Abstract
The CCAAT/enhancer-binding protein (C/EBP) family consists of transcription factors essential for hematopoiesis. The defining feature of the C/EBPs is a highly conserved carboxy-terminal bZIP domain that is necessary and sufficient for dimerization and DNA binding, whereas their amino-terminal domains are unique. This study reports a novelc/ebp gene (c/ebp1) from zebrafish that encodes a protein homologous to mammalian C/EBPs within the bZIP domain, but with an amino terminus lacking homology to any C/EBP or to any known sequence. In zebrafish embryos, c/ebp1 expression was initially observed in cells within the yolk sac circulation valley at approximately the 16-to 18-somite stage, and at 24 hours postfertilization (hpf), also in circulating cells. Mostc/ebp1+cells also expressed a known early macrophage marker, leukocyte-specific plastin (l-plastin). Expression of both markers was lost in cloche, a mutant affecting hematopoiesis at the level of the hemangioblast. Expression of both markers was retained in m683 andspadetail, mutants affecting erythropoiesis, but not myelopoiesis. Further, c/ebp1 expression was lost in a mutant with defective myelopoiesis, but intact erythropoiesis. These data suggest that c/ebp1 is expressed exclusively in myeloid cells. In electrophoretic mobility shift assays, c/ebp1 was able to bind a C/EBP consensus DNA site. Further, a chimeric protein containing the amino-terminal domain of c/ebp1 fused to the DNA-binding domain of GAL4 induced a GAL4 reporter 4000-fold in NIH3T3 cells. These results suggest that c/ebp1 is a novel member of the C/EBP family that may function as a potent transcriptional activator in myeloid cells.
Introduction
Differentiation of hematopoietic cells is regulated by coordinate activation and repression of transcription factors.1 Multiple members of the CCAAT/enhancer-binding protein (C/EBP) family are essential for myeloid development.2 Six mammalian members have been identified to date and have been named C/EBPα, β, γ, δ, ε, and ζ.3,4 C/EBPs all have a highly conserved carboxy-terminal DNA-binding basic region, followed by a leucine zipper responsible for homodimerization and heterodimerization. C/EBPα was the first described leucine zipper protein and consists of 5 leucine repeats in a heptad periodicity.5 In addition to the C/EBP family, other proteins containing a leucine zipper include Myc, Fos, Jun, and the yeast protein GCN4. Together, the basic region and leucine zipper are termed the bZIP domain. The amino-terminal domains of each C/EBP are unique and define the transcriptional activation or repression characteristics of the proteins.
In vitro analyses and murine targeted deletion models have shown that C/EBPα, β, and ε all play essential roles in myelopoiesis.2,4 C/EBPα-deficient mice exhibit an early block in granulocytic maturation as well as severe hepatic and adipocyte abnormalities.6-8 C/EBPα has been shown to up-regulate genes encoding granulocyte-specific proteins such as granulocyte colony-stimulating factor (G-CSF) receptor, secondary granule proteins, and C/EBPε. Further, expression of C/EBPα in cells capable of granulocytic or monocytic differentiation induces neutrophilic differentiation while blocking the monocytic program.9 C/EBPβ is expressed in multiple tissues and acts as a regulator of acute-phase proteins and cytokines. A C/EBPβ null mouse model was shown to have defective macrophage activity, immunodeficiency, and a lymphoproliferative disorder.10 11
Among the C/EBP family members, C/EBPε is unique in demonstrating a very restricted expression pattern. It is expressed in myeloid and lymphoid cells only with the highest level of expression in promyelocytic and late myeloblastic cell lines.12-14C/EBPε−/− mice do not develop functional neutrophils and eosinophils and succumb to opportunistic infections or myelodysplasia with proliferation of atypical granulocytes in the bone marrow.15 Thus, C/EBPε appears to be involved in terminal differentiation of granulocytes, whereas C/EBPα blocks early granulocyte development.
The zebrafish, Danio rerio, offers a powerful model system in which to study hematopoiesis due to its external fertilization, transparent embryos, and rapid embryonic development.16Erythrocytes, myeloid cells, T lymphocytes, and thrombocytes have been identified in the zebrafish.17-21 Recently, macrophage development was shown to originate in the ventrolateral mesoderm anterior to the cardiac field.17 Macrophages were shown to develop and stream along the yolk sac to enter the circulation.
Twenty-six hematopoietic mutants have been identified to date.18,19 A number of bloodless mutants have been described and are characterized by few or no blood cells at the onset of circulation. The mutation in the bloodless mutant,cloche, has been shown to affect both hematopoiesis and vasculogenesis and is believed to occur at the level of the “hemangioblast”22-24; characterization of other bloodless mutants has shown that the mutations affect later steps along the erythroid pathway.25 However, data on mutants that specifically affect the myeloid lineage have not yet been published.
To facilitate analysis of myelopoiesis in D rerio, a zebrafish kidney complementary DNA (cDNA) library was screened forc/ebp cDNAs using a probe encoding the conserved bZIP region of human C/EBPε. A gene encoding a c/ebp family member was identified that has no known ortholog in any other organism and was namedc/ebp1. The c/ebp1 gene was mapped within the zebrafish genome and its expression was analyzed by RNA in situ hybridization both in wild-type and hematopoietic mutant embryos. The c/ebp1 protein was shown to bind DNA containing a canonical mammalian C/EBP binding site motif. Further, c/ebp1 contains a novel amino-terminal domain that, although apparently unrelated to any known activation domain, acted as a potent activator of transcription in a mammalian system.
Materials and methods
Library screening and sequence analysis of cDNAs
Random-primed and oligo-dT primed zebrafish adult kidney cDNA libraries in the phagemid pBKCMV24 were screened using a probe encoding the bZIP domain of human C/EBPε generated via polymerase chain reaction (PCR). The probe was amplified from a plasmid containing human C/EBPε (courtesy of K. G. Xanthopoulos)14 using the forward primer, 5′-AAGGGCAAGAAGGCAGTGAAC-3′ (nucleotides [nt] 583-603), and the reverse primer 5′-TCAGCTGCAACCCCCCACGCC-3′ (nt 846-826). The amplified fragment (264 bp) was labeled using 32P-dCTP with the Rediprime kit (Amersham-Pharmacia, Piscataway, NJ) according to the manufacturer's instructions. Positive clones were isolated and sequenced in one direction with a T7 primer (performed by ACGT, Northbrook, IL). Sequences were placed into a contig map using the Sequencing Project Manager program within DNAstar. A single clone encompassing the open reading frame was sequenced in both directions. The sequence has been submitted to GenBank and the accession number is AF306857.
Radiation hybrid mapping
The c/ebp1 gene was mapped within the zebrafish genome using a 94-clone radiation hybrid panel generated from a zebrafish primary fibroblast cell line, *AB.9, and a hamster fibroblast cell line, WG3H.26 The primers used to amplify a portion of c/ebp1 were 5′-GCATTAATACGAAGTTTCAGG-3′ (nt 364-383) and 5′-ATCCAGATGACCCAGCAGAG-3′ (nt 476-457). PCR results on the panel of 94 radiation hybrid DNAs and 2 control DNAs from the zebrafish and hamster parental lines were submitted to http://www.eb.tuebingen.mpg.de/abt.3/for analysis and placement on the zebrafish map.27
Zebrafish maintenance and breeding
Zebrafish were maintained and bred essentially as described28 under an approved animal use protocol of the National Institutes of Health. After breeding, embryos were maintained in egg water (0.006% Instant Ocean in distilled water) with 2 parts per million methylene blue to prevent fungal growth.
Whole-mount in situ hybridization
Embryos used for in situ hybridization were obtained from breedings of the wild-type EK strain (Ekkwill)29 and 3 hematopoietic mutants, cloche(clo39 ),22spadetail(sptb104),30 andm683.25 Embryos were staged as described.28 When growing embryos for harvest at more than 24 hours postfertilization (hpf), embryos were grown in 0.003% 1-phenyl-2-thiourea (Sigma, St Louis, MO) to prevent melanization. Embryos were dechorionated in 2 mg/mL pronase followed by extensive washing in 30% Danieau solution (58 mM NaCl, 0.67 mM KCl, 0.4 mM MgSO4, 0.6 mM Ca(NO3)2, and 4.5 mM HEPES, pH 7.5) and killed in 0.2% 3-amino benzoic acidethylester (Tricaine). Whole-mount in situ hybridization was performed essentially as described with the following modifications. Embryos less than 24 hpf were not treated with proteinase K. Embryos between 24 and 36 hpf were treated for 5 minutes in 10 μg/mL proteinase K and embryos 36 hpf to 2 days postfertilization (dpf) were treated for 10 to 20 minutes. Hybridization and washing were performed at 55°C for all probes. RNA antisense probes were synthesized according to the manufacturer's instructions with either UTP-digoxigenin (c/ebp1) or UTP-fluorescein (l-plastin) (Boehringer Mannheim, Indianapolis, IN). Probes labeled with digoxigenin were visualized with BM-purple (Boehringer Mannheim) and probes with fluorescein with fast red (Boehringer Mannheim). The c/ebp1 cDNA used for in situ hybridization started at nt +3 and included over 600 bp of 3′ untranslated sequence. The plasmid was digested with EcoRI and T7 RNA polymerase was used to synthesize the antisense probe. The plasmid containing l-plastin (kindly provided by B. Thisse)17 was linearized with NotI and transcribed with T7 RNA polymerase. Embryos were examined with an Olympus dissecting microscope or a Nikon Microphot-FXA compound microscope and photographed with a Spot (Diagnostic Instruments, Sterling Heights, MI) or Quantix (Photometrics, Tucson, AZ) CCD camera. In some cases, images focused in 2 different planes were merged using Adobe Photoshop to allow both yolk sac and tail regions to be visualized in the same image.
Electrophoretic mobility shift assays
For DNA-binding assays, a double-stranded probe containing an optimal C/EBP binding site was prepared by annealing a self-complementary oligonucleotide with a GATC overhang (shown in bold), 5′-GATCTGCAGATTGCGCAATCTGCA-3′12 and labeling using Klenow polymerase and 32P-dCTP. pBKCMV-c/ebp1 with a T3 promoter upstream ofc/ebp1, and pCMV-C/EBPε containing a T7 promoter upstream of human C/EBPε,31 were used to in vitro transcribe and translate proteins (TNT reticulocyte lysate system, Promega, Madison, WI) according to manufacturer's directions. Products from 40 ng of each plasmid were combined with 1 μg poly[dI-dC], 1 ng labeled probe (1 × 105 cpm), and varying amounts of unlabeled probe as indicated. A probe with a mutated C/EBP binding site,125′-GATCTGCAGAGACTAGTCTCTGCA-3′ (the GATC overhang is shown in bold and changes from the wild-type probe are underlined) was used as a negative control. DNA-binding assay samples were separated on a 4% polyacrylamide/0.25 × TBE (tris borate EDTA) gel. 35S-methionine-labeled proteins were analyzed on a 4% to 12% bis-tris gradient gel (Invitrogen, Carlsbad, CA), fixed, treated with Amplify (Amersham-Pharmacia), and dried followed by autoradiography.
Transfection and activation assays
The cDNAs encoding putative activation domains were placed in a pcDNA3-based vector containing the region encoding the DNA-binding domain (DBD) of GAL4 (amino acids [aa] 1-147) (plasmid kindly provided by N. Perkins, University of Dundee). A PCR fragment encoding the region amino terminal of the bZIP domain of c/ebp1 was ligated downstream of the GAL4 DBD using EcoRI and BamHI. The primers used to amplify the region of c/ebp1 were 5′-CCGGAATTCATGTCGGTGTCTGACAACATC-3′ (nt 1-21,EcoRI site in bold letters) and 5′-CGCGGATCCGCGCACAGGCGGAGCGCAGACG-3′ (nt 262-241,BamHI site in bold letters). PCR fragments encoding the first 194 aa and the first 102 aa of human C/EBPε were amplified from a human C/EBPε plasmid and placed downstream of the GAL4 DBD with EcoRI and BamHI. Both PCR products were obtained using the forward primer: 5′-CCGGAATTCATGTCCCACGGGACCTACTCA-3′ (nt 1-21,EcoRI site in bold letters). The product encoding aa 1-194 was amplified with the reverse primer: 5′-CGCGGATCCCTTGTGTAAGGGGCCAGCCGG-3′ (nt 586-565,BamHI site in bold letters) and the product encoding aa 1-102 was amplified with the reverse primer: 5′-CGCGGATCCCAGCGCCTTCCTGTCTGGGCC-3′ (nt 306-286,BamHI site in bold letters). The amplified regions and the restriction site junctions were sequenced in both directions for all constructs. NIH3T3 cells were grown in Dulbecco modified Eagle medium containing 10% fetal calf serum (Life Technologies, Rockville, MD) at 37°C under 5% CO2. Approximately 5 × 105cells were cotransfected with 125 to 250 ng of a cytomegalovirus promoter-driven β-galactosidase expression vector and the indicated quantities of plasmids in 6-well dishes using Superfect (Qiagen, Valencia, CA) according to the manufacturer's instructions. Transfected cells were harvested 24 to 48 hours after transfection for luciferase and β-galactosidase assays. Cells were lysed by the addition of 500 μL reporter lysis buffer (Promega) into each well. Cells were scraped from each well and lysates were incubated at room temperature with gentle shaking for 10 minutes followed by centrifugation at 14 000 rpm for 5 minutes to pellet debris. Twenty microliters of lysate was used to measure luminescence from luciferase or from a chemiluminescent β-galactosidase assay in a Tropix TR717 microplate reader (Applied Biosystems, Foster City, CA) following the manufacturer's protocol (Promega). Luminescence units were normalized for transfection efficiency using β-galactosidase activity.
Results
Isolation and mapping of c/ebp1 in zebrafish
A zebrafish kidney cDNA library was screened with a 264-bp PCR product encoding the conserved bZIP domain of human C/EBPε to identify C/EBP family members from zebrafish. One c/ebpcDNA, represented by 25% of the clones, was not orthologous to any mammalian C/EBP genes and was namedc/ebp1.
The start of the open reading frame for c/ebp1 was determined based on the rules of Kozak.32c/ebp1 encodes a 170-aa protein including a bZIP domain containing 4 leucines in a heptad periodicity. The bZIP domain of c/ebp1 was between 26% and 50% identical and 66% and 79% conserved when compared to the bZIP domains of the human C/EBP family members. However, the amino-terminal region of c/ebp1 showed no significant homology with any of the human C/EBPs (Figure1A). Further, BLAST analysis did not reveal homology between the amino-terminal region of c/ebp1 and any open reading frame in GenBank. To clarify the evolutionary relationship of c/ebp1 to other C/EBP family members, a phylogenetic tree was generated using an alignment of the conserved bZIP region of c/ebp1 with those of all the human C/EBP family members and the recently cloned zebrafish c/ebpa, c/ebpb,c/ebpd, and c/ebpg genes (S.E.L. et al, manuscript in preparation) (Figure 1B). In this analysis, c/ebp1 was excluded from the branches containing the human C/EBP family members and thus does not appear to have a human ortholog.
Alignment and phylogenetic tree of c/ebp1with human C/EBP family members.
(A) Zebrafish (Dr) c/ebp1 was aligned with known human (Hs) C/EBP sequences using CLUSTAL within the GCG program (http://molbio.info.nih.gov/molbio/gcglite/). Black shading shows identities and gray shading shows conserved amino acid substitutions seen in at least 3 of 7 sequences. The leucines that make up the leucine zipper in c/ebp1 are marked with asterisks and the additional leucine present in the leucine zippers of C/EBPα, β, and δ is marked with a filled circle. The bZIP domain5 of c/ebp1 is underlined and the region encoded by the probe used to screen the zebrafish cDNA library is delineated with arrows. The GenBank accession number is AF306857. (B) A phylogenetic tree was generated from an alignment of the bZIP regions of the zebrafish (Dr) c/ebps and the human (Hs) C/EBPs using CLUSTAL within the DNAstar Megalign program.
Alignment and phylogenetic tree of c/ebp1with human C/EBP family members.
(A) Zebrafish (Dr) c/ebp1 was aligned with known human (Hs) C/EBP sequences using CLUSTAL within the GCG program (http://molbio.info.nih.gov/molbio/gcglite/). Black shading shows identities and gray shading shows conserved amino acid substitutions seen in at least 3 of 7 sequences. The leucines that make up the leucine zipper in c/ebp1 are marked with asterisks and the additional leucine present in the leucine zippers of C/EBPα, β, and δ is marked with a filled circle. The bZIP domain5 of c/ebp1 is underlined and the region encoded by the probe used to screen the zebrafish cDNA library is delineated with arrows. The GenBank accession number is AF306857. (B) A phylogenetic tree was generated from an alignment of the bZIP regions of the zebrafish (Dr) c/ebps and the human (Hs) C/EBPs using CLUSTAL within the DNAstar Megalign program.
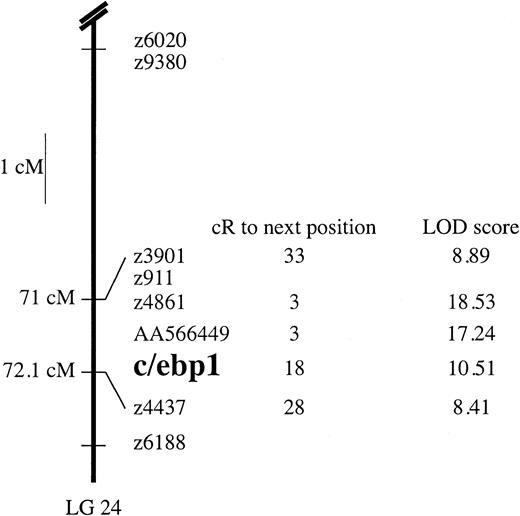
The c/ebp1 gene was mapped to the zebrafish genome to aid future mutant screening analyses. PCR primers within the coding sequence of c/ebp1 were used to type the Goodfellow radiation hybrid panel.26 The PCR primers used did not amplify other zebrafish c/ebps or sequences in hamster genomic DNA (data not shown). The c/ebp1 gene was mapped to linkage group 24 (LG24) between 71 and 72.1 cM from the top of LG24 with a LOD score of 10.51 (Figure 2).
Radiation hybrid mapping of c/ebp1.
The c/ebp1 gene was mapped using PCR primers to amplifyc/ebp1 from a 94-clone zebrafish-hamster radiation hybrid panel. The placement of c/ebp1 relative to other zebrafish markers is shown with the mapped position in centimorgans (cM) from the top of the linkage group, position in centirays (cR), and the LOD scores as shown.
Radiation hybrid mapping of c/ebp1.
The c/ebp1 gene was mapped using PCR primers to amplifyc/ebp1 from a 94-clone zebrafish-hamster radiation hybrid panel. The placement of c/ebp1 relative to other zebrafish markers is shown with the mapped position in centimorgans (cM) from the top of the linkage group, position in centirays (cR), and the LOD scores as shown.
Expression of c/ebp1 in normal embryos
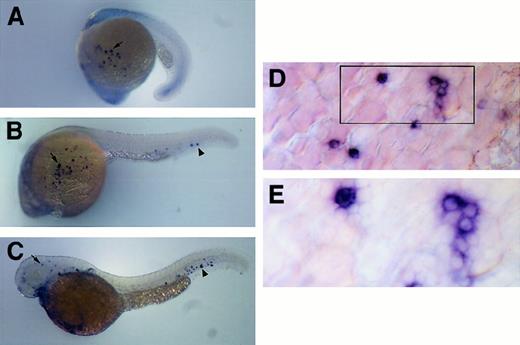
Expression of c/ebp1 throughout embryonic development was analyzed by RNA in situ hybridizations using digoxigenin-labeled antisense RNA. At approximately the 16- to 18-somite (17-18 hpf) stage of development, expression was first detected in a few cells overlying the yolk sac in the embryo (data not shown). By the 20- to 21-somite stage (19.5 hpf), approximately 25 cells located between the epithelial layer and the yolk sac mass were stained (Figure3A). At approximately 24 hpf, 25 to 50 stained cells could be seen overlying the yolk sac (Figure 3B,D) and stained cells could be seen within the caudal portion of the axial vein. The exact locations of the c/ebp1+ cells over yolk sac and in the axial vein are not fixed, because they are cells in circulation. The stained cells appeared small and round, differing from the yolk syncytial layer underneath, consistent with circulating hematopoietic cells (Figure 3D). The majority of cells appeared to have a scant amount of cytoplasm with a large, circular nucleus (Figure 3E). At 2 dpf, stained cells were still visible within the circulation and the surrounding mesenchyme around the caudal part of the axial vein (Figure 3C). Further, cells were seen in the mesenchyme of the head. There is minimal staining at 3.5 days, with only a few circulating cells visible (data not shown).
Expression pattern of c/ebp1 in zebrafish embryos.
RNA in situ hybridization was performed with a digoxigenin-labeledc/ebp1 RNA antisense probe. (A) Lateral view of 20-somite embryo with arrow indicating c/ebp1-expressing cells overlying yolk sac. (B) Lateral view of 24-hpf embryo withc/ebp1-expressing cells on the yolk sac (arrow) and in the axial vein (arrowhead). (C) Lateral view of 2-dpf embryo withc/ebp1-expressing cells in the axial vein and surrounding mesenchyme (arrowhead) and in mesenchyme of the head (arrow). (D) Yolk sac at 20 × magnification in a 24-hpf embryo using a Microphot AX compound microscope. Boxed area is shown in higher magnification in panel E. (E) Yolk sac at 40 × magnification in a 24-hpf embryo using a Microphot AX compound microscope.
Expression pattern of c/ebp1 in zebrafish embryos.
RNA in situ hybridization was performed with a digoxigenin-labeledc/ebp1 RNA antisense probe. (A) Lateral view of 20-somite embryo with arrow indicating c/ebp1-expressing cells overlying yolk sac. (B) Lateral view of 24-hpf embryo withc/ebp1-expressing cells on the yolk sac (arrow) and in the axial vein (arrowhead). (C) Lateral view of 2-dpf embryo withc/ebp1-expressing cells in the axial vein and surrounding mesenchyme (arrowhead) and in mesenchyme of the head (arrow). (D) Yolk sac at 20 × magnification in a 24-hpf embryo using a Microphot AX compound microscope. Boxed area is shown in higher magnification in panel E. (E) Yolk sac at 40 × magnification in a 24-hpf embryo using a Microphot AX compound microscope.
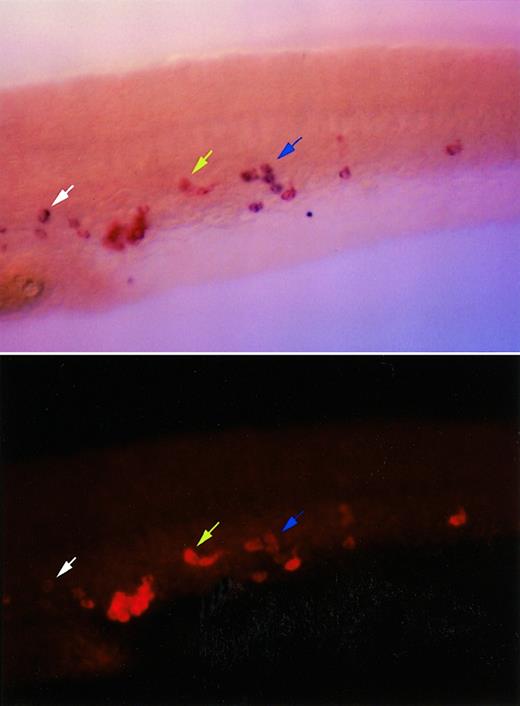
The expression pattern appeared similar to that seen with a known myeloid marker, l-plastin, which is expressed in circulating cells along the yolk sac initially at the 16- to 18-somite stage (17-18 hpf) (data not shown) and subsequently in the axial vein and the head.17 It is difficult to compare expression patterns between 2 embryos hybridized separately with l-plastin andc/ebp1 probes, due to the fact that cells expressing either of these 2 genes are in circulation, and therefore, likely to change their locations from embryo to embryo. However, when embryos at 24 hpf were double-labeled with the c/ebp1 and l-plastinprobes, the majority of cells visualized expressed bothc/ebp1 and l-plastin (Figure4). Individual cells appeared to express different ratios of l-plastin and c/ebp1, as shown by arrows in Figure 4. These data suggest that c/ebp1is expressed specifically in myeloid cells from the 16- to 18-somite stage through 2 dpf stages of development.
Coexpression pattern of c/ebp1 andl-plastin in zebrafish embryos.
RNA in situ hybridization was performed using a digoxigenin-labeled c/ebp1 RNA antisense probe developed with BM purple, and a fluorescein-labeled l-plastin RNA antisense probe developed with fast red. Lateral views of a tail region immediately caudal to the yolk sac extension oriented with the head to the left are shown. In the upper panel a bright field view of an in situ hybridized 24-hpf embryo shows l-plastin staining in red and c/ebp1 staining in purple. The lower panel shows the same view with l-plastin staining visualized with a rhodamine filter under fluorescence, photographed with a Quantix CCD camera, and pictured with red pseudocoloring using IP lab software. The blue arrow shows cells expressing both c/ebp1 andl-plastin. The yellow arrow indicates a cell with undetectable expression of c/ebp1, but strong expression ofl-plastin. The white arrow shows a cell with high expression of c/ebp1, but minimal l-plastinexpression.
Coexpression pattern of c/ebp1 andl-plastin in zebrafish embryos.
RNA in situ hybridization was performed using a digoxigenin-labeled c/ebp1 RNA antisense probe developed with BM purple, and a fluorescein-labeled l-plastin RNA antisense probe developed with fast red. Lateral views of a tail region immediately caudal to the yolk sac extension oriented with the head to the left are shown. In the upper panel a bright field view of an in situ hybridized 24-hpf embryo shows l-plastin staining in red and c/ebp1 staining in purple. The lower panel shows the same view with l-plastin staining visualized with a rhodamine filter under fluorescence, photographed with a Quantix CCD camera, and pictured with red pseudocoloring using IP lab software. The blue arrow shows cells expressing both c/ebp1 andl-plastin. The yellow arrow indicates a cell with undetectable expression of c/ebp1, but strong expression ofl-plastin. The white arrow shows a cell with high expression of c/ebp1, but minimal l-plastinexpression.
Expression of c/ebp1 in hematopoietic mutant embryos
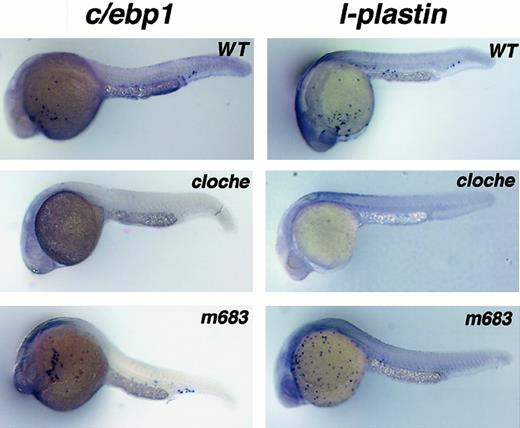
To determine if c/ebp1 gene expression is altered in hematopoietic mutants, RNA in situ hybridization was performed with mutant embryos. The bloodless mutant, cloche, has been shown to be affected at a very early stage in both hematopoietic and vascular development.22 The mutation occurs upstream ofscl, a gene encoding a transcription factor required for hematopoietic stem cell differentiation.24 Expression ofc/ebp1 was absent in cloche embryos as wasl-plastin expression (Figure5).
Analysis of c/ebp1 and l-plastinexpression in hematopoietic mutant embryos.
RNA in situ hybridization with c/ebp1 andl-plastin RNA antisense probes using wild-type,cloche, and m683 embryos at 24 hpf. Stained cells are seen over the yolk sac in wild-type and m683 embryos. Individual sampled embryos have variable staining patterns with a single probe (either c/ebp1 or l-plastin) because the stained cells were freely flowing over the yolk sac and in the circulation before sacrifice and fixation of the embryos. Lack of staining using c/ebp1 or l-plastin probes was observed in 25% of the cloche embryos from at least 3 independent heterozygote in-crosses.
Analysis of c/ebp1 and l-plastinexpression in hematopoietic mutant embryos.
RNA in situ hybridization with c/ebp1 andl-plastin RNA antisense probes using wild-type,cloche, and m683 embryos at 24 hpf. Stained cells are seen over the yolk sac in wild-type and m683 embryos. Individual sampled embryos have variable staining patterns with a single probe (either c/ebp1 or l-plastin) because the stained cells were freely flowing over the yolk sac and in the circulation before sacrifice and fixation of the embryos. Lack of staining using c/ebp1 or l-plastin probes was observed in 25% of the cloche embryos from at least 3 independent heterozygote in-crosses.
In a bloodless mutant, m683, scl expression was intact, but gata-1 expression was absent, consistent with a defect beyond the stem cell level.25 In this mutant, expression of both c/ebp1 and l-plastin was normal, suggesting a defect in the erythroid pathway with normal myeloid development. Therefore, c/ebp1 expression parallelsl-plastin expression in hematopoietic mutants, consistent with a common lineage for the cells marked by c/ebp1 andl-plastin. Another mutant, spadetail(sptb104), which affects somitic mesoderm specification,30,33 has defects in erythropoiesis with normal vascular development34 and normal expression of pu.1, another early myeloid marker.35 Thec/ebp1 gene was expressed later than pu.1 and was maintained in the spadetail mutant (sptb104) (data not shown), again consistent with c/ebp1 as a marker of the myeloid lineage.
Finally, a mutant was recently isolated fromN-nitroso-N-ethylurea (ENU) mutagenesis screening that has lost expression of l-plastin (S.E.L. et al, results to be described elsewhere). In this mutant, c/ebp1expression was also lost while expression of the stem cell marker,cbfb,25 and the erythroid marker,gata1, was maintained. Characterization of this mutant suggests a specific defect in the myeloid pathway with intact hematopoietic progenitor and erythroid pathways. Therefore, the loss ofc/ebp1 expression in this mutant points strongly toward expression of c/ebp1 specifically in cells of myeloid lineage.
Analysis of c/ebp1 as a transcriptional activator
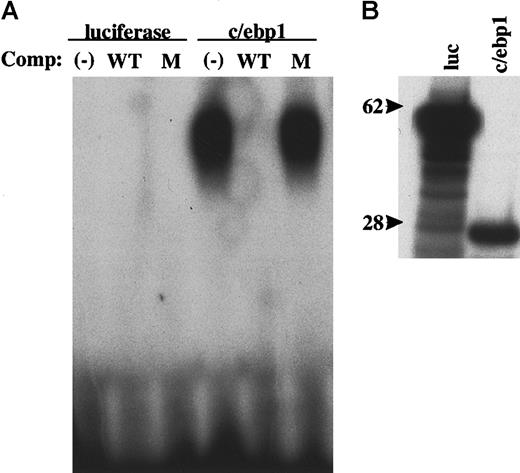
Sequence comparison between c/ebp1 and mammalian C/EBP family members revealed significant homology within the bZIP domain, but no homology at the amino terminus. Consistent with the high homology within the bZIP domain, full-length c/ebp1 was able to bind a palindromic C/EBP binding site by an electrophoretic mobility shift assay (Figure 6). This binding was specific because it was competed away with addition of an excess amount of unlabeled self DNA, but not with a mutated site probe. In the same experiment, human C/EBPε also bound the C/EBP site specifically (data not shown).
DNA-binding ability of c/ebp1.
Electrophoretic mobility shift assay was performed using in vitro transcribed and translated proteins and a 32P-labeled C/EBP optimal binding site probe. (A) Unlabeled in vitro translation control protein (luciferase) or c/ebp1 was incubated with a32P-labeled C/EBP site probe and, where indicated, with 50-fold excess of unlabeled competitor self probe (WT) or mutant probe (M) and samples were then separated on a 4% TBE gel. The gel was dried and autoradiography was performed. (B) 35S-labeled in vitro translation control protein (luc) and c/ebp1 were separated on a 4% to 12% bis-tris gradient gel. The processed gel was then exposed to film for autoradiography. Size markers of 62 kd and 28 kd are indicated.
DNA-binding ability of c/ebp1.
Electrophoretic mobility shift assay was performed using in vitro transcribed and translated proteins and a 32P-labeled C/EBP optimal binding site probe. (A) Unlabeled in vitro translation control protein (luciferase) or c/ebp1 was incubated with a32P-labeled C/EBP site probe and, where indicated, with 50-fold excess of unlabeled competitor self probe (WT) or mutant probe (M) and samples were then separated on a 4% TBE gel. The gel was dried and autoradiography was performed. (B) 35S-labeled in vitro translation control protein (luc) and c/ebp1 were separated on a 4% to 12% bis-tris gradient gel. The processed gel was then exposed to film for autoradiography. Size markers of 62 kd and 28 kd are indicated.
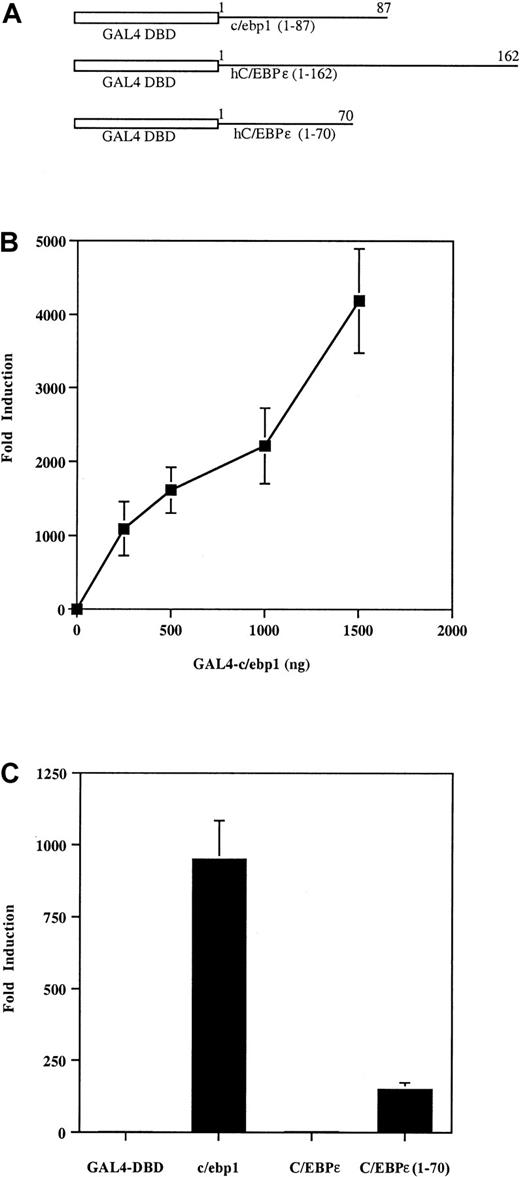
To assess whether the amino terminus of c/ebp1 could function as a transcriptional activator, a chimera (GAL4-c/ebp1 [1-87]) containing the DBD of GAL4 fused to the region of c/ebp1 amino terminal to the bZIP domain (aa 1-87) was used to activate a GAL4 DNA recognition site–driven luciferase reporter gene (GAL4-luc) (Figure7A,B). The GAL4-c/ebp1 (1-87) expression construct induced the GAL4-luc reporter up to 4000-fold in NIH3T3 cells relative to induction of the GAL4-luc reporter by the GAL4 DBD without an activation domain (Figure 7B). A dose response was seen when increasing amounts of GAL4-c/ebp1 (1-87) were used to induce the GAL4-luc reporter (Figure 7B). To compare the transcriptional activation activity of c/ebp1 to human C/EBPε, the amino terminus of human C/EBPε was fused to the GAL4-DBD and tested for transcriptional activation ability. The chimera including the GAL4-DBD and the amino-terminus of human C/EBPε resulted in only a 2-fold induction compared to a 900-fold induction of GAL4-luc by the GAL4-c/ebp1 (1-87) protein under the same conditions (Figure 7C). The human C/EBPε protein has been shown to contain a repression region between aa 115 and 162, whereas its activation domain resides in aa 1-70.36 Therefore, a chimera containing the GAL4 DBD and the first 70 aa of C/EBPε was also compared to c/ebp1. GAL4-C/EBPε (1-70) activated approximately 170-fold, suggesting that the amino-terminus of c/ebp1 is a more potent activator than the activation domain of C/EBPε. Taken together, these data indicate that c/ebp1 may act as a potent transcriptional activator of genes with a C/EBP DNA binding site motif.
Transcriptional activation activity of the amino terminus of c/ebp1.
(A) The GAL4 DNA-binding domain (GAL4 DBD) from yeast was fused to the cDNAs encoding regions of c/ebp1 or human C/EBPε, as indicated. (B) Transfection with increasing amounts ofGAL4-c/ebp1 (1-87) as indicated. (C) Transfection with 500 ngGAL4-c/ebp1 (1-87): c/ebp1; human C/EBPε (1-162): C/EBPε; or human C/EBPε (1-70): C/EBPε (1-70). NIH3T3 cells were transfected with 1 μg GAL4-luc reporter and various expression constructs. Transfections were normalized by cotransfection with a β-galactosidase expression vector and samples were all transfected in duplicate. The experiments shown in panels B and C are representative of 3 independent transfections. The y-axes represent fold inductions by the expression constructs relative to induction by a GAL4 DBD vector without an activation domain.
Transcriptional activation activity of the amino terminus of c/ebp1.
(A) The GAL4 DNA-binding domain (GAL4 DBD) from yeast was fused to the cDNAs encoding regions of c/ebp1 or human C/EBPε, as indicated. (B) Transfection with increasing amounts ofGAL4-c/ebp1 (1-87) as indicated. (C) Transfection with 500 ngGAL4-c/ebp1 (1-87): c/ebp1; human C/EBPε (1-162): C/EBPε; or human C/EBPε (1-70): C/EBPε (1-70). NIH3T3 cells were transfected with 1 μg GAL4-luc reporter and various expression constructs. Transfections were normalized by cotransfection with a β-galactosidase expression vector and samples were all transfected in duplicate. The experiments shown in panels B and C are representative of 3 independent transfections. The y-axes represent fold inductions by the expression constructs relative to induction by a GAL4 DBD vector without an activation domain.
Discussion
In this report, we have identified a novel myeloid-restricted member of the C/EBP family in zebrafish containing a highly conserved bZIP domain and a unique amino-terminal transactivation domain. The existence of 2 copies of many mammalian genes in zebrafish has led to the belief that an additional genome-wide duplication occurred during evolution of teleost fish compared to other vertebrates.37For example, each HOX-bearing chromosome in humans has 2 paralogous chromosomes in zebrafish.38 However, c/ebp1 is not likely to be the result of this tetraploidization event or a tandem duplication based on its sequence divergence from the other c/ebp family members. No c/ebp1 ortholog has been found in the nearly completed human genome, and therefore, the mammalian ortholog ofc/ebp1 may have been lost in humans while being maintained in the zebrafish.
The myeloid specificity of c/ebp1 expression in zebrafish embryos is supported by the following observations: (1) c/ebp1is only expressed in a fraction of circulating cells over the yolk sac and the axial vein; (2) most of the c/ebp1+cells also express the myeloid marker, l-plastin, another myeloid marker; (3) its expression is abolished in cloche, a mutant affecting hematopoiesis at the level of stem cells; and (4) its expression is retained in mutants affecting specifically the erythroid lineage, whereas it is lost in a mutant affecting specifically the myeloid lineage. It is possible that this gene is also expressed in lymphoid and megakaryocytic lineages, although the location and the timing of the expression make such possibilities unlikely.20,39 40
Most of the cells expressing c/ebp1 also express the macrophage marker, l-plastin. However, there are cells that express varying ratios of c/ebp1 and l-plastin. The cells with these different expression patterns may represent myeloid cells at slightly different stages of development or may represent separate subsets of myeloid cells. Further studies with additional hematopoietic markers, as they become available, will help to elucidate the identity of the cells observed in this analysis.
In our transcriptional activation assay, the amino-terminal 87 aa of c/ebp1 act as a strong transcriptional activation domain, far more potent than that of human C/EBPε.36 In mammals, C/EBPα, β, δ, and ε contain amino-terminal activation domains and act as transcriptional activators. In contrast, C/EBPγ and ζ lack these domains and usually act as trans-dominant repressors.4,41 Deletion and mutation analysis of C/EBPα has shown that it contains multiple activation regions that act cooperatively.42 One of these regions is proline rich, but the prolines do not appear to be required for activity.43In C/EBPβ an acidic, hydrophobic region has been demonstrated to have transactivation ability.44 Many transactivation domains are acidic, proline rich, glutamine rich, or in some cases, serine/threonine rich. The activation domain of c/ebp1 contains 26% serines/threonines, but does not appear to be proline or glutamine rich. Further, although the activation domain carries a net charge of −5, the region is not significantly acidic in comparison to described activation domains such as VP16 with a charge of −18. Much is still not understood about the sequence and structural requirements of transactivation domains, and activation domains defined by deletion and mutation analysis do not always fall into the classic categories. Further studies on the amino-terminal region of c/ebp1 will be required to assess the structure of the c/ebp1 activation domain and the elements of the region (aa 1-87) that are essential for transactivation.
c/ebp1 binds a consensus C/EBP site with high affinity and its amino terminus acts as a potent transcriptional activation domain in a mammalian system. Therefore, c/ebp1 likely acts as a transcriptional activator in vivo. Like the mammalian C/EBPs, c/ebp1 may heterodimerize with other c/ebps, such as c/ebpα and c/ebpβ, or other bZIP proteins to act on different DNA targets. Further, as for other c/ebps, additional cofactors such as c-myb might be required for optimal transactivation.45 46 Only a few myeloid-specific genes have been isolated in the zebrafish to date, and none of their promoters have been characterized, but it is likely that many will contain c/ebp sites. Differential affinity of these sites for the c/ebps and their partners would be expected to determine gene expression. However, analysis of the c/ebp1 target genes will have to await further characterization of zebrafish promoters.
The myeloid-specific expression pattern of c/ebp1 is similar to the expression of mammalian C/EBPε, which is seen primarily in myeloid and lymphoid cells. To date, an ortholog of mammalian C/EBPε has not been discovered in zebrafish (S.E.L. et al, unpublished results). However, c/ebp1 may perform some or all of the functions of the mammalian C/EBPε. Alternatively, if a zebrafish ortholog to C/EBPε does exist, c/ebp1 and the zebrafish C/EBPε ortholog may share the functions performed by the mammalian C/EBPε. Given the myeloid expression pattern ofc/ebp1, it may play a role in cell-type specification or maturation of myeloid cells.
In this study, we have identified a novel member of the c/ebp family with myeloid-specific expression. This factor has a previously undescribed amino-terminal domain with potent transcriptional activation properties. Our findings suggest that c/ebp1 may play a role in myelopoiesis in the zebrafish. In the near future, the use ofc/ebp1 as a marker in directed mutagenesis screens should lead to the identification and characterization of myeloid mutants and thus to the identification of genes crucial to myeloid development.
We thank Maria Anderson and the physical mapping core for radiation hybrid mapping; Brant Weinstein, P. J. Bennett, and Amy Chin for hematopoietic mutant embryos; Bernard Thisse, Neil Perkins, and Julie Lekstrom-Himes for constructs; and Lin Lei, Trevor Blake, and the laboratories of Brant Weinstein and Ajay Chitnis for helpful discussions and technical assistance.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Pu Paul Liu, NHGRI, NIH, 49 Convent Dr, Rm 3A18, Bethesda, MD 20892; e-mail: pliu@nhgri.nih.gov.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal