Abstract
In transplant patients, Kaposi sarcoma (KS)-associated herpesvirus or human herpesvirus-8 (HHV-8) infection is associated with the development of KS, primary effusion lymphoma and Castleman disease. Whether HHV-8 is either reactivated in the recipient or transmitted by the donor has been investigated so far only by serologic studies. Thus, we addressed the issue of HHV-8 transmission in the transplantation setting by molecular methods. We exploited the high level variability of the orf-K1 gene and the polymorphism of theorf-73 gene of the HHV-8 genome to assess the genetic relatedness of the HHV-8 strains identified in the posttransplant KS lesions that developed, simultaneously, 20 months after transplantation, in 2 recipients of twin kidneys from the same cadaver donor. The 100% identity of nucleotide sequence of the most variable viral region and the presence of the same, singleorf-73 type in both patients provides strong molecular evidence of organ-related transmission of HHV-8 in the setting of transplantation.
Introduction
Kaposi sarcoma–associated herpesvirus (KSHV) or human herpesvirus-8 (HHV-8) has been implicated in the pathogenesis of Kaposi sarcoma (KS), primary effusion lymphoma (PEL), and multicentric Castleman disease (CD) of plasma cell type.1,2 HHV-8 is not ubiquitous in the general population, with seroprevalence rates being very low in the UK and US and higher only in certain geographic areas of the world (the Mediterranean area and Africa) with a known high incidence of classic/endemic KS.1,2 HHV-8 is spread sexually, but nonsexual routes of transmission are likely to occur in HHV-8 endemic areas, where this herpesviral infection is acquired early in childhood.1 2
In the setting of transplantation, HHV-8 infection has been associated with an increased risk of developing posttransplant KS,3-10 and few cases of HHV-8–associated PEL, CD, and plasmacytic proliferations have been also described in recipients of solid organs.11-13 It has been recently proposed that posttransplant KS is primarily due to HHV-8 reactivation in endemic areas and to primary infection in nonendemic areas.6However, all previous reports have focused only on the use of serologic assays to investigate whether HHV-8 infection is either preexisting in the recipient or transmitted by the donor.3-10
In this report, we used molecular methods to address the issue of HHV-8 transmission in the unique clinical setting of 2 renal transplant patients, who had cutaneous KS develop, simultaneously, 20 months after receiving twin kidneys from the same cadaver donor. HHV-8 genome contains, at the left end, the orf-K1 gene, encoding a transmembrane protein that exhibits much more sequence variability than the rest of the viral genome.14,15 The variability in this gene among different isolates has so far been exploited to study the molecular epidemiology of HHV-8 obtained from individuals in different parts of the world.14,15 Moreover, molecular polymorphism has been recently found within the internal repeat domain of theorf-73 gene of the HHV-8 genome.16 The size of the orf-73 internal repeat domain, which is dependent of the variable number of repeats, is characteristic of each individual HHV-8 genotype or isolate, and has been used to develop a new genotyping technique.16 Thus, we exploited the high-level variability in the orf-K1 gene and the polymorphism of theorf-73 gene to assess the degree of genetic relatedness of the HHV-8 strains detected in the posttransplant KS lesions of the 2 renal transplant patients.
Study design
In August 1994, 2 Italian women, 46 and 45 years old, respectively, living in different cities, received renal grafts from the same cadaver donor, a 53-year-old Italian man. Both patients received immunosuppression with cyclosporin A (CsA) and methylprednisolone (MP), but, during the posttransplantation course, both were also treated with MP pulses for an episode of acute rejection. Azathioprine was also added to the immunosuppressive regimen in one case. Twenty months after transplantation, cutaneous KS of the lower extremities developed simultaneously in both patients. A detailed description of the clinical histories of the 2 transplant patients has been reported elsewhere.17 Reduction of immunosuppressive therapy has led to KS regression in both patients, which still persists.
Frozen biopsy specimens from the cutaneous KS lesions of both patients, as well as the Ficoll separated peripheral blood mononuclear cells (PBMCs) collected about 4 years after the initial diagnosis of KS, were available for molecular analysis. DNA was purified after proteinase digestion and phenol chloroform extraction. HHV-8 DNA was amplified on the same material by polymerase chain reaction (PCR) with primers specific for the orf-K1 gene, as we previously described.15 The PCR products were subjected to direct automated sequence analysis, and phylogenetic analysis of theK1 gene was performed as we previously reported.15 PCR amplification of the fragment of theorf-73 internal repeat domain was performed as described by Gao et al.16 To avoid false-positive results, all procedures were performed in strict adherence to the recommendations of Kwok and Higuchi.18 Negative controls consisting of all reagents, except sample DNA, were also present during the DNA extraction and equalled or exceeded the number of samples assayed.
Results and discussion
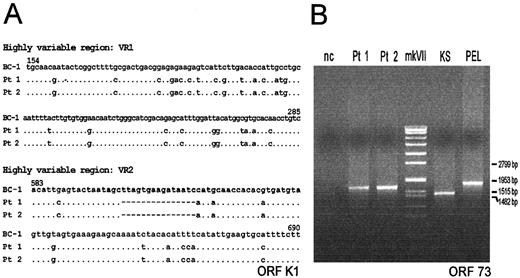
HHV-8 DNA was detected by PCR in the cutaneous posttransplant KS lesion from both renal recipients. Sequence analysis of the 2 highly variable regions of the orf-K1 gene from the 2 patients showed 100% identity (Figure 1A) and phylogenetic analysis showed that the infecting strain belonged to clade C, which is rather common in Italy.15 Theorf-K1 gene sequence results were also the same in the PBMCs collected from the 2 patients about 4 years after the initial diagnosis of KS. PCR of the orf-73 internal repeat domain showed a single band of the same size in both patients (Figure 1B). In the same assay, a single band of different size was detected in one classic KS and in one PEL specimen, indicating the occurrence of different isolates in these latter cases (Figure 1B).
Alignment of the nucleotide sequences and genotyping of the internal repeat domain detected in the KS lesions.
(A) Alignment of the nucleotide sequences of the 2 highly variable regions (VR1 and VR2) of the orf-K1 gene. The alignment includes a prototype K1 sequence (clade A) from the BC-1 PEL cell line (GeneBank accession number U75698), and the K1 sequences (clade C) detected in the KS lesions from each of the 2 renal recipients (Pt 1 and Pt 2), after transplantation. Dots indicate identity with the first sequence in the alignment. (B) Genotyping of the polymorphic internal repeat domain of orf-73 gene detected in the KS lesions from each of the 2 renal recipients (Pt 1 and Pt 2), after transplantation, in the cutaneous KS lesion from an elderly Italian patient with classic KS (KS), and in the pleural PEL specimen from an elderly HIV negative Italian patient (PEL). nc, negative control, represented by the DNA from HHV-8 negative Raji cell line; mk VII, molecular weight marker (Boehringer Mannheim, Germany).
Alignment of the nucleotide sequences and genotyping of the internal repeat domain detected in the KS lesions.
(A) Alignment of the nucleotide sequences of the 2 highly variable regions (VR1 and VR2) of the orf-K1 gene. The alignment includes a prototype K1 sequence (clade A) from the BC-1 PEL cell line (GeneBank accession number U75698), and the K1 sequences (clade C) detected in the KS lesions from each of the 2 renal recipients (Pt 1 and Pt 2), after transplantation. Dots indicate identity with the first sequence in the alignment. (B) Genotyping of the polymorphic internal repeat domain of orf-73 gene detected in the KS lesions from each of the 2 renal recipients (Pt 1 and Pt 2), after transplantation, in the cutaneous KS lesion from an elderly Italian patient with classic KS (KS), and in the pleural PEL specimen from an elderly HIV negative Italian patient (PEL). nc, negative control, represented by the DNA from HHV-8 negative Raji cell line; mk VII, molecular weight marker (Boehringer Mannheim, Germany).
Sera collected before and after transplantation from 28 recipients who had posttransplant KS develop, have so far been examined for anti–HHV-8 antibodies in 8 independent studies.3-10Twenty-three patients, most of whom originated from endemic areas, were infected with HHV-8 before the graft, suggesting that KS was mainly due to virus reactivation.3,5-10 In the remaining 5 patients who seroconverted after transplantation, transmission of HHV-8 from the organ donor to the recipient was suggested.3,4 However, for 4 of these patients, originating from nonendemic areas, the source of infection was not determined, as serum samples from their paired donors were not available.4 For one patient, originating from an endemic area (southern Italy), serum from the living-related donor was seropositive for HHV-8, providing the only clear available example of organ-related transmission of HHV-8, although the serum was tested more than 2 years after transplantation (Table1).3
Updated review of serologic studies for anti–HHV-8 antibodies in patients with posttransplant Kaposi sarcoma
| No. of patients with anti–HHV-8 Ab before transplantation . | No. of patients seroconverting to HHV-8 after transplantation . | Reference no. . |
|---|---|---|
| 10 | 1 | (3) |
| 0 | 2 | (4) |
| 4 | 0 | (5) |
| 1 | 2 | (6) |
| 5 | 0 | (7) |
| 1 | 0 | (8) |
| 1 | 0 | (9) |
| 1 | 0 | (10) |
| Total no. of patients | ||
| 23 | 5 |
| No. of patients with anti–HHV-8 Ab before transplantation . | No. of patients seroconverting to HHV-8 after transplantation . | Reference no. . |
|---|---|---|
| 10 | 1 | (3) |
| 0 | 2 | (4) |
| 4 | 0 | (5) |
| 1 | 2 | (6) |
| 5 | 0 | (7) |
| 1 | 0 | (8) |
| 1 | 0 | (9) |
| 1 | 0 | (10) |
| Total no. of patients | ||
| 23 | 5 |
HHV-8 indicates human herpesvirus-8; Ab, antibodies.
In our study, the finding of a 100% identity of nucleotide sequence of the most variable region of HHV-8 genome and the presence of the same, single orf-73 type in the posttransplant KS lesions of the 2 patients receiving twin kidneys from the same cadaver donor, provide strong molecular evidence of organ-related transmission of HHV-8. Although among populations with high HHV-8 seroprevalence, the development of posttransplant KS has been found to be mainly associated with HHV-8 reactivation, the occurrence of HHV-8 transmission from the donor should not be underestimated. Analysis of the genetic relatedness in the highly variable orf-K1 gene and the polymorphicorf-73 gene of HHV-8 isolates from the transplant recipients and their paired donors represents a useful tool to obtain a molecular tracing of HHV-8 transmission in this clinical setting.
M.L., P.B., and G.S. equally contributed to the study.
Supported by a grant from Associazione Italiana per la Ricerca sul Cancro (A.I.R.C.), Milan, Italy, and the European Concerted Action on HHV-8/KSHV (M.L.), a grant from M.U.R.S.T. (G.T.), and a grant from NHS Biomedical R&D Fund (RDO/22/09) (T.F.S.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Giuseppe Torelli, Department of Medical Sciences, Section of Hematology, Policlinico, Via del Pozzo 71, 41100, Modena, Italy; e-mail: gtorelli@unimo.it.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal