, β492–96, α1104–116, β5117–122, α2128–141, α3146–158, α4166–170, β6176–180, β7182–185, α5196–201, α6212–220, β8224–234, β9239–247, β10263–271, α7278–290, α8292–300, α9304–316, β11318–323, and α10328–347. (B) Ribbon diagram of cdb3 monomer, with blue corresponding to the peripheral protein binding domain and red to the dimerization arm. (C) Stereo drawing of the α-carbon trace for the cdb3 monomer in approximately the same orientation as in panel B. Colors change from blue at the N-terminus to red at the C-terminus.
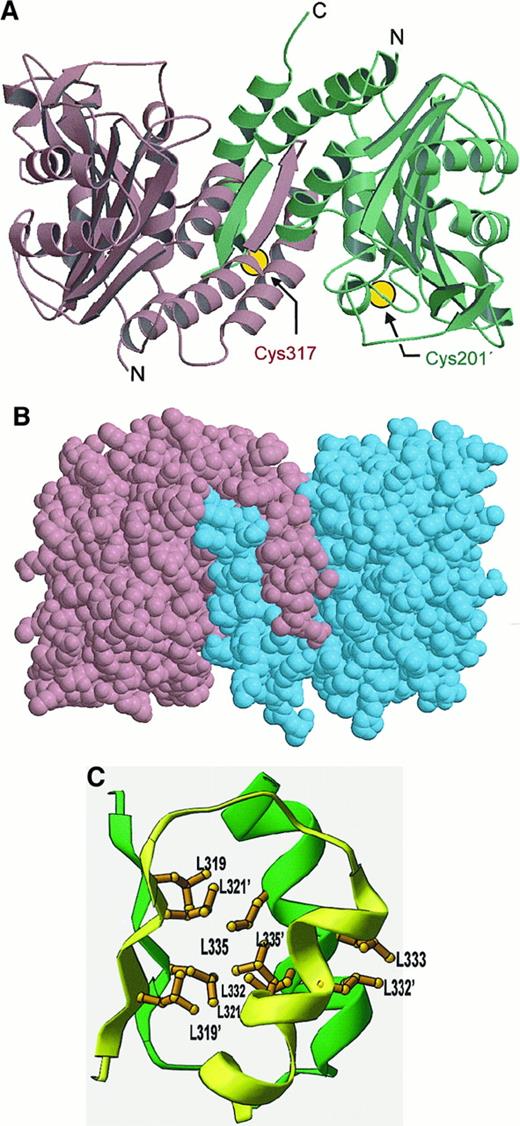
Structure of cdb3 dimer.
(A) Ribbon diagram of cdb3 dimer showing interactions between interlocking dimerization arms. The positions of the 2 cysteines (Cys 201A and Cys 317B) that form an inter-subunit disulfide bond upon treatment with the oxidizing agent, o-phenanthroline/CuSO4(pH at least 7.5), are also shown. Because these cysteines cannot form a disulfide in the low pH conformation of cdb3, they are not expected to reside near each other in the low pH structure shown here. (B) Space filling model of cdb3 dimer. (C) Cluster of 9 leucine residues (extending from β11 and α10 of each subunit) that stabilize the dimerization domain.
Structure of cdb3 dimer.
(A) Ribbon diagram of cdb3 dimer showing interactions between interlocking dimerization arms. The positions of the 2 cysteines (Cys 201A and Cys 317B) that form an inter-subunit disulfide bond upon treatment with the oxidizing agent, o-phenanthroline/CuSO4(pH at least 7.5), are also shown. Because these cysteines cannot form a disulfide in the low pH conformation of cdb3, they are not expected to reside near each other in the low pH structure shown here. (B) Space filling model of cdb3 dimer. (C) Cluster of 9 leucine residues (extending from β11 and α10 of each subunit) that stabilize the dimerization domain.
Residues 1-54, 202-211, and 357-379 were not observed or were poorly defined by the electron density. Flexibility was indeed expected in the first 54 residues because this region is strongly anionic, containing 20 acidic residues, no basic residues, and a blocked N-terminal methionine.42,43 The region is also involved in most peripheral protein interactions,44 suggesting that its conformation may be adaptable to a variety of protein ligands. The N-terminal segment has, in fact, been shown by solution NMR and x-ray crystallography to bind aldolase45 and deoxyhemoglobin21 in different extended conformations. The inability to resolve residues 202-211 and 357-379 is also assumed to be due to segmental flexibility. Physical studies of cdb3 have shown the protein to be a highly flexible molecule.46 47
The positions of the N- and C-terminal segments are important in defining the interactions of cdb3. The C-terminal residues of cdb3 connect to the membrane-spanning domain, and hence, must orient toward the lipid bilayer. The N-terminal segment, in contrast, must interact with peripheral proteins, eg, the glycolytic enzymes, hemoglobin, protein 4.1, ankyrin, and hemichromes.21,23 48-50 To bind these proteins without steric interference from the bilayer, the N-termini must presumably face away from the membrane. The locations of the last resolvable residues at the N- and C-termini of cdb3 are marked in Figures 1B and 2A. How these locations impact the various peripheral protein interactions will be evaluated below.
Dimerization of cdb3
Isolated cdb3 has been shown to exist as a dimer over a range of protein concentrations and solution conditions.51,52 This strong preference for dimerization is readily explained by the crystal structure. Within the asymmetric unit are 2 independent but equivalent dimers, P-Q and R-S (Figure 3A), that are essentially 2-fold symmetric and interlocked by extensive interactions within an intermonomeric dimerization domain (Figure 2A). To build this domain each monomer contributes a dimerization arm (residues 314-344) that extends away from the major globular domain and interdigitates with the equivalent arm from the second monomer (Figure 2B). Among the extensive interactions at this interface are 8 intermonomeric backbone-to-backbone hydrogen bonds contributed in part by the 2 strands of the intermonomeric antiparallel β-sheet. Subunit association is further stabilized by a hydrophobic core of 9 interacting leucine residues (Figure 2C). Approximately 520 nm (5200 Å)2 of exposed surface is buried at the dimer interface, and 370 nm (3700 Å)2 of the buried surface is associated with nonpolar side chain atoms. The 2 markers for the C-termini lie near the center of the same face of the dimer, where they could easily attach to adjacent membrane-spanning domains. The 2 markers for the N-termini locate on the lateral surfaces of the dimer, where they would allow extension of the nonresolved N-terminal residues away from the membrane. Figure 2B also shows the general shape of the dimer, which fits within a rectangular prism of approximate dimensions 7.5 × 5.5 × 4.5 nm (75 × 55 × 45 Å).
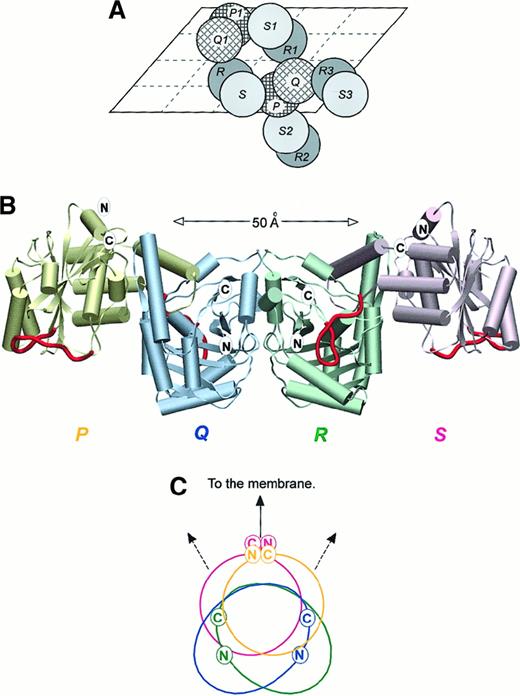
Structure of cdb3 tetramer.
(A) The packing of selected cdb3 dimers in the crystal illustrated schematically by projection onto an a, c plane of the unit cell. The unit cell includes 4 monomers of types P, Q, R, and S. (B) The association of cdb3 subunits in the likely physiological tetramer, with each potential ankyrin binding loop (residues 176-190) highlighted in red. The plane of the bilayer is believed to lie in a horizontal plane at the top of the diagram. (C) Relative orientation of the truncated N- and C-termini of the tetramer, as viewed from one end of the tetramer. Color coding is the same as in Figure 3B. Panels A and B are rotated with respect to each other by 180° about a horizontal axis in the plane of the page.
Structure of cdb3 tetramer.
(A) The packing of selected cdb3 dimers in the crystal illustrated schematically by projection onto an a, c plane of the unit cell. The unit cell includes 4 monomers of types P, Q, R, and S. (B) The association of cdb3 subunits in the likely physiological tetramer, with each potential ankyrin binding loop (residues 176-190) highlighted in red. The plane of the bilayer is believed to lie in a horizontal plane at the top of the diagram. (C) Relative orientation of the truncated N- and C-termini of the tetramer, as viewed from one end of the tetramer. Color coding is the same as in Figure 3B. Panels A and B are rotated with respect to each other by 180° about a horizontal axis in the plane of the page.
Conformational equilibrium in cdb3 expansion of the dimer at high pH
A remarkable property of cdb3 is its reversible, pH-dependent conformational equilibrium, which is readily detected by intrinsic fluorescence, sedimentation analysis, differential scanning calorimetry, fluorescence energy transfer, gel filtration chromatography, native gel electrophoresis, and a variety of functional assays.44,46,47,52,53 Thus, as the pH is raised from 6.5 to 9.5, the Stokes radius of cdb3 enlarges reversibly by 1.1 nm (11 Å), and the intrinsic fluorescence, which is highly quenched at a lower pH, more than doubles. A disulfide bond that can form between cysteine (Cys) 201 of one subunit and Cys 317 of the paired subunit at a pH of at least 7.5 can no longer be generated below pH 7.0, as seen in locations of participating cysteines in low pH conformation in Figure 2A, and cdb3-associated hemoglobin pivots away from the membrane.53 Because ankyrin cannot bind to the high pH form, but avidly associates with the low pH form,25 54 the nature of this conformational equilibrium is important to understanding the regulation of membrane-skeleton interactions.
We have examined the structure of cdb3 for a possible hinge around which adjacent regions of cdb3 might pivot, thereby generating the conformational change. Both the peripheral protein binding domain and dimerization arm are tightly folded structures, and therefore, we doubt that the conformational change can arise solely within one of these regions. Two short segments between these domains, however, could accommodate the anticipated bending. The first, between helices α7 and α8, is centered at glutamic acid (Glu) 291, and the second, between α8 and α9, is centered at glutamine (Gln) 302. A change in the backbone conformation at either or both locations would permit considerable expansion of cdb3 while leaving the dimer tightly coupled through the unmodified dimerization domain. Such a pivoting motion would also explain why the aforementioned disulfide bond cannot form below pH 7.0 and why its formation at pH 8.0 prevents ankyrin association.54
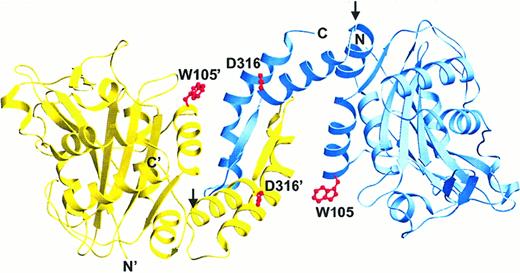
Although crystallographic data were not available at a high pH, we rotated the respective domains between helices α8 and α9 (at glycine [Gly] 305) to visualize a possible high pH conformation of the dimer. As seen in Figure 4, this operation not only elongated the dimer, but it also disjoined a hydrogen bond connecting tryptophan (Trp) 105 of one subunit to aspartic acid (Asp) 316 of the other. Because this H bond would be expected to quench the Trp fluorescence,55 this structural change could account for both the increase in Trp fluorescence and cdb3 elongation.47 The decrease in thermostability of the high pH form might also be explained by disruption of this and other interactions present in the more compact (low pH) structure. The decline in cooperativity of thermal denaturation with increasing pH might further derive from the separation of tightly coupled domains into noninteracting domains.52
Possible expansion of cdb3 dimer at elevated pH.
To envision a possible high pH conformation of cdb3, the 2 peripheral protein binding domains were rotated away from their shared dimerization domain by allowing them to pivot at Gly 305 (indicated by arrows), a plausible site for rotation to occur. The resulting protein would be expected to remain dimeric, but also to become more elongated and fluorescent as the domains separate and the quenching H bonds between Asp 316 and Trp 105 (shown in red) disjoin.
Possible expansion of cdb3 dimer at elevated pH.
To envision a possible high pH conformation of cdb3, the 2 peripheral protein binding domains were rotated away from their shared dimerization domain by allowing them to pivot at Gly 305 (indicated by arrows), a plausible site for rotation to occur. The resulting protein would be expected to remain dimeric, but also to become more elongated and fluorescent as the domains separate and the quenching H bonds between Asp 316 and Trp 105 (shown in red) disjoin.
The cdb3 tetramer
In its membrane environment, intact band 3 exists primarily as a mixture of dimers and tetramers.56-59 Because cdb3 is thought to be essential for tetramer formation60-62 and because tetramer formation is critical for ankyrin association,54,56 63-65 we examined the interactions within the crystal for contacts that might correspond to the tetramer interface in vivo. The crystal structure revealed 3 possible tetramers defined by association of different R-S dimers with a central P-Q dimer (Figure 3A). These are R2-S2:P-Q or P-Q:R3-S3 (tetramer I), P-Q:R-S (tetramer II), and P-Q:R1-S1 (tetramer III). Remarkably, 2 of the tetramers (I and II) are approximately 2-fold symmetric, and the third (tetramer III) involves a 2-fold rotation coupled to a moderate translation.
Although identification of the physiological tetramer cannot be established from crystallographic data, 4 lines of evidence favor tetramer I as the likely native oligomer. First, locations of contacts among subunits in tetramers II and III prohibit bending and/or pivoting of the cdb3 dimerization arm relative to its peripheral protein binding domain, thereby blocking the likely conformational change in cdb3 seen both in situ and in vitro.44 This restriction is not seen in tetramer I. Second, the closed tetramers in models II and III cannot elongate into hexamers and higher oligomers, even though such extended forms of band 3 have been observed.29,51,63,66-68 Tetramer I, in contrast, is capable of head-to-tail extension. Third, binding of ankyrin to its likely site at residues 176-190 would obstruct the interfacial contacts in tetramers II and III (eg, residues 187-190), thereby preventing rather than promoting the ankyrin-induced tetramer formation seen in situ.56,69 70 No such obstruction exists in tetramer I. And finally, the noncontiguous regions of cdb3 involved in ankyrin binding are proximal in tetramer I but further apart in tetramers II and III, as indicated below.
Tetramer I is formed by 2 crystallographically independent but structurally identical associations of P-Q and R-S dimers, ie, R2-S2:P-Q and P-Q:R3-S3 (Figure 3A). In projection the tetramer is approximately 14.0 nm (140 Å) in the longest dimension and 4.5 nm (45 Å) in the cross-section. Only 2 monomers, S with P or Q with R, are in contact at the dimer:dimer interface, and the amino acids in contact comprise residues 208-225 and 252-261 from both subunits. Approximately 200 nm (2000 Å)2 of exposed surface is buried at this interface, and 170 nm (1700 Å)2 is associated with nonpolar side chain atoms.
Figure 3B shows the shape and subunit orientations in the above tetramer, and Figures 3B and 3C indicate the relative locations of the last resolvable residues at the N- and C-termini of each polypeptide. The C-terminal markers (locations of serine [Ser] 356) project onto an arc 130° wide and are thus approximately on the same face of cdb3, thereby allowing simultaneous communication with the membrane-spanning domains via the unresolved sequences, residues 357-399 (Figure 3B). Two of the N-terminal markers (locations of histidine [His] 55) also extend from a face of the tetramer that orients toward the membrane, whereas the other 2 are on lateral surfaces and fully accessible. Although this tetramer is consistent with much published data, it will still be important to verify its organization through biochemical studies in the near future.
Ankyrin association with cdb3
Virtually all investigators agree that ankyrin binds primarily to the tetramer of band 3, with little or no affinity for the dimer.54,55,63-65,69,70 The addition of ankyrin to RBCM forces band 3 to a tetramer.53,55,69,70As a result, it is not unlikely that the ankyrin binding site comprised sequences which were contributed by 2 separate dimers. The addition of ankyrin tethers these dimers together, perhaps via association with 2 distinct band 3 binding domains on the ankyrin.71Importantly, the specific residues on cdb3 involved in ankyrin binding have been partially mapped to at least 2 noncontiguous regions. Antibody competition studies, site-directed mutagenesis, chimera analysis, and phosphorylation inhibition experiments all implicate the unresolved N-terminal segment in this interaction.49,72-75Similar methods plus proteolysis protection studies further demonstrate involvement of residues 176-190, with additional contributions possible from adjacent residues. As illustrated in Figures 1A and 3B, residues 176-190 lie near the middle of a largely exposed and convoluted 100-residue segment located between strands β5 and β8 of the main sheet. Residues 177-185 include strands β6 and β7 and take the form of a well-known β-hairpin structure.76 Evidence that this hairpin structure may indeed constitute an ankyrin binding site also stems from recent crystallographic results which show that the putative ankyrin docking segment from the sodium/potassium–adenosine 5′-triphosphatase (Na+/K+-ATPase) also forms a stalk-and-loop structure when fused to the C-termini of glutathione S transferase.77
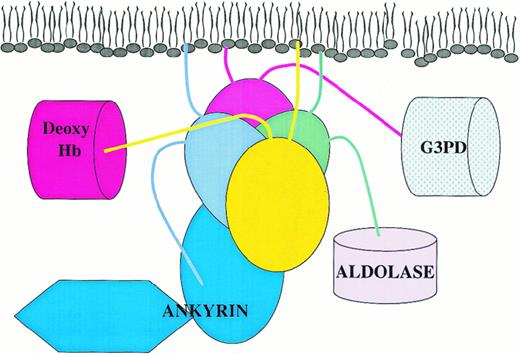
As noted above, an N-terminal segment must also contribute to the high affinity binding site for ankyrin. In tetramer I the last resolvable N-terminal amino acid from 1 dimer (His 55) lies in reasonable proximity, 3.1-4.1 nm (31-41 Å), and on the same surface of the tetramer as residues 176-190 from the other dimer (Figure 3B). Not only does this attachment site orient ankyrin toward the cytoplasm, where it can easily bridge to the spectrin skeleton, but it also necessitates tetramer formation because juxtaposition of an N-terminal segment and an ankyrin binding loop requires the association of 2 dimers. In contrast, the distances in tetramer II range from 5.9-7.6 nm (59-76 Å), and the 2 segments implicated in ankyrin binding lie on different faces of the tetramer. A possible arrangement of ankyrin on the extended band 3 tetramer is depicted in Figure5.
Cartoon displaying the possible organization of a peripheral protein complex on a cdb3 tetramer in situ.
Protein 4.1 is not displayed in the diagram because it competes with ankyrin for an overlapping site. Deoxy Hb indicates deoxyhemoglobin; G3PD, glyceraldehyde-3-phosphate dehydrogenase.
Cartoon displaying the possible organization of a peripheral protein complex on a cdb3 tetramer in situ.
Protein 4.1 is not displayed in the diagram because it competes with ankyrin for an overlapping site. Deoxy Hb indicates deoxyhemoglobin; G3PD, glyceraldehyde-3-phosphate dehydrogenase.
Association of cdb3 with other peripheral proteins
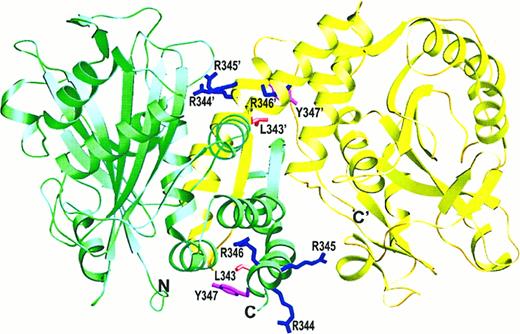
Protein 4.1 is the only peripheral protein known to associate partially with residues in the dimerization arm of cdb3. In addition to the required contact at the N-termini,50 protein 4.1 also binds to an LRRRY sequence that forms part of a helix near the C-termini (residues 343-34678; Figure6). Based on biochemical studies, the LRRRY sequence is believed to interact with an oppositely charged LEEDY peptide in protein 4.1. However, because the side chains of I343, R346, and Y347 are on less accessible surfaces of the helix, any model that specifies direct pairing of IRRRY with LEEDY would require either partial unfolding of the helix or only delocalized interactions with the more buried residues. Importantly, the marker for each subunit's N-terminus lies sufficiently close to its LRRRY sequence to allow binding of 4.1 to a single monomer. Further, the corresponding sites on the 2 monomers of a cdb3 dimer appear sufficiently separated to allow both monomers to bind a different protein 4.1 simultaneously. While association of protein 4.1 with the extended cdb3 tetramer may also be possible, steric hindrance from the membrane should afford preference to complexes with cdb3 N-termini pointing away from the bilayer. This would place protein 4.1 in direct competition with ankyrin for the N-termini of subunits Q and R in the cdb3 tetramer. Competition between protein 4.1 and ankyrin for cdb3 has, in fact, been reported by several laboratories.16,50 79
Location of the protein 4.1 binding site on the cdb3 dimer.
The side chain orientations of the LRRRY sequence are shown, and the location of the proximal N-terminal marker is labeled to allow visualization of the protein 4.1 binding surface.
Location of the protein 4.1 binding site on the cdb3 dimer.
The side chain orientations of the LRRRY sequence are shown, and the location of the proximal N-terminal marker is labeled to allow visualization of the protein 4.1 binding surface.
Information on the binding site of protein 4.2 on cdb3 is limited. Natural mutations at Glu 40, Gly 130, and proline (Pro) 327 of cdb3 lead to a protein 4.2 deficiency and hereditary spherocytosis.28,30 31 Unfortunately, none of these mutations colocalize to a common region of cdb3. Pro 327 lies in the cdb3 dimerization domain and hence its mutation to arginine (Arg) could impact self association. Gly 130 is situated in an α-helix near the ankyrin binding loop and its replacement with Arg could affect local folding. The Glu 40 to lysine (Lys) charge reversal could compromise electrostatic interactions with the N-termini. Further studies will obviously be necessary to resolve which if any of these mutations identifies the binding site of protein 4.2.
The glycolytic enzymes (GAPDH, aldolase, and phosphofructokinase), deoxyhemoglobin, and hemichromes all bind to the extreme N-terminal segment of cdb3.21,23,48,80 These interactions, which are largely electrostatic in nature, lead to inhibition of glycolysis, reduced hemoglobin-oxygen affinity, and the clearance of RBCs from circulation, respectively.8,21,26 Because p72syk also associates with band 3 and phosphorylates tyrosine (Tyr) 8,24 81 it may also interact within the same region. Given that 2 of the N-termini of the tetramer must exit the tightly folded core of the oligomer on the membrane-apposed surface, at least a fraction of the above enzyme binding sites can be anticipated to lie close to the bilayer (Figure 5).
Noting that many associations involve the N-terminal segment of cdb3, it might seem that competition for binding would be a significant factor in regulating the assembly of complexes on cdb3. However, there are sufficient molecules of band 3 in RBCs to bind all copies of ankyrin, protein 4.1, and the aforementioned glycolytic enzymes simultaneously.44 Not all peripheral proteins can, however, associate with the same oligomer of band 3 simultaneously (Figure 5).
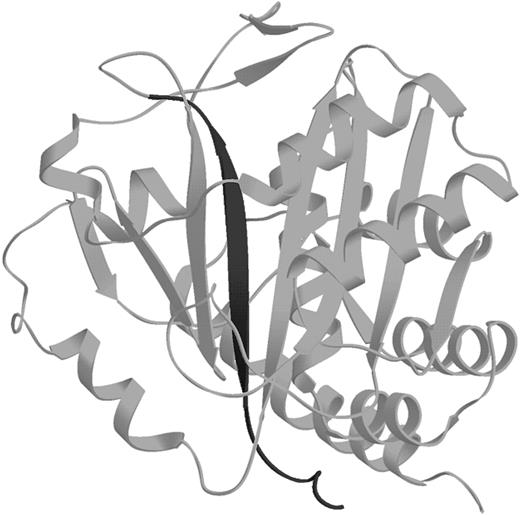
The structure of kidney cdb3
The kidney band 3 is identical to the RBC band 3 except that translation of the polypeptide in the kidney band 3 begins at exon 5, resulting in the absence of residues 1-65. As seen in Figure7, this alternative initiation site also causes deletion of a central strand in the major β-sheet. How the remaining folded structure of cdb3 accommodates removal of this middle strand is intriguing. However, it should not be overlooked that the functional differences between erythrocyte and kidney cdb3 are not minor because kidney cdb3 no longer binds ankyrin, protein 4.1, or the glycolytic enzymes.74 75 Thus, the N-terminal 65 residues of cdb3 may be critical to band 3–specific functions.
Ribbon diagram of the cdb3 monomer.
The dark strand in the middle is the β-strand that is deleted from the kidney cdb3.
Ribbon diagram of the cdb3 monomer.
The dark strand in the middle is the β-strand that is deleted from the kidney cdb3.
Discussion
The significance of the cdb3 structure and function to erythrocyte properties is emphasized by the fact that missense mutations in cdb3 can result in global changes in RBC shape and deformability.28,30-32 Although the deleterious effects of 1 or 2 of these amino acid substitutions can possibly be explained by localization to the binding site of an important peripheral protein,28,30 others appear more likely to derive from their potential impact on cdb3 conformation. Indeed, 3 mutations with a significant effect on erythrocyte morphology reside in conserved regions of the secondary structure that are far removed from assumed docking sites of peripheral proteins.32 Thus, band 3 Tuscaloosa (Pro 327 → Arg), band 3 Boston (alanine [Ala] 285 → Asp), and band 3 Nachod (GTVLL 117-121 deleted) all contain mutations that localize to helices or sheets which may be critical to the correct folding of the polypeptide (Figures 1 and 2). The effects of these mutations on erythrocyte morphology would argue that the native conformation of cdb3 is essential to the structure and function of the entire erythrocyte.
In contrast to the above mutations, amino acid substitutions in unstructured regions of cdb3 may not cause alteration in cell morphology. An example of this class of polymorphism is found in band 3 Memphis,82 where Lys 56 is replaced by Glu directly upstream of the first strand of the central 8–stranded β-pleated sheet. In this instance the substitution results in a 20% reduction in the rate of anion transport across the membrane.82 From the position of the Lys → Glu substitution near the attachment of cdb3 to the membrane-spanning domain, we speculate that the change in electrostatic charge might reduce the concentration of anions near their transport site at the membrane surface.
The erythrocyte band 3 has been sequenced from a variety of organisms including human, cow, rat, mouse, chicken, and trout.42,43 83-86 A comparison of these sequences reveals extended regions of sequence homology interconnected by regions of moderate divergence. When the regions of homology are aligned with the 3-dimensional map of cdb3, it is observed that they correspond with high fidelity to the secondary structural elements displayed in Figure1. This suggests that the general fold of cdb3 has been conserved among organisms examined to date, even though the erythrocytes of birds and fish are nucleated and stabilized by a classical actin- and tubulin-based cytoskeleton. Of further relevance, the sequences conserved in AE2 and AE3 (ie, other members of the anion exchanger gene family) also correspond to the above regions of secondary structure, which suggests a related fold for these more divergent proteins. In fact, the sequences of greatest divergence among all isoforms of band 3 constitute loops of different lengths connecting the well-defined segments of the protein secondary structure.
One of the more surprising features to emerge from the crystal structure was the overall shape of cdb3. As noted above, the pH 4.8 crystal form of the cdb3 dimer has dimensions of approximately 7.5 × 5.5 × 4.5 nm (75 × 55 × 45 Å). In contrast, hydrodynamic measurements of the solution form of the same dimer yield Stokes radii estimated at 4.65 nm (46.5 Å; pH 8)51 to 5.3 nm (53 Å; pH 7.4).52 Further, sedimentation measurements at pH 8 yield an estimate of P = 1.5 (approximately), which in turn predicts an axial ratio of 10:1 for an equivalent prolate ellipsoid of revolution.51 In contrast, the atomic coordinates of the interlocked dimer yield a calculatedP = 1.04 (approximate) and an axial ratio of 1.9:1.
Various experimental factors could obviously account for these discrepant results, but 3 explanations deserve further consideration. First, the hydrodynamic estimates could be flawed. Although reasonably similar Stokes radii have been reported by 2 independent labs,51,52 major assumptions were still required in these measurements, and these assumptions could have lead to distorted estimates. Second, the crystallized conformation of cdb3 could be nonphysiological. However, while minor differences between cdb3 in situ and the crystallized structure are not unlikely, the ability of the crystal structure to explain many previous functional and structural observations argue against this interpretation; eg, the strong preference of cdb3 to dimerize, the ability of protein 4.1 to simultaneously dock at both the N- and C-termini of cdb3, the presence of a bendable hinge between 2 independent domains in cdb3, the inability of kidney cdb3 to bind any of erythrocyte cdb3 peripheral protein ligands, the tendency of ankyrin to force band 3 tetramer formation, and the nature of the conserved sequences in cdb3. Still, one must recognize that cdb3 undergoes a reversible structural change as the pH is lowered, becoming increasingly more compact and thermally stable.44,46,47,52,53 So it is not unreasonable to expect some differences between the conformation described here and the predominant form at pH 7.4. Further studies guided by the crystal data will obviously be required to resolve this issue. Finally, neither the free N-termini nor free C-termini are seen in the crystal structure. Thus, if the unresolved N-terminal 54 and C-terminal 23 amino acids were to form highly extended structures, the axial ratio derived from the hydrodynamic experiments would be significantly greater than the dimensions measured in the crystal. The observations that the N-terminus of cdb3 binds both hemoglobin21 and aldolase45 in an extended conformation would favor this interpretation.
With the tertiary structure of the low pH conformation of cdb3 now established, it should be possible to construct a more predictive model of the architecture of the erythrocyte membrane. It is already apparent that the site of the ankyrin attachment to the membrane cannot be much more than approximately 6.0 nm (60 Å) from the lipid surface87 because the cross-sectional dimension of the cdb3 orthogonal to the membrane is only approximately 5.0 nm (50 Å), not 25.0 nm (250 Å) as previously supposed.44 The position of protein 4.1 binding is also more accurately defined because the LRRRY sequence resides in the dimerization arm near the interface between the subunits. Glycolytic enzymes, in contrast, may form a ring around each cdb3 oligomer as they associate with available radiating N-termini (Figure 5). It is even conceivable that an enclosed metabolite compartment, such as that envisioned by Hoffman,88 could assemble on the cdb3 oligomer if interactions among the glycolytic enzymes were correctly arranged. Finally, the crystal structure of cdb3 now provides a central framework on which to build a more elaborate membrane model as further peripheral protein structures are solved. In the absence of a technology that allows resolution of complex noncrystallizable structures, the assembly of an intact structure from its well-defined parts may constitute the best approach available.
Acknowledgments
Christopher L. Colbert is thanked for his assistance in the diffraction experiments. John E. Johnson (while at Purdue University, West Lafayette, IN) is thanked for his guidance in growing cdb3 crystals and searching for heavy atom derivatives. Stephen Hending is thanked for discussions of hydrodynamic measurements.
Supported by grant GM24417 from the National Institutes of Health (NIH), Bethesda, MD. Use of the Advanced Photon Source was supported by contract W-31-109-Eng-38 from the U.S. Department of Energy, Washington, DC. Use of BioCARS Sector 14 was supported by grant RR-07707 from the NIH, National Center for Research Resources, Bethesda, MD.
Submitted March 13, 2000; accepted June 23, 2000.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Philip S. Low, Department of Chemistry, 1393 Brown Bldg, Purdue University, West Lafayette, IN 47907-1393; e-mail:plow@purdue.edu., or Jeffrey T. Bolin, Department of Biological Sciences, 1392 Lilly Hall, Purdue University, West Lafayette, IN 47907-1392; e-mail: jtb@cc.purdue.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal