Abstract
Monocyte differentiation induced by 1,25-dihydroxyvitamin D3 (1,25(OH)2D3) is interrupted during the course of acute promyelocytic leukemia (APL). One form of APL is associated with the translocation t(11;17), which joins the promyelocytic leukemia zinc finger (PLZF) and retinoic acid receptor α (RARα) genes. Because PLZF is coexpressed in the myeloid lineage with the vitamin D3 receptor (VDR), the interplay between PLZF and VDR was examined. It was found that PLZF interacts directly with VDR. This occurred at least partly through contacts in the DNA-binding domain of VDR and the broad complex, tram-trak, bric-a-brac/pox virus zinc finger (BTB/POZ) domain of PLZF. Moreover, PLZF altered the mobility of VDR derived from nuclear extracts when bound to its cognate binding site, forming a slowly migrating DNA-protein complex. Overexpression of PLZF in a monocytic cell line abrogated 1,25(OH)2D3 activation from both a minimal VDR responsive reporter and the promoter of p21WAF1/CIP1, a target gene of VDR. Deletion of the BTB/POZ domain significantly relieved PLZF-mediated repression of 1,25(OH)2D3-dependent activation. In addition, stable, inducible expression of PLZF in U937 cells inhibited the ability of 1,25(OH)2D3 to induce surface expression of the monocytic marker CD14 and morphologic changes associated with differentiation. These results suggest that PLZF may play an important role in regulating the process by which 1,25(OH)2D3 induces monocytic differentiation in hematopoietic cells.
Introduction
Steroid and vitamin hormones play an integral part in the maturation and differentiation of hematopoietic cells. Members of the steroid and nuclear receptor superfamily, including the retinoic acid receptor (RAR), have an important role in this process, presumably through the ligand-dependent activation of genes that initiate or facilitate differentiation.1 The hormone 1,25-dihydroxyvitamin D3(1,25(OH)2D3), a metabolite of vitamin D, can induce differentiation of hematopoietic cell lines such as HL-60 and U937 along a macrophage-monocyte lineage, including a decrease or cessation in their proliferation.2-6 The effects of 1,25(OH)2D3 on differentiation are mediated through its transcriptional modulation of key target genes by heterodimers consisting of the vitamin D3 receptor (VDR) and the retinoid X receptor.7-10 During differentiation of hematopoietic cells, 1,25(OH)2D3 strongly induces expression of macrophage-monocyte markers, such as CD14, the lipopolysaccharide receptor, and the family of β2leukocyte integrins including CD11a, CD11b, and CD11c.11 12
In a search for target genes that might mediate differentiation of myelomonocytic cells by 1,25(OH)2D3, our laboratory previously identified the cell-cycle inhibitor p21WAF1/CIP1 and the homeobox geneHoxA10 as direct transcriptional targets of VDR.7,9 Furthermore, exogenous expression of these genes in U937 cells was sufficient to induce differentiation.8Ligand-dependent activation of such target genes is thought to occur through recruitment of coactivator complexes, including p160–CREB-binding protein (CBP) and VDR interacting protein (DRIP).13
Repression of VDR-dependent gene activation is not as well understood. Clearly, proper control of ligand-dependent expression of genes that mediate differentiation and cell-cycle arrest is an essential component of the function of normal, noncancerous cells. Aberrations in hormonal control of the differentiation process in hematopoietic cells occur in such cancers as acute promyelocytic leukemia (APL).
In APL, the M3 subtype of acute myelogenous leukemia, differentiation of cells is blocked during an intermediate promyelocyte stage of myeloid differentiation.14-18 These malignant promyelocytes can be induced to differentiate into mature polymorphonuclear leukocytes by providing pharmacologic doses of all-trans retinoic acid (ATRA). In contrast, these cancers are resistant to the effects of 1,25(OH)2D3. APL is thought to be caused by one of several reciprocal translocations involving RARα.19 One of these is t(11;17), which joins the promyelocytic leukemia zinc finger (PLZF) gene to RARα to create the PLZF-RARα fusion protein. PLZF–RAR-α expression is sufficient to block differentiation by 1,25(OH)2D3 or ATRA in both U937 and HL-60 cells.20
Several groups21-24 characterized the molecular mechanism of PLZF-RARα resistance to ATRA. However, the mechanism by which VDR-mediated effects are interrupted has not been described. Thus, we investigated the function of the untranslocated PLZF protein with respect to 1,25(OH)2D3-dependent transcription and differentiation of the histiocytic lymphoma cell line U937 to gain a more thorough understanding of the pathogenesis of t(11;17)-derived APL.
The PLZF gene encodes a 673–amino acid protein that contains a broad complex, tram-trak, bric-a-brac/pox virus zinc finger (BTB/POZ) domain—regions thought to be critical for transcriptional repression by PLZF25 —and 9 Krüppel-like, Cys2His2 zinc fingers at its C-terminal that form the DNA-binding domain (DBD).26 The 118-amino acid BTB/POZ domain was shown previously to mediate protein-protein interaction and repression.21,27,28 PLZF is highly expressed in CD34+ bone marrow progenitor cells, but it is expressed only at low levels, if at all, in mature hematopoietic cells such as peripheral blood mononuclear cells or lineage-restricted, partly differentiated hematopoietic cell lines.26,29Accordingly, expression of PLZF is down-regulated in promyelocytic cells on differentiation treatment with retinoic acid.26PLZF has been shown to cause growth arrest and cell-cycle inhibition in myeloid cell lines.30 31
Because 1,25(OH)2D3 is a potent inducer of monocytic differentiation32 and VDR is widely expressed in hematopoietic cells, we wondered whether PLZF might oppose this effect. We found that PLZF and VDR physically interact and that PLZF expression both abrogates the ability of VDR to activate transcription and impairs its ability to mediate differentiation of U937 cells. This study provides the first characterization of a protein that may regulate 1,25(OH)2D3-dependent differentiation of hematopoietic cells by blocking transcriptional activation by the receptor.
Materials and methods
Cell culture and transient transfections
The histiocytic lymphoma cell line U93733 and its stably transfected derivative, PLZF45, were maintained as follows. Cells were grown at 37°C in 5% carbon dioxide in RPMI 1640 tissue culture medium (Gibco, Rockville, MD) supplemented with 10% fetal-calf serum (FCS; Gibco), 1 mM sodium pyruvate, 2 mM L-glutamine, and 100 units/mL penicillin and 100 μg/mL streptomycin. The stable cell line PLZF45 was maintained in this medium supplemented with 0.5 μg/mL puromycin, 0.1 μg/mL tetracycline, and 1.0 mg/mL G418. The U937 cells were used in all transient transfections as described previously.34 After transfection, ligand (1,25(OH)2D3 [1 × 10−8 M], ATRA [1 × 10−6 M]), or trichostatin A (TSA [150 ng/mL]), or a combination of these, was added for 24 hours. Luciferase assays were done with 10 μg whole-cell extract prepared from harvested cells by using the Promega (Madison, WI) luciferase assay system. Where applicable, luciferase values were normalized to β-galactosidase activity as described previously.35
Construction of PLZF-inducible cell lines
U937T cells, a line of U937 cells stably expressing tet-virion protein (VP) 16 under the control of the tet-operator promoter,36 were used to produce a tetracycline-inducible PLZF cell line.37 Briefly, PLZF(B) complementary DNA26 was cloned into the EcoRI site of pTRE (pUHD10-3) to yield pUHD-PLZF. Ten micrograms pUHD-PLZF and 1 μg pIND (Invitrogen, Carlsbad, CA) as a neomycin-selection marker were linearized and cotransfected into 2 × 107 U937T cells by electroporation (Bio-Rad, Hercules, CA) in 400 μL additive-free RPMI at 960 μF and 0.17 kV. After 24 hours, cells were plated into methylcellulose containing 0.1 μg/mL tetracycline, 0.5 μg/mL puromycin, and 10% FCS with 1 mg/mL G418, and resistant clones were isolated.
In vitro interaction assay
Nuclear receptor proteins or domains were fused to glutathione-S-transferase (GST) and expressed inEscherichia coli. Putative interacting partners were translated in vitro (TNT kit; Promega) in the presence of sulfur 35 (35S)–methionine (Amersham, Piscataway, NJ). GST fusions and translated proteins were incubated in buffer A (170 mM potassium chloride [KCL], 20 mM Tris [pH 7.9], 20% glycerol, 0.2 mM EDTA, 4 mg/mL bovine serum albumin [BSA], 0.05% NP-40, 0.1 mM phenylmethylsulfonyl fluoride [PMSF], and 1 mM dithiothreitol [DTT]) for 3 hours before 3 washes in buffer B (same as buffer A but without BSA). Beads were resuspended in sodium dodecyl sulfate (SDS) sample buffer, separated by electrophoresis through 7.5% to 10% SDS polyacrylamide gels, dried, and exposed overnight at room temperature to Bio-Max film (Kodak, Rochester, NY).
Immunoprecipitations
Nuclear extracts were prepared from U937-PLZF45 cells as described previously.38 Then, 200 μg nuclear extract was resuspended in buffer A.1 (170 mM KCl, 20 mM Tris [pH 7.9], 10% glycerol, 0.2 mM EDTA, 0.05% NP-40, 0.1 mM PMSF, and 1 mM DTT) to a final volume of 445 μL. Extracts were precleared with normal rabbit serum (Santa Cruz Biotechnology, Santa Cruz, CA) and 50 μL protein A/G plus agarose beads (Santa Cruz Biotechnology) for 1 hour and then combined with 50 μL protein A/G plus beads, antibody, and either ethanol or 1,25(OH)2D3 (final concentration, 1 × 10−7 M) per reaction). Reactions were carried out at 4°C for 2 hours and were followed by washing in buffer A.1 to 200 mM KCl. Washed pellets of beads were resuspended in SDS sample buffer, separated by electrophoresis through 7.5% to 10% SDS polyacrylamide gels, dried, and transferred to nitrocellulose filter paper. The filters were subsequently immunoblotted for various proteins by using standard procedures. SKNO-1 cells were cultured in RPMI 1640 with 10% FCS. Cells (1 × 107) were lysed for each immunoprecipitation experiment. A PLZF mouse monoclonal antibody (IgG2a isotype)30,39 and isotype control preimmune mouse monoclonal IgG2a (Jackson Immunoresearch, West Grove, PA) were used at a concentration of 1 mg/mL. Subsequent steps in the immunoprecipitation were done as described previously.40 The precipitated proteins were electrophoresed through a 12% SDS-polyacrylamide gel and transferred to an Immobilon P membrane (Millipore, Bedford, MA). Immunoblotting was done by using standard methods.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assay (EMSA) was done as described previously41 by using 5 μg nuclear extract as described above. Cold specific and nonspecific competitor oligonucleotides were added to the reactions in 10 times the concentration of specific radioactive oligonucleotide. Antibody supershifts were done by adding anti-VDR monoclonal antibody or anti-PLZF polyclonal serum to the reactions. The vitamin D3 response element (VDRE) from the mouse osteopontin gene (OPN; also called Spp1) was generated as complementary oligonucleotides of the sequence 5′-GATCCACAAGGTTCACGAGGTTCACG-3′ (top strand). The oligonucleotide used as the nonspecific control was a glucocorticoid response element of the top strand sequence 5′-GATCCGACCGAGAACAAGATGTTCTGTCGAG-3′.
Differentiation assay and flow cytometry
PLZF45 cells were washed in phosphate-buffered saline (PBS), transferred to medium without tetracycline, and allowed to grow for 16 hours before addition of 1,25(OH)2D3(10−8 M), ATRA (10−6 M), or ethanol and dimethyl sulfoxide. At various times, 1 × 106 cells were obtained for flow cytometric analysis. Each sample was washed twice in ice-cold PBS and 1% BSA and incubated in 150 μL cold PBS and 1% BSA with 1.5 μL of either IgG1–fluorescein isothiocyanate conjugated (FITC) and IgG1– R-phycoerythrin or IgG1-FITC anti-CD14 or anti-CD11c (Caltag, Burlingame, CA) for 45 minutes. After incubation, the cells were washed in PBS and 1% BSA 3 times; resuspended in 400 μL PBS, 1% BSA, and 0.5% paraformaldehyde; and stored at 4°C. Flow cytometry was done on a fluorescence-activated cell-sorter scanner (FACS Calibur; Becton Dickinson, Mountain View, CA).
Morphometric analysis
At various times during the PLZF-1,25(OH)2D3-ATRA induction experiment, 2 × 105 cells were removed from each growth condition, cytospun onto microscope slides at 600 rpm for 10 minutes, air dried, and fixed in 100% methanol for 5 minutes. They were then stained with Giemsa staining (Hema-tek 2000; Bayer Corp, Pittsburgh, PA). Several fields on each slide were examined, and the cell dimensions were measured from digitized images by using National Institutes of Health Image 1.62 software. Total-cell and nuclear areas were determined, and the percentage of each cell occupied by the nucleus was derived for each cell. Ten cells were measured, and the average nuclear-area percentage was calculated for each condition.
Results
PLZF blocks 1,25(OH)2D3-activated transcription
PLZF-RARα expression can block 1,25(OH)2D3-mediated monocyte differentiation.20 To determine the possible molecular mechanism for this activity, we used PLZF-RARα, PLZF, and VDR to transfect U937 cells, along with a reporter gene containing a consensus VDRE, a 6–base pair direct repeat with a spacing of 3 nucleotides between the half sites. As shown in Figure1A, exogenous expression of either PLZF or PLZF-RARα inhibited VDR-mediated activation of a luciferase reporter (lane 1 versus lanes 2 and 3), regardless of the presence of the histone deacetylase (HDAC) inhibitor, TSA (lane 4 versus lanes 5 and 6). PLZF-RARα and PLZF were expressed at similar levels (data not shown).
Effects of PLZF and PLZF-RARα on VDR, VP-16, and RARα-mediated transactivation.
(A) Transient overexpression of PLZF or PLZF-RARα represses 1,25(OH)2D3-dependent activation of a VDR responsive reporter (lanes 1-3). This repression was not affected by the addition of 150 ng/mL TSA to the culture medium (lanes 4-6). Plasmids containing complementary DNAs for PLZF, PLZF-RARα, VDR, and a minimal VDR responsive reporter containing 2 copies of a VDRE from the OPN gene, cloned approximately 100 bp upstream of the E1b TATA box and luciferase gene, were used to electroporate U937 cells with the indicated amounts of DNAs. The cells were left for 2 hours before the addition of 1,25(OH)2D3(10−8 M), TSA, or vehicle (ethanol). Data are from a representative experiment done in triplicate. (B) VP16-dependent activation is not affected by PLZF expression. Transfection of U937 cells was carried out as described in the legend for panel A. A GAL4-VP16 fusion was coexpressed with PLZF and a GAL responsive reporter, with the indicated amounts of DNA. (C) PLZF did not repress ATRA-dependent activation of an ATRA responsive reporter by RARα. The reporter used here contained the RARE from the RARβ gene promoter positioned upstream of the Tk promoter and luciferase gene. Transfection was done as described above, and the cells were left for 2 hours before the addition of ATRA (10−6 M) or vehicle (dimethyl sulfoxide [DMSO]). (D) PLZF associates with VDR but not RARα in a ligand-independent manner. In vitro–translated35S-labeled PLZF was incubated at 4°C for 2 hours with affinity-purified GST-VDR or GST-RARα in the presence (+) or absence (−) of 1,25(OH)2D3 or ATRA (10−7M and 10−6 M, respectively). The resulting complexes were washed, separated by SDS–polyacrylamide gel electrophoresis (PAGE), dried, and exposed to film. Lane 1 represents 10% of the labeled PLZF input (I). PLZF did not interact with GST alone (lane 2). PLZF interacted with GST-VDR independent of ligand (lanes 3 and 4) but not with GST- RARα (lanes 5 and 6). All transfections were done at least 3 times in triplicate each time. The histograms show results from a representative experiment from this series. LUC indicates luciferase reporter gene; TK, thymidine kinase promoter; RLU, relative light unit.
Effects of PLZF and PLZF-RARα on VDR, VP-16, and RARα-mediated transactivation.
(A) Transient overexpression of PLZF or PLZF-RARα represses 1,25(OH)2D3-dependent activation of a VDR responsive reporter (lanes 1-3). This repression was not affected by the addition of 150 ng/mL TSA to the culture medium (lanes 4-6). Plasmids containing complementary DNAs for PLZF, PLZF-RARα, VDR, and a minimal VDR responsive reporter containing 2 copies of a VDRE from the OPN gene, cloned approximately 100 bp upstream of the E1b TATA box and luciferase gene, were used to electroporate U937 cells with the indicated amounts of DNAs. The cells were left for 2 hours before the addition of 1,25(OH)2D3(10−8 M), TSA, or vehicle (ethanol). Data are from a representative experiment done in triplicate. (B) VP16-dependent activation is not affected by PLZF expression. Transfection of U937 cells was carried out as described in the legend for panel A. A GAL4-VP16 fusion was coexpressed with PLZF and a GAL responsive reporter, with the indicated amounts of DNA. (C) PLZF did not repress ATRA-dependent activation of an ATRA responsive reporter by RARα. The reporter used here contained the RARE from the RARβ gene promoter positioned upstream of the Tk promoter and luciferase gene. Transfection was done as described above, and the cells were left for 2 hours before the addition of ATRA (10−6 M) or vehicle (dimethyl sulfoxide [DMSO]). (D) PLZF associates with VDR but not RARα in a ligand-independent manner. In vitro–translated35S-labeled PLZF was incubated at 4°C for 2 hours with affinity-purified GST-VDR or GST-RARα in the presence (+) or absence (−) of 1,25(OH)2D3 or ATRA (10−7M and 10−6 M, respectively). The resulting complexes were washed, separated by SDS–polyacrylamide gel electrophoresis (PAGE), dried, and exposed to film. Lane 1 represents 10% of the labeled PLZF input (I). PLZF did not interact with GST alone (lane 2). PLZF interacted with GST-VDR independent of ligand (lanes 3 and 4) but not with GST- RARα (lanes 5 and 6). All transfections were done at least 3 times in triplicate each time. The histograms show results from a representative experiment from this series. LUC indicates luciferase reporter gene; TK, thymidine kinase promoter; RLU, relative light unit.
Because PLZF alone was capable of significantly blocking VDR-dependent transactivation, we subsequently focused on PLZF. To test whether the repression of VDR activity by PLZF was a result of general transcriptional repression, PLZF was cotransfected with GAL-VP16 and a GAL operator–containing responsive reporter in U937 cells. As shown in Figure 1B, activation of the GAL reporter by VP16 was not affected by exogenous expression of PLZF, indicating that PLZF does not generally squelch transcription. Additionally, coexpression of PLZF with VDR did not affect VDR expression levels (data not shown). For comparison, PLZF was also cotransfected with RARα and a reporter containing the RARβ2 RARE. Figure 1C shows that exogenous expression of PLZF did not affect ATRA-dependent activation of this reporter. Thus, we found that PLZF repression of transactivation is selective for VDR.
We addressed the question of whether the PLZF-mediated repression of VDR function was due to a physical interaction between the proteins by conducting in vitro pull-down assays to determine whether35S-labeled PLZF could bind to GST-VDR or GST-RARα in the presence and absence of 1,25(OH)2D3 or ATRA, respectively (Figure 1D). In vitro, PLZF associated with VDR independent of ligand (lanes 3 and 4) but did not associate with GST-RARα in either the presence or absence of ATRA (lanes 5 and 6). These findings show that the ability of VDR and PLZF to interact is correlated with the selective loss of VDR-mediated transactivation in the presence of PLZF.
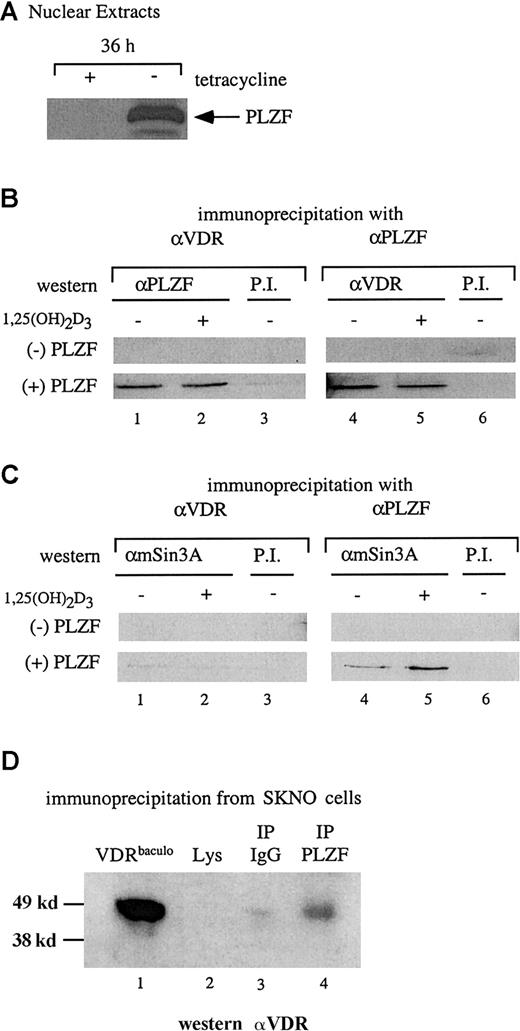
PLZF and VDR interact in cellular extracts
The results of the in vitro interaction assay were confirmed by coimmunoprecipitation studies. Stable U937 cells capable of induced expression of PLZF on removal of tetracycline were used to assess an interaction between endogenous VDR and PLZF in a nuclear extract. Figure 2A shows the induction of PLZF expression after 36 hours of culture in medium without tetracycline. Nuclear extracts were prepared from cells in the presence and absence of tetracycline (the absence and presence of PLZF, respectively) and used in a coimmunoprecipitation assay with antibodies to both VDR and PLZF. The extracts (with and without PLZF) were precipitated with either anti-VDR or anti-PLZF antibodies and immunoblotted with the opposite antibody. As shown in Figure 2B, in the absence of PLZF, neither antibody precipitated the reciprocal protein. However, in the presence of PLZF, anti-VDR and anti-PLZF coprecipitated PLZF and VDR, respectively. As in the in vitro pull-down assays, the interaction between PLZF and VDR occurred independently of 1,25(OH)2D3.
VDR and PLZF interact in U937 nuclear extracts.
(A) Immunoblot for expression of PLZF in nuclear extracts. Nuclear extracts from the tetracycline-regulated stable line U937-PLZF45 were prepared from cells grown for 36 hours in the presence (− PLZF) or absence (+ PLZF) of tetracycline and immunoblotted for PLZF expression by using an anti-PLZF monoclonal antibody. (B) Coimmunoprecipitation of PLZF and VDR from nuclear extracts. Immunoprecipitations were done by using nuclear extracts from U937-PLZF45 cells with either anti-VDR or anti-PLZF monoclonal antibodies and protein A/G agarose. Precipitated complexes were washed in propidium iodide buffer containing 200 mM KCl, separated by SDS-PAGE, and immunoblotted for either PLZF (lanes 1-3) or VDR (lanes 4-6). Preimmune serum and protein A/G beads were used as a negative control immunoprecipitation (lanes 3 and 6). (C) Coimmunoprecipitation of PLZF and mSin3A. Antibodies directed against PLZF (lanes 4 and 5) but not those against VDR (lanes 1 and 2) coimmunoprecipitated Sin3A from PLZF45 extracts. Coimmunoprecipitation was performed as in panel B, except that the complexes were immunoblotted for mSin3A with an antibody that cross-reacts with human Sin3A. Preimmune serum and protein A/G beads were used as a control (lanes 3 and 6). (D) Coimmunoprecipitation of PLZF and VDR in SKNO cells. Lysates of these cells were subjected to direct immunoblotting (lane 2) or blotting after precipitation with PLZF antibody (lane 4) or an isotype control (lane 3). A positive control (lane 1), confirming the efficacy of the antibody and size of the precipitated species, consisted of 25 ng purified baculovirus-derived VDR (VDRbaculo). Direct immunoblotting was done with 100 μg lysate from SKNO-1 cells. PI indicates preimmune serum; IP, immunoprecipitation; Lys, lysate.
VDR and PLZF interact in U937 nuclear extracts.
(A) Immunoblot for expression of PLZF in nuclear extracts. Nuclear extracts from the tetracycline-regulated stable line U937-PLZF45 were prepared from cells grown for 36 hours in the presence (− PLZF) or absence (+ PLZF) of tetracycline and immunoblotted for PLZF expression by using an anti-PLZF monoclonal antibody. (B) Coimmunoprecipitation of PLZF and VDR from nuclear extracts. Immunoprecipitations were done by using nuclear extracts from U937-PLZF45 cells with either anti-VDR or anti-PLZF monoclonal antibodies and protein A/G agarose. Precipitated complexes were washed in propidium iodide buffer containing 200 mM KCl, separated by SDS-PAGE, and immunoblotted for either PLZF (lanes 1-3) or VDR (lanes 4-6). Preimmune serum and protein A/G beads were used as a negative control immunoprecipitation (lanes 3 and 6). (C) Coimmunoprecipitation of PLZF and mSin3A. Antibodies directed against PLZF (lanes 4 and 5) but not those against VDR (lanes 1 and 2) coimmunoprecipitated Sin3A from PLZF45 extracts. Coimmunoprecipitation was performed as in panel B, except that the complexes were immunoblotted for mSin3A with an antibody that cross-reacts with human Sin3A. Preimmune serum and protein A/G beads were used as a control (lanes 3 and 6). (D) Coimmunoprecipitation of PLZF and VDR in SKNO cells. Lysates of these cells were subjected to direct immunoblotting (lane 2) or blotting after precipitation with PLZF antibody (lane 4) or an isotype control (lane 3). A positive control (lane 1), confirming the efficacy of the antibody and size of the precipitated species, consisted of 25 ng purified baculovirus-derived VDR (VDRbaculo). Direct immunoblotting was done with 100 μg lysate from SKNO-1 cells. PI indicates preimmune serum; IP, immunoprecipitation; Lys, lysate.
The physiologic nature of the PLZF-VDR interaction was explored further in the M2 leukemia cell line SKNO,42 in which both PLZF and VDR are expressed endogenously. VDR could not be detected by direct immunoblotting from SKNO lysates (Figure 2D, lane 2). However, coimmunoprecipitates with the PLZF monoclonal antibody reacted positively (6.1-fold increase over lysate alone with the IgG signal subtracted) when probed with an anti-VDR antibody (Figure 2D, lane 4). These results show that antibodies to PLZF and VDR can reciprocally immunoprecipitate both proteins and that this interaction occurs even at the low endogenous protein levels in SKNO cells.
Because of the apparent ligand-independent nature of the interaction between PLZF and VDR, and given that PLZF can interact with the corepressors SMRT–N-CoR, HDAC-1, and Sin3A,21,28,43 44 we wondered whether VDR associates with components of a corepressor complex through its association with PLZF. As shown in Figure 2C, endogenous Sin3A was clearly immunoprecipitated by anti-PLZF antibodies in the presence of PLZF (lanes 4 and 5) but not by anti-VDR antibodies (lanes 1 and 2). Similar results were observed when VDR coimmunoprecipitations were blotted for SMRT and HDAC-1 (data not shown). Thus, although VDR interacted with PLZF and PLZF interacted with Sin3A and other repressors, a 3-way interaction was not detected.
Delineation of interacting domains
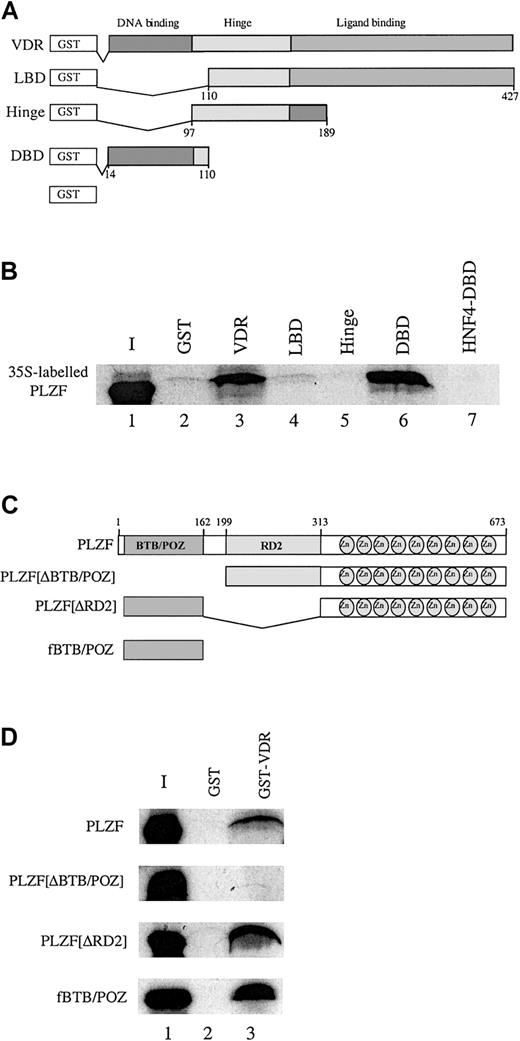
To determine how PLZF might block transcriptional activation by VDR, an in vitro protein affinity assay was used to identify the domains in PLZF and VDR responsible for the interaction. Figure3A depicts a series of GST fusions containing all or part of VDR. The 35S-labeled, in vitro–translated PLZF was incubated with the individual VDR derivatives (Figure 3B). In vitro–translated PLZF interacted to a significant extent with full-length VDR (Figure 3B, lane 3) but also with the GST-VDR DBD (GST-VDRDBD; Figure 3B, lane 6). It did not interact with GST alone, the GST-VDR ligand-binding domain (GST-VDRLBD, or the hinge region (GST-VDRhinge) (lanes 2, 4, and 5). Consistent with the inability of PLZF to interact with GST-VDRLBD, a GAL-VDRLBD construct that strongly activated transcription from a GAL-UAS reporter in response to 1,25(OH)2D3 was not affected by coexpression of PLZF in U937 cells (data not shown).
PLZF associates with the VDR DBD in vitro by means of the BTB/POZ domain.
(A) Schematic diagram of the GST-VDR fusions used to assess an in vitro interaction between PLZF and VDR. These included the full-length VDR, the LBD (amino acids 110-427), the hinge region (amino acids 97-189), the DBD (amino acids 14-110), and GST alone. The VDR derivatives were affinity purified after expression in Escherichia coli on GST Sepharose. The expression levels of the GST-VDR derivatives were assessed by SDS-PAGE analysis of expressed proteins (data not shown). (B) PLZF interacts selectively in vitro with full-length GST-VDR and GST-VDRDBD (lanes 3 and 6, respectively) but not with GST-VDRLBD, GST-VDRhinge, or the orphan receptor HNF4DBD fused to GST (lanes 4, 5, and 7). PLZF was translated in the presence of 35S-methionine and incubated at 4°C with the various VDR derivatives depicted in panel A. Interacting complexes were washed and subjected to SDS-PAGE analysis, dried, and exposed to film. Similar amounts of specific protein were used in each experiment. Lane 1 represents 10% of the labeled PLZF input (I). PLZF runs slightly differently in the input lane because of the lack of BSA (which was added to stabilize interactions in the other lanes). Approximately 5 × 10−13 mM PLZF was used in each reaction. (C) Schematic diagram of PLZF derivatives. PLZFΔBTB/POZ (Δ amino acids 1-162), PLZFΔRD2 (Δ amino acids 199-313), and fBTB/POZ (flag-tagged amino acids 1-162) were used in subsequent in vitro GST pull-down assays. (D) The BTB/POZ domain is necessary and sufficient for PLZF interaction with VDR. GST-VDR on GST agarose beads (lane 3) was incubated with 35S-labeled, in vitro–translated PLZF, PLZFΔBTB/POZ, PLZFΔRD2, or fBTB/POZ at 4°C. Resulting complexes were washed and analyzed by SDS-PAGE and autoradiography. None of the derivatives interacted with GST alone (lane 2). Approximately 5 × 10−13 mM translated protein was used in each reaction. Pull-down experiments were done at least 3 times, and a representative film is shown.
PLZF associates with the VDR DBD in vitro by means of the BTB/POZ domain.
(A) Schematic diagram of the GST-VDR fusions used to assess an in vitro interaction between PLZF and VDR. These included the full-length VDR, the LBD (amino acids 110-427), the hinge region (amino acids 97-189), the DBD (amino acids 14-110), and GST alone. The VDR derivatives were affinity purified after expression in Escherichia coli on GST Sepharose. The expression levels of the GST-VDR derivatives were assessed by SDS-PAGE analysis of expressed proteins (data not shown). (B) PLZF interacts selectively in vitro with full-length GST-VDR and GST-VDRDBD (lanes 3 and 6, respectively) but not with GST-VDRLBD, GST-VDRhinge, or the orphan receptor HNF4DBD fused to GST (lanes 4, 5, and 7). PLZF was translated in the presence of 35S-methionine and incubated at 4°C with the various VDR derivatives depicted in panel A. Interacting complexes were washed and subjected to SDS-PAGE analysis, dried, and exposed to film. Similar amounts of specific protein were used in each experiment. Lane 1 represents 10% of the labeled PLZF input (I). PLZF runs slightly differently in the input lane because of the lack of BSA (which was added to stabilize interactions in the other lanes). Approximately 5 × 10−13 mM PLZF was used in each reaction. (C) Schematic diagram of PLZF derivatives. PLZFΔBTB/POZ (Δ amino acids 1-162), PLZFΔRD2 (Δ amino acids 199-313), and fBTB/POZ (flag-tagged amino acids 1-162) were used in subsequent in vitro GST pull-down assays. (D) The BTB/POZ domain is necessary and sufficient for PLZF interaction with VDR. GST-VDR on GST agarose beads (lane 3) was incubated with 35S-labeled, in vitro–translated PLZF, PLZFΔBTB/POZ, PLZFΔRD2, or fBTB/POZ at 4°C. Resulting complexes were washed and analyzed by SDS-PAGE and autoradiography. None of the derivatives interacted with GST alone (lane 2). Approximately 5 × 10−13 mM translated protein was used in each reaction. Pull-down experiments were done at least 3 times, and a representative film is shown.
An interaction with VDR by means of its DBD could explain why repression by PLZF is ligand independent. To address whether the interaction with the VDR DBD was specific, we conducted a pull-down study using the VDR DBD and another nuclear receptor DBD, that from the orphan receptor HNF4. PLZF interacted with GST VDRDBD but not with GST-HNF4DBD (Figure 3B, lanes 6 and 7), thereby indicating that the interaction is specific for VDR and not the result of promiscuous interaction with a hormone receptor zinc finger region.
We used a similar analysis to determine the domains of PLZF protein that facilitate VDR interaction. Figure 3C depicts a series of PLZF mutants containing deletions covering the BTB/POZ domain (PLZFΔBTB/POZ]), a second repression domain (PLZFΔRD2]), and both the RD2 and zinc finger regions (BTB/POZ). The PLZF BTB/POZ domain was implicated as being responsible for protein-protein interactions in several other systems.27 We translated PLZF and derivatives and allowed them to interact with GST-VDR. Deletion of either the RD2 domain or both the RD2 and zinc finger domains of PLZF did not affect binding to VDR (Figure 3D). In contrast, deletion of the BTB/POZ domain completely abrogated binding of PLZF to VDR. Thus, it appears that the major surface responsible for VDR interaction lies within the BTB/POZ domain of PLZF.
We correlated the ability of various PLZF mutants to interact with VDR with their ability to repress VDR-dependent transactivation of cotransfected reporter constructs in U937 cells. Deletion of the BTB/POZ domain abrogated the ability of PLZF to repress VDR transactivation (Figure 4A, lanes 4 and 5) relative to full-length PLZF (lanes 2 and 3). Thus, we found that, in vivo, the BTB/POZ domain plays an important role in the functional repression of VDR, presumably through its ability to physically associate with the receptor. Interestingly, an epitope-tagged version of the BTB/POZ domain did not repress VDR-dependent transcription (Figure 4B). Furthermore, deletion of the zinc finger region of PLZF partly relieved the repression of VDR transactivation (Figure 4C), whereas a deletion of the RD2 domain retained the ability to repress VDR (Figure 4D). Therefore, we found that although the BTB/POZ domain is necessary and sufficient for the interaction of PLZF with VDR, it cannot on its own repress transcription, a result that suggests the existence of additional contributions from other moieties of PLZF.
The PLZF BTB/POZ domain is necessary but not sufficient for repression of VDR-mediated transactivation.
(A) Deletion of the PLZF BTB/POZ domain relieves repression mediated by PLZF of 1,25(OH)2D3-dependent activation. U937 cells were transfected with the indicated amounts of either PLZF (lanes 2 and 3) or PLZF(ΔBTB/POZ) (lanes 4 and 5). Levels of expression of each protein in transfected extracts were similar (data not shown). (B) Expression of the BTB/POZ domain alone is not sufficient to repress 1,25(OH)2D3-dependent activation. A construct containing an epitope (flag)-tagged version of the PLZF BTB/POZ domain was cotransfected with VDR in U937 cells in the indicated DNA amounts. The fBTB/POZ protein was detectable in transfected extracts (data not shown). U937 cells were transfected as described previously and treated with ligand for 24 hours before harvesting. (C) Deletion of the 9 carboxyl zinc fingers in PLZF (PLZFΔZF1-9]) partly relieves PLZF-mediated repression of 1,25(OH)2D3-dependent activation. Protein amounts determined by Western blotting were similar for PLZF and PLZFΔZF1-9 (data not shown). (D) Deletion of the central RD2 domain of PLZF does not affect the ability of PLZF to repress 1,25(OH)2D3-dependent activation. All experiments shown are representative and were done in triplicate.
The PLZF BTB/POZ domain is necessary but not sufficient for repression of VDR-mediated transactivation.
(A) Deletion of the PLZF BTB/POZ domain relieves repression mediated by PLZF of 1,25(OH)2D3-dependent activation. U937 cells were transfected with the indicated amounts of either PLZF (lanes 2 and 3) or PLZF(ΔBTB/POZ) (lanes 4 and 5). Levels of expression of each protein in transfected extracts were similar (data not shown). (B) Expression of the BTB/POZ domain alone is not sufficient to repress 1,25(OH)2D3-dependent activation. A construct containing an epitope (flag)-tagged version of the PLZF BTB/POZ domain was cotransfected with VDR in U937 cells in the indicated DNA amounts. The fBTB/POZ protein was detectable in transfected extracts (data not shown). U937 cells were transfected as described previously and treated with ligand for 24 hours before harvesting. (C) Deletion of the 9 carboxyl zinc fingers in PLZF (PLZFΔZF1-9]) partly relieves PLZF-mediated repression of 1,25(OH)2D3-dependent activation. Protein amounts determined by Western blotting were similar for PLZF and PLZFΔZF1-9 (data not shown). (D) Deletion of the central RD2 domain of PLZF does not affect the ability of PLZF to repress 1,25(OH)2D3-dependent activation. All experiments shown are representative and were done in triplicate.
PLZF is present in a VDR DNA-binding complex
Our observation of a physical interaction between the VDR DBD and PLZF led us to consider that PLZF may affect the ability of VDR to bind its hormone response element or affect the nature of the DNA-bound complex so that it cannot activate transcription. To explore this issue, nuclear extracts prepared from U937-PLZF cells, grown in either the presence or absence of tetracycline to regulate expression of PLZF, were used in an EMSA (Figure 5). The probe used in this assay was a direct repeat with a spacing of 3 nucleotides (DR-3) derived from the VDRE of the mouse OPNgene promoter and was identical to the canonical DR-3 in the reporter construct used in the transfection experiments. As shown in Figure 5A, addition of extract yielded at least 4 shifted species (lane 2), which showed specific competition with the addition of cold excess VDRE but not a nonspecific competitor (lanes 3 and 4). In extract containing PLZF, there was a significant change in shift mobility (lane 5). This shift showed competition with the specific oligo (lane 6) and was not affected by the nonspecific oligo (lane 7). Addition of an anti-VDR antibody confirmed that these shifted species contained VDR. Anti-VDR antibodies could compromise binding (open arrow) or alter the mobility of several of the major species on the gel and induce a supershift (asterisk in lane 9). In the presence of PLZF, addition of anti-VDR antibody disrupted the large lower shift (solid arrow) and a high-molecular-weight shift appeared (2 asterisks in lane 11). The disappearance of a significant specific shift in the presence of the anti-VDR antibody in lanes 9 and 11 indicates that, in both cases, VDR was present in the complex but the complex was altered by the presence of PLZF. The most supershifted complexes in lanes 9 and 11 appear to have different mobilities, with lane 11 showing slightly more delay, a finding consistent with the presence of PLZF in the DNA-protein complex.
PLZF and VDR comigrate in a DNA-bound complex.
(A) Endogenous VDR from U937-PLZF45 nuclear extracts binds to a VDRE with an altered mobility in the presence of PLZF. Nuclear extracts were prepared from U937-PLZF45 cells and incubated in the presence or absence of tetracycline (− PLZF or + PLZF). Then, 5μg nuclear extract was combined with a phosphorus 32–labeled VDRE probe, incubated for 20 minutes at room temperature, and electrophoresed on a nondenaturing polyacrylamide gel. The VDRE probe used in this assay was a DR-3 from the mouse OPN gene promoter. Lane 1 is free probe and lanes 2 to 7 show the specificity of the shifts observed. Lane 2 depicts the 4 major species found in U937 nuclear extracts without PLZF expression, which competed with unlabeled (10-fold excess) specific oligonucleotide (spec) (lane 3) but not a nonspecific oligonucleotide (non-spec) (a glucocorticoid response element [GRE]; lane 4). Lanes 5 through 7 show controls similar to those in lanes 2 to 4 but with use of extracts expressing PLZF. There was a significant change in mobility in the presence of PLZF (lane 5). This shift competed with the specific oligonucleotide (lane 6) and was not affected by the nonspecific oligonucleotide (lane 7). Lanes 8 through 11 show the effects of the addition of antibodies to VDR on the mobility shift. The presence of control IgG antibody had no effect on the shifted species (lane 8). The addition of anti-VDR antibody caused a loss of the major species (open arrow) and a supershift (asterisk) indicating that the complex contained VDR. In the presence of PLZF, another major species (solid arrow) was supershifted (2 asterisks) by anti-VDR antibodies (lane 10 versus lane 11). (B) PLZF is present in a DNA-bound complex containing VDR. EMSA was done as shown in panel A. The addition of anti-PLZF antibody did not affect the shift pattern observed when PLZF was absent (lanes 12 and 13 versus 19). The addition of anti-VDR antibodies confirmed the presence of VDR in a DNA-bound complex. VDR complexes were abrogated by the addition of anti-PLZF antibody (lanes 16 and 17; arrow), indicating that both PLZF and VDR exist in the bound species. The addition of preimmune serum did not affect the shifts in the absence (lane 18) or presence (lane 19) of PLZF.
PLZF and VDR comigrate in a DNA-bound complex.
(A) Endogenous VDR from U937-PLZF45 nuclear extracts binds to a VDRE with an altered mobility in the presence of PLZF. Nuclear extracts were prepared from U937-PLZF45 cells and incubated in the presence or absence of tetracycline (− PLZF or + PLZF). Then, 5μg nuclear extract was combined with a phosphorus 32–labeled VDRE probe, incubated for 20 minutes at room temperature, and electrophoresed on a nondenaturing polyacrylamide gel. The VDRE probe used in this assay was a DR-3 from the mouse OPN gene promoter. Lane 1 is free probe and lanes 2 to 7 show the specificity of the shifts observed. Lane 2 depicts the 4 major species found in U937 nuclear extracts without PLZF expression, which competed with unlabeled (10-fold excess) specific oligonucleotide (spec) (lane 3) but not a nonspecific oligonucleotide (non-spec) (a glucocorticoid response element [GRE]; lane 4). Lanes 5 through 7 show controls similar to those in lanes 2 to 4 but with use of extracts expressing PLZF. There was a significant change in mobility in the presence of PLZF (lane 5). This shift competed with the specific oligonucleotide (lane 6) and was not affected by the nonspecific oligonucleotide (lane 7). Lanes 8 through 11 show the effects of the addition of antibodies to VDR on the mobility shift. The presence of control IgG antibody had no effect on the shifted species (lane 8). The addition of anti-VDR antibody caused a loss of the major species (open arrow) and a supershift (asterisk) indicating that the complex contained VDR. In the presence of PLZF, another major species (solid arrow) was supershifted (2 asterisks) by anti-VDR antibodies (lane 10 versus lane 11). (B) PLZF is present in a DNA-bound complex containing VDR. EMSA was done as shown in panel A. The addition of anti-PLZF antibody did not affect the shift pattern observed when PLZF was absent (lanes 12 and 13 versus 19). The addition of anti-VDR antibodies confirmed the presence of VDR in a DNA-bound complex. VDR complexes were abrogated by the addition of anti-PLZF antibody (lanes 16 and 17; arrow), indicating that both PLZF and VDR exist in the bound species. The addition of preimmune serum did not affect the shifts in the absence (lane 18) or presence (lane 19) of PLZF.
To determine whether PLZF was present in the DNA-bound complex, we conducted similar antibody additions using a monoclonal antibody to PLZF (Figure 5B). Addition of the anti-PLZF antibody did not affect the shift pattern observed when PLZF was not present (lanes 12 and 13). In the presence of PLZF, increasing amounts of anti-PLZF antibody abrogated both the upper and lower prominent shifts observed in the absence of the antibody (lanes 16 and 17 versus lane 14). The data indicate that PLZF is present in the VDR-VDRE complexes. When coupled with the data on in vitro interaction and transfection described earlier, these findings suggest that PLZF and VDR from nuclear extracts can form complexes on a VDRE and that these complexes could be responsible for the inability of VDR to transactivate in the presence of PLZF.
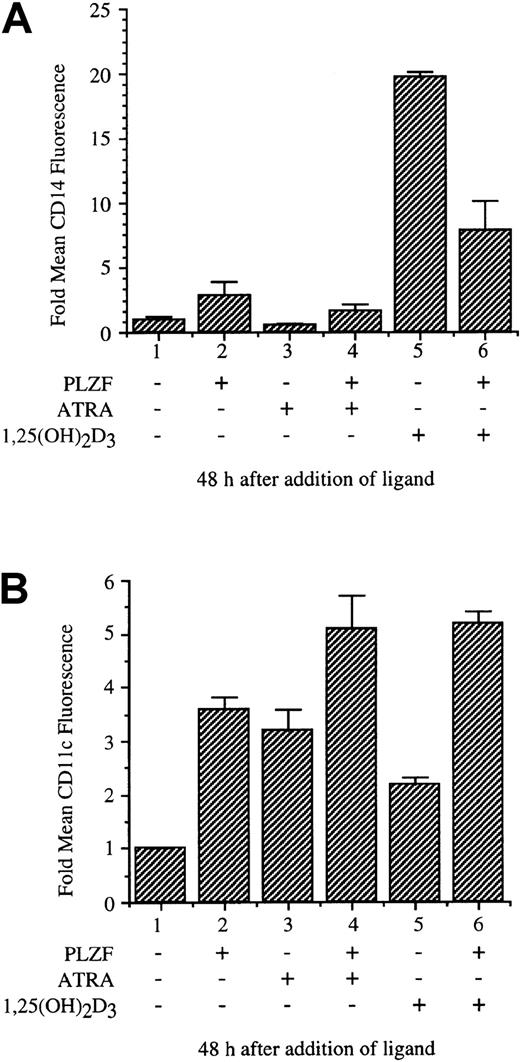
Monocytic differentiation induced by 1,25(OH)2D3 is inhibited by PLZF
The stable, tetracycline-inducible U937 cell line PLZF45 was used to assay the effects of PLZF expression on monocytic differentiation induced by 1,25(OH)2D3 and, in comparison, ATRA. Both 1,25(OH)2D3 and ATRA induce differentiation of hematopoietic precursor cells as measured by surface-antigen expression and changes in cellular morphologic features. PLZF45 cells were cultured in medium without tetracycline, inducing PLZF expression; treated with 1,25(OH)2D3 or ATRA for 48 hours; and harvested for analysis of cell-surface antigens by flow cytometry (Figure6). The fold change in mean fluorescence of the monocytic differentiation marker CD14 was measured as a function of PLZF expression and treatment with 1,25(OH)2D3 or ATRA (Figure 6A). PLZF alone minimally induced expression of CD14 over background levels (lane 2). ATRA did not induce expression of CD14 (lane 3). CD14 expression was induced by 1,25(OH)2D3, to 20 times the levels in uninduced cells, and this induction was substantially reduced by PLZF (Figure 6A, lane 5 versus lane 6). In contrast, PLZF expression did not block ATRA-induced differentiation, as measured by the monocyte-granulocyte marker CD11c after 48 hours of treatment with ATRA (Figure 6B, lane 3 versus lane 4) or alone (lane 1 versus lanes 2 and 6); instead, it appeared to contribute to induction with ATRA. PLZF also did not block induction of CD11c family members CD11b and CD11a or of CD18 (data not shown). Thus, PLZF appears, possibly through a specific block of VDR function, to alter CD14 but not CD11c surface-marker expression. These results suggest that PLZF may alter very specific 1,25(OH)2D3-induced monocytic differentiation pathways.
PLZF expression blocks 1,25(OH)2D3-inducible cell-surface expression of CD14.
The U937-PLZF45 cell line was cultured in the presence or absence of tetracycline (− PLZF and + PLZF). After 16 hours, 1,25(OH)2D3 or ATRA was added, and samples were collected 48 hours later. The samples were fixed and stained with FITC anti-CD14 (A), CD11c (B), or nonspecific FITC antibodies. These samples were then analyzed for surface-marker expression by flow cytometry. FITC nonspecific antibodies did not stain cells (−) or (+) PLZF and were used to set detection levels and size variables for the flow cytometry analyses. Cell viability for all samples was greater than 95% (data not shown). The error shown for induction of cell-surface markers is the result of combining data from 3 separate experiments done in triplicate.
PLZF expression blocks 1,25(OH)2D3-inducible cell-surface expression of CD14.
The U937-PLZF45 cell line was cultured in the presence or absence of tetracycline (− PLZF and + PLZF). After 16 hours, 1,25(OH)2D3 or ATRA was added, and samples were collected 48 hours later. The samples were fixed and stained with FITC anti-CD14 (A), CD11c (B), or nonspecific FITC antibodies. These samples were then analyzed for surface-marker expression by flow cytometry. FITC nonspecific antibodies did not stain cells (−) or (+) PLZF and were used to set detection levels and size variables for the flow cytometry analyses. Cell viability for all samples was greater than 95% (data not shown). The error shown for induction of cell-surface markers is the result of combining data from 3 separate experiments done in triplicate.
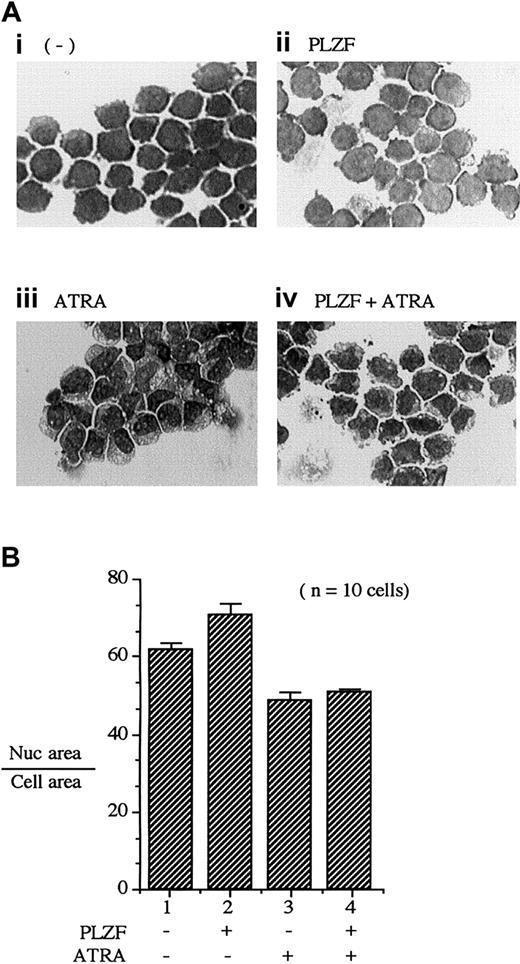
As multipotent precursor cells progress through monocytic differentiation, they undergo morphologic changes. One morphologic indicator of differentiation is a reduction in the volume of the nucleus volume relative to that of the cytoplasm. We used the PLZF45 cell line to assess whether PLZF expression would alter 1,25(OH)2D3-dependent changes in cell morphologic features (Figure 7A). For U937T-PLZF45 cells without either 1,25(OH)2D3treatment or PLZF expression, nuclei occupied 71.5% ± 2.0% of total-cell area (Figure 7B, lane 1). Expression of PLZF (Figure 7A, panel ii) did not cause a significant decrease in nuclear size. In contrast, treatment with 1,25(OH)2D3resulted in a striking decrease in nuclear area, to 45% ± 3.0%, indicative of a differentiating cell (Figure 7A, panel iii). Expression of PLZF during treatment with 1,25(OH)2D3blocked the shrinking of nuclear volume (Figure 7A, panel iv, versus Figure 7A panel iii, and Figure 7B; 61.0% ± 3.0% versus 45.0 ± 3.0%, respectively). Unlike results observed with 1,25(OH)2D3 treatment, PLZF expression had little effect on the morphologic changes resulting from ATRA treatment (Figure 8A). Treatment with ATRA resulted in a 13% reduction in nucleus-to-cytoplasm area (Figure 8B, lanes 2 and 3), a change that was not affected by the presence of PLZF (Figure 8B, lanes 3 and 4). These results, along with the observed effects of PLZF expression on 1,25(OH)2D3-induced CD14 surface expression, strongly suggest that PLZF is a potent and specific regulator of 1,25(OH)2D3-dependent monocytic differentiation.
PLZF expression blocks morphologic changes induced by 1,25(OH)2D3.
Morphometric analysis was done by comparing the nuclear (Nuc) with total-cell area as a function of 1,25(OH)2D3 treatment. (A) Giemsa staining of PLZF45 cells. U937-PLZF45 cells were cultured in the presence or absence of tetracycline (− PLZF and + PLZF) with the indicated treatments. Ligand (1,25(OH)2D3[10−8 M]) was added 16 hours after tetracycline was removed to induce PLZF expression. Total-cell area and nuclear area were calculated on the basis of horizontal and vertical measurements of the diameter of the cell nucleus. Ratios were determined for 10 cells/field. Fields shown were representative of those counted. Original magnifications × 200. (B) Quantitation of the ratios of nuclear-to-cell area in the cells depicted in panel A. The nuclear-to-cell volumes were calculated for 10 separate cells/field shown in panel A, and the results are plotted in panel B (± SD). The x-axis numbers in panel B correspond to the label numbers in panel A.
PLZF expression blocks morphologic changes induced by 1,25(OH)2D3.
Morphometric analysis was done by comparing the nuclear (Nuc) with total-cell area as a function of 1,25(OH)2D3 treatment. (A) Giemsa staining of PLZF45 cells. U937-PLZF45 cells were cultured in the presence or absence of tetracycline (− PLZF and + PLZF) with the indicated treatments. Ligand (1,25(OH)2D3[10−8 M]) was added 16 hours after tetracycline was removed to induce PLZF expression. Total-cell area and nuclear area were calculated on the basis of horizontal and vertical measurements of the diameter of the cell nucleus. Ratios were determined for 10 cells/field. Fields shown were representative of those counted. Original magnifications × 200. (B) Quantitation of the ratios of nuclear-to-cell area in the cells depicted in panel A. The nuclear-to-cell volumes were calculated for 10 separate cells/field shown in panel A, and the results are plotted in panel B (± SD). The x-axis numbers in panel B correspond to the label numbers in panel A.
PLZF expression fails to block morphologic changes induced by ATRA.
Morphometric analysis was done as described in the legend to Figure 7. In this case, however, U937-PLZF45 cells were treated with ATRA (10−6 M) for 48 hours. (A) Giemsa staining of PLZF45 cells with the indicated treatments. Original magnifications × 200. (B) Quantitation of nuclear area as a percentage of total-cell area. The x-axis numbers in panel B correspond to the panel numbers in panel A.
PLZF expression fails to block morphologic changes induced by ATRA.
Morphometric analysis was done as described in the legend to Figure 7. In this case, however, U937-PLZF45 cells were treated with ATRA (10−6 M) for 48 hours. (A) Giemsa staining of PLZF45 cells with the indicated treatments. Original magnifications × 200. (B) Quantitation of nuclear area as a percentage of total-cell area. The x-axis numbers in panel B correspond to the panel numbers in panel A.
We were concerned that the well-documented ability of PLZF to cause cell-cycle arrest might affect the differentiation results shown in Figures 6 to 8. Table 1 shows the cell-cycle profile of PLZF45 cells during the 1,25(OH)2D3-dependent treatments illustrated in those figures. At the times described, PLZF induced a G1arrest similar to that induced by 1,25(OH)2D3alone (71.7% versus 72.6%). However, PLZF did not induce CD14, nor did it change the nucleus-to-cytoplasm ratio. PLZF expression also did not inhibit ATRA induction of CD11c or ATRA-mediated morphologic changes. These results strongly suggest that, although PLZF does induce a G1 arrest, it neither blocks all hormone-induced differentiation nor induces general differentiation on its own.
Effects of promyelocytic leukemia zinc finger (PLZF) and 1,25-dihydroxyvitamin D3 (1,25(OH)2D3) on U937 cell cycle
| Treatment . | Cell-cycle phase . | |||
|---|---|---|---|---|
| G1 . | S . | G2/M . | Sub G1 . | |
| None | 54.7 | 31.5 | 12.9 | 0.7 |
| PLZF | 71.7 | 15.3 | 10.8 | 2.3 |
| 1,25(OH)2D3 | 72.6 | 17.0 | 9.9 | 0.6 |
| PLZF and 1,25(OH)2D3 | 81.3 | 9.1 | 8.6 | 0.6 |
| Treatment . | Cell-cycle phase . | |||
|---|---|---|---|---|
| G1 . | S . | G2/M . | Sub G1 . | |
| None | 54.7 | 31.5 | 12.9 | 0.7 |
| PLZF | 71.7 | 15.3 | 10.8 | 2.3 |
| 1,25(OH)2D3 | 72.6 | 17.0 | 9.9 | 0.6 |
| PLZF and 1,25(OH)2D3 | 81.3 | 9.1 | 8.6 | 0.6 |
Values are the average percentage of cells in a given cell-cycle phase. At 48 hours after the beginning of the PLZF–1,25(OH)2D3–all-trans retinoic acid induction experiment, 2 × 105 cells were removed from each growth condition, fixed in ice-cold 70% ethanol, and resuspended in 10 μg/mL propidium iodide and 200 μg/mL ribonuclease A in phosphate-buffered saline. After a 30-minute incubation at 37°C, 1.5 × 103 cells for each condition were analyzed by flow cytometry.
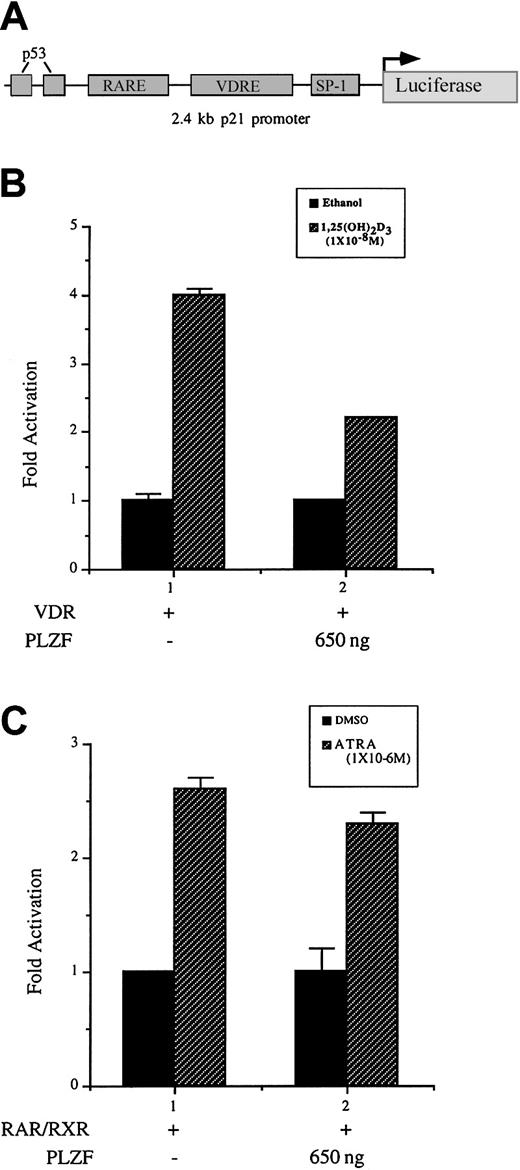
Our study demonstrated that PLZF blocks transcriptional activation of a canonical VDRE reporter construct, revealed a physical interaction between VDR and PLZF, and showed physiologic effects on VDR-dependent processes by PLZF. To more clearly define the role of PLZF in VDR-mediated differentiation, we examined its effect on a direct VDR target gene. Our laboratory previously identified the cyclin-dependent kinase inhibitor p21WAF1/CIP1 gene as a direct transcriptional target of VDR.7 Expression of p21 was also shown to be sufficient to begin differentiation of U937 cells along a macrophage-monocyte lineage. Using a 2.4-kb fragment of the p21 promoter cloned upstream of the luciferase gene (Figure9A), we here observed that coexpression of VDR and PLZF resulted in a reproducible attenuation of 1,25(OH)2D3-mediated activation of p21 transcription (Figure 9B). RARα also can directly transactivate through the p21 promoter.45 Expression of PLZF did not significantly affect the activation of the p21 promoter in response to ATRA and RARα. These data support the results of earlier transfection experiments and protein-protein interaction assays, thereby indicating that PLZF has a selective role in VDR-dependent activation. The findings show a strong correlation between p21 activation and possibly other VDR-dependent target genes and their attenuation by PLZF that results in an alteration of the differentiation course of a cell.
PLZF expression inhibits 1,25(OH)2D3-dependent activation but not ATRA-dependent activation of the p21 promoter in U937 cells.
(A) Schematic diagram of the 2.4-kb fragment of the p21 promoter illustrated in panels B and C. The p21 promoter contained VDREs and a RARE located approximately 800 bp and 1200 bp from the start site of transcription.7 45 (B) PLZF expression reduces VDR-mediated activation of the p21 promoter. The p21 promoter cloned upstream of the luciferase gene was cotransfected with VDR and PLZF in U937 cells. Results are graphed as fold activation. (B) PLZF does not appreciably affect RARα-mediated p21 transcription. U937 cells were cotransfected with RARα, retinoid X receptor (RXR), and the p21 promoter and reporter. Results are graphed as fold activation. RXR was used here because strong ATRA-dependent activation of this promoter required its coexpression.
PLZF expression inhibits 1,25(OH)2D3-dependent activation but not ATRA-dependent activation of the p21 promoter in U937 cells.
(A) Schematic diagram of the 2.4-kb fragment of the p21 promoter illustrated in panels B and C. The p21 promoter contained VDREs and a RARE located approximately 800 bp and 1200 bp from the start site of transcription.7 45 (B) PLZF expression reduces VDR-mediated activation of the p21 promoter. The p21 promoter cloned upstream of the luciferase gene was cotransfected with VDR and PLZF in U937 cells. Results are graphed as fold activation. (B) PLZF does not appreciably affect RARα-mediated p21 transcription. U937 cells were cotransfected with RARα, retinoid X receptor (RXR), and the p21 promoter and reporter. Results are graphed as fold activation. RXR was used here because strong ATRA-dependent activation of this promoter required its coexpression.
Discussion
In this study, we found that PLZF is a potent repressor of VDR-dependent transcriptional activation. At the molecular level, this repression appears to be due to a selective physical interaction between VDR and PLZF. We showed that PLZF and VDR can be coimmunoprecipitated from a hematopoietic precursor cell line and that this interaction appears to occur through the DBD of VDR. Consistent with these results, PLZF was detected in a VDR-VDRE–bound complex in vitro, suggesting that the PLZF-mediated block of VDR transactivation might be due to a PLZF-induced alteration in the nature of the receptor-DNA complex that is unfavorable for VDR-dependent transcriptional activation.
The BTB/POZ domain of PLZF plays an essential role in both the interaction of PLZF and VDR and subsequent repression of transcription.46 In studies in Drosophila, proteins containing BTB/POZ were shown to regulate gene expression through local control of nucleosome positioning and remodeling.47 The BTB/POZ domain of PLZF-RARα APL fusion protein was also implicated in transcriptional repression of ATRA-induced genes.22-24,44 PLZF is highly expressed in immature but not differentiated hematopoietic cells and may play a role in the maintenance of uncommitted precursor cells.19,29-31VDR is widely expressed in hematopoietic cells,32 and we showed that endogenous PLZF and VDR can interact in an undifferentiated leukemic cell line. This suggests that the interaction of PLZF and VDR may reflect a facet of normal cellular physiologic characteristics. Therefore, we propose that, in undifferentiated cells, PLZF could repress VDR function through the BTB/POZ domain.
The BTB/POZ domain is able to associate with several corepressors, notably Sin3A, HDAC1, SMRT, and N-CoR.21-24 28 Many researchers have found that constitutive association of these repressors with PLZF-RARα is responsible for the resistance to ATRA therapy in patients with t(11;17)-derived APL. Surprisingly, our data suggest that repression of VDR is not due to the association of Sin3A, HDAC1, or SMRT (Figure 2 and data not shown). In supporting of this idea is our finding that repression of VDR by PLZF was not sensitive to the addition of TSA (Figure 1A). We could not coimmunoprecipitate Sin3A with anti-VDR antibodies by means of a bridging PLZF molecule, despite the fact that anti-PLZF antibodies coimmunoprecipitated Sin3A effectively (Figure 2C). This result indicates that the fraction of PLZF associated with VDR may not be able to also associate with Sin3A. Our data indicate that deletion of the BTB/POZ domain abolishes the interaction of PLZF with VDR and that the BTB/POZ domain alone can interact with VDR. It is conceivable that VDR interacts with the same or an overlapping region of the BTB/POZ domain as the corepressors and that is why we could not show a 3-way interaction among VDR, PLZF, and the corepressors. We conclude that the mechanism of inhibition of VDR by PLZF does not involve corepressors that work though HDAC mechanisms.
The BTB/POZ domain of PLZF is an obligate homodimer,25,46and we found that when bound to its cognate site, PLZF forms a high-molecular-weight complex that may contain as many as 4 PLZF molecules.48 This information suggests that several molecules of PLZF could form a complex with the VDR when the 2 proteins interact. Hence, PLZF may not block VDR action by recruitment of corepressors but may instead work by steric blockade of the VDR.
PLZF, like many of members of its family, contains not only a BTB/POZ domain but also 9 Krüppel-like zinc fingers (Cys2His2). We showed previously that PLZF can bind a specific DNA site and repress transcription of reporter genes.48,49 Our data suggest that PLZF is present in a DNA-bound complex with VDR. Although PLZF did not bind the VDRE probe used in the assay (data not shown), the transfection data suggest that the zinc fingers are partly responsible for repression of VDR transactivation (Figure 4C). In a previous study,50 the more N-terminal zinc fingers of PLZF-RAR were found to be required for the dominant negative effects of the fusion protein on RARα, thereby implicating these zinc fingers in protein-protein interactions. We conclude that the combined actions of the BTB/POZ domain and zinc finger region of PLZF are responsible for repression of VDR-mediated transcriptional activation.
Through DNA binding to target promoters, VDR activates target genes, affecting a variety of biologic processes, including differentiation of hematopoietic precursors. Many studies have examined this phenomenon by assessing induction of surface-marker antigens known to indicate a particular cell type. CD14 is a marker of 1,25(OH)2D3-mediated monocytic differentiation. We found that contemporaneous expression of PLZF dramatically reduced the number of CD14 molecules expressed per cell, reflecting an alteration in monocytic differentiation. CD14 and CD11c have traditionally been regarded as having similar function (inflammatory and phagocytic response to lipopolysaccharide stimulation) and cell-type expression patterns. However, studies have shown that CD14, but not CD11c, plays a significant role in noninflammatory phagocytosis of apoptotic cells.51 Our data suggest that PLZF selectively regulates the ability of VDR to induce CD14 compared with CD11c (and its family members). Although this idea is unproved, we speculate that PLZF may function to regulate highly specific lineage commitments in precursor hematopoietic cells in response to hormonal signals.
We also showed that morphologic changes associated with monocytic differentiation that occurred when U937 cells were treated with 1,25(OH)2D3 failed to occur when PLZF was present. PLZF may inhibit essential VDR target-gene activation, resulting in an inability to differentiate. Indeed, we also observed that PLZF attenuated 1,25(OH)2D3-induced activation of p21, a factor with an important role during initiation of differentiation in hematopoietic cells. Interruption of transcription of p21 and other VDR target genes9 could result in a strong block of the ability of VDR to mediate differentiation of precursor hematopoietic cells. In contrast, PLZF did not interact with RARα, and it did not inhibit RARα transactivation or the ability of ATRA to induce differentiation of U937 cells.
PLZF was shown previously to be growth suppressive in tumor cell models and may participate in the establishment and maintenance of a state of quiescence in developing mammalian tissues, including maintaining immature hematopoietic precursors.29,30,49,52,53 PLZF has been thought to show this growth-suppressive effect by repressing specific growth-promoting target genes that contain a specific DNA-binding site.48,49 54 We found that PLZF physically interacts with the nuclear-localized transcription factor VDR and that PLZF expression inhibits the ability of 1,25(OH)2D3 to induce specific hematopoietic differentiation. We suggest that, through a combination of these mechanisms, PLZF participates in the establishment of distinct populations of monocytic hematopoietic cells in response to the presence of external endocrine signals. We hope that additional studies will determine whether PLZF-mediated regulation of VDR, or an aberration thereof, participates in APL leukemogenesis.
We thank C. Bromleigh, C. Rachez, G. Farmer, and T. Staeva for helpful discussions; A. Ward for manuscript preparation; the members of the Flow Cytometry Core Facility of Memorial Sloan-Kettering Cancer Center and the Mount Sinai Flow Cytometry Shared Research Facility for their assistance; and V. Richon and F. Sladek for antibodies against mSin3A and the HNF-4 DBD, respectively.
Supported by a grant from the Jack and Susan Rudin foundation (J.O.W), National Institutes of Health grants DK52621 (L.P.F.) and CA 59936 (J.D.L.), American Cancer Society Grant DHP 160, and the Chemotherapy Foundation (J.D.L.).
J.O.W. and M.J.M. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Leonard P. Freedman, Cell Biology Program, Box 470, Memorial Sloan-Kettering Cancer Center, 1275 York Ave, New York, NY 10021; e-mail: l-freedman@ski.mskcc.org.

![Fig. 1. Effects of PLZF and PLZF-RARα on VDR, VP-16, and RARα-mediated transactivation. / (A) Transient overexpression of PLZF or PLZF-RARα represses 1,25(OH)2D3-dependent activation of a VDR responsive reporter (lanes 1-3). This repression was not affected by the addition of 150 ng/mL TSA to the culture medium (lanes 4-6). Plasmids containing complementary DNAs for PLZF, PLZF-RARα, VDR, and a minimal VDR responsive reporter containing 2 copies of a VDRE from the OPN gene, cloned approximately 100 bp upstream of the E1b TATA box and luciferase gene, were used to electroporate U937 cells with the indicated amounts of DNAs. The cells were left for 2 hours before the addition of 1,25(OH)2D3(10−8 M), TSA, or vehicle (ethanol). Data are from a representative experiment done in triplicate. (B) VP16-dependent activation is not affected by PLZF expression. Transfection of U937 cells was carried out as described in the legend for panel A. A GAL4-VP16 fusion was coexpressed with PLZF and a GAL responsive reporter, with the indicated amounts of DNA. (C) PLZF did not repress ATRA-dependent activation of an ATRA responsive reporter by RARα. The reporter used here contained the RARE from the RARβ gene promoter positioned upstream of the Tk promoter and luciferase gene. Transfection was done as described above, and the cells were left for 2 hours before the addition of ATRA (10−6 M) or vehicle (dimethyl sulfoxide [DMSO]). (D) PLZF associates with VDR but not RARα in a ligand-independent manner. In vitro–translated35S-labeled PLZF was incubated at 4°C for 2 hours with affinity-purified GST-VDR or GST-RARα in the presence (+) or absence (−) of 1,25(OH)2D3 or ATRA (10−7M and 10−6 M, respectively). The resulting complexes were washed, separated by SDS–polyacrylamide gel electrophoresis (PAGE), dried, and exposed to film. Lane 1 represents 10% of the labeled PLZF input (I). PLZF did not interact with GST alone (lane 2). PLZF interacted with GST-VDR independent of ligand (lanes 3 and 4) but not with GST- RARα (lanes 5 and 6). All transfections were done at least 3 times in triplicate each time. The histograms show results from a representative experiment from this series. LUC indicates luciferase reporter gene; TK, thymidine kinase promoter; RLU, relative light unit.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/12/10.1182_blood.v98.12.3290/6/m_h82311804001.jpeg?Expires=1763680402&Signature=amdjuaAXgpI3qenm9j53JMwCWCt7~EcOrojStTT1SvEyJdpdJTOG~GHNwL07irov6B6IWymVo7MqmTF62~neLxWga9dTn751QTnI6abq3OrNwHOJkEuJxVZJimAVn6U0woZUm95a-HwFWTtcEJb76ftHMh4WGcvwmC8DhRHoPDCAT1GtjEsXS66dfM0VfbhAlchgjBCOZ4UBOYy4fQwDtNbJE5FggnQU9d9T65t9xxQKGIAPXPWgGm5ucj5AxgExw~s3MnsrG4OljsWDrDzQ1RruOtYFOzloBoJ3udJI5AZy7kU7LKH6oY4qY9mubUTbcRieB2lMLJD-zinf1UXBaQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 4. The PLZF BTB/POZ domain is necessary but not sufficient for repression of VDR-mediated transactivation. / (A) Deletion of the PLZF BTB/POZ domain relieves repression mediated by PLZF of 1,25(OH)2D3-dependent activation. U937 cells were transfected with the indicated amounts of either PLZF (lanes 2 and 3) or PLZF(ΔBTB/POZ) (lanes 4 and 5). Levels of expression of each protein in transfected extracts were similar (data not shown). (B) Expression of the BTB/POZ domain alone is not sufficient to repress 1,25(OH)2D3-dependent activation. A construct containing an epitope (flag)-tagged version of the PLZF BTB/POZ domain was cotransfected with VDR in U937 cells in the indicated DNA amounts. The fBTB/POZ protein was detectable in transfected extracts (data not shown). U937 cells were transfected as described previously and treated with ligand for 24 hours before harvesting. (C) Deletion of the 9 carboxyl zinc fingers in PLZF (PLZFΔZF1-9]) partly relieves PLZF-mediated repression of 1,25(OH)2D3-dependent activation. Protein amounts determined by Western blotting were similar for PLZF and PLZFΔZF1-9 (data not shown). (D) Deletion of the central RD2 domain of PLZF does not affect the ability of PLZF to repress 1,25(OH)2D3-dependent activation. All experiments shown are representative and were done in triplicate.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/12/10.1182_blood.v98.12.3290/6/m_h82311804004.jpeg?Expires=1763680402&Signature=nDZfPuO14d4vhOI2oVi2TDoNzlRFF~OQjSte4dCt2SYNNyBIg6nNiAJA6EaJD1W7KIYEGT3mGNHucDKR2c9KmZdjdSxTwnims523BolM~uGW4U4BpmLWwoIzaflQwnAsx2CtqWj5klnIpM7VFiyaBoWKUOls31B-Qdmh5LkJGn52JPAKTX0SCAleBkOPHzgvOjjWGMXeZSqncZ33ECnWtfk4iFH9RNPaxElKWRkq815EwDtv26MXhy1fQx0~BZwopT8VUysU8vvXDnaltT1VsOevGML~HCe1QxeTZA7tJjU98k31u9Cs0Zuo8e4xJBPa5RMAT~dzlezgB7KDHNwg-g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. PLZF and VDR comigrate in a DNA-bound complex. / (A) Endogenous VDR from U937-PLZF45 nuclear extracts binds to a VDRE with an altered mobility in the presence of PLZF. Nuclear extracts were prepared from U937-PLZF45 cells and incubated in the presence or absence of tetracycline (− PLZF or + PLZF). Then, 5μg nuclear extract was combined with a phosphorus 32–labeled VDRE probe, incubated for 20 minutes at room temperature, and electrophoresed on a nondenaturing polyacrylamide gel. The VDRE probe used in this assay was a DR-3 from the mouse OPN gene promoter. Lane 1 is free probe and lanes 2 to 7 show the specificity of the shifts observed. Lane 2 depicts the 4 major species found in U937 nuclear extracts without PLZF expression, which competed with unlabeled (10-fold excess) specific oligonucleotide (spec) (lane 3) but not a nonspecific oligonucleotide (non-spec) (a glucocorticoid response element [GRE]; lane 4). Lanes 5 through 7 show controls similar to those in lanes 2 to 4 but with use of extracts expressing PLZF. There was a significant change in mobility in the presence of PLZF (lane 5). This shift competed with the specific oligonucleotide (lane 6) and was not affected by the nonspecific oligonucleotide (lane 7). Lanes 8 through 11 show the effects of the addition of antibodies to VDR on the mobility shift. The presence of control IgG antibody had no effect on the shifted species (lane 8). The addition of anti-VDR antibody caused a loss of the major species (open arrow) and a supershift (asterisk) indicating that the complex contained VDR. In the presence of PLZF, another major species (solid arrow) was supershifted (2 asterisks) by anti-VDR antibodies (lane 10 versus lane 11). (B) PLZF is present in a DNA-bound complex containing VDR. EMSA was done as shown in panel A. The addition of anti-PLZF antibody did not affect the shift pattern observed when PLZF was absent (lanes 12 and 13 versus 19). The addition of anti-VDR antibodies confirmed the presence of VDR in a DNA-bound complex. VDR complexes were abrogated by the addition of anti-PLZF antibody (lanes 16 and 17; arrow), indicating that both PLZF and VDR exist in the bound species. The addition of preimmune serum did not affect the shifts in the absence (lane 18) or presence (lane 19) of PLZF.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/12/10.1182_blood.v98.12.3290/6/m_h82311804005.jpeg?Expires=1763680402&Signature=OjsJuE6L1L2CseUZiMfJbVx4IkLbsMzHuNuNvgUA-7cQB~smzh-LOiCmQ1qcBIPA3PsBBpjdsk-WZaafiBtTuabgwTKhKqFwnpHUjp0wprOCzs~gHCqAhRPhzg359AzPHjmBCBDSP7xzE8xzt80xCd9Fs6R7IZSc51Nq8zVciR4VC0KgOpBdFkLrCdTCsjmi4BdtNUbQErNdjvitjG4t2Izis940vXKtSz~1n47xT69eKiUp7lIakC2GK9P~xsLshBTjCCDWFldzicvvzopMTheubFMcKTF7CDm4LjsD8qnjbLWuKoUwUIoSYMzU7YDpV8Z2Y4vKCE-UO8czckiAaA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 7. PLZF expression blocks morphologic changes induced by 1,25(OH)2D3. / Morphometric analysis was done by comparing the nuclear (Nuc) with total-cell area as a function of 1,25(OH)2D3 treatment. (A) Giemsa staining of PLZF45 cells. U937-PLZF45 cells were cultured in the presence or absence of tetracycline (− PLZF and + PLZF) with the indicated treatments. Ligand (1,25(OH)2D3[10−8 M]) was added 16 hours after tetracycline was removed to induce PLZF expression. Total-cell area and nuclear area were calculated on the basis of horizontal and vertical measurements of the diameter of the cell nucleus. Ratios were determined for 10 cells/field. Fields shown were representative of those counted. Original magnifications × 200. (B) Quantitation of the ratios of nuclear-to-cell area in the cells depicted in panel A. The nuclear-to-cell volumes were calculated for 10 separate cells/field shown in panel A, and the results are plotted in panel B (± SD). The x-axis numbers in panel B correspond to the label numbers in panel A.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/12/10.1182_blood.v98.12.3290/6/m_h82311804007.jpeg?Expires=1763680402&Signature=LPbVZuJoBi6TZYRjSHkM7teUXV8KVPjgaufxtvqf28ISQF7EcFUrN2vSO3gc7cE56dAFtm8CkMrxuPFEBXPbLxOsj44XKHJbz6y2c6SEp1ttE~3ZXPMKK~Aa3ibaMcCyQpo6OCQ5-r~5gQ1~ml93aEmz0GeU4hCdQiCy8p86gAgCDdjWK2htzSUrJZbDNGPb9GCAM6eoGuqceh4qlcrHMmLEbjaOTJUaN4sG1QU4n68geD3XtAWOoCu3nm3BhNjWyZ35thhdRWsFZgm95~T6D23owN1-nSG3pgyEP8s5Ywsx7AVaaRuMPVHQ79PLQcqiHSEq~vPnhiH20zy3BBxbnQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal