Mast cells (MCs) and eosinophils are thought to play important roles in evoking allergic inflammation. Cell-type–specific gene expression was screened among 12 000 genes in human MCs and eosinophils with the use of high-density oligonucleotide probe arrays. In comparison with other leukocytes, MCs expressed 140 cell-type–specific transcripts, whereas eosinophils expressed only 34. Among the transcripts for expected MC-specific proteins such as tryptase, major basic protein (MBP), which had been thought to be eosinophil specific, was ranked fourth in terms of amounts of increased MC-specific messenger RNA. Mature eosinophils were almost lacking this transcript. MCs obtained from 4 different sources (ie, lung, skin, adult peripheral blood progenitor–derived and cord blood progenitor–derived MCs, and eosinophils) were found to have high protein levels of MBP in their granules with the use of flow cytometric and confocal laser scanning microscopic analyses. The present finding that MCs can produce abundant MBP is crucial because many reports regarding allergic pathogenesis have been based on earlier findings that MBP was almost unique to eosinophils and not produced by MCs.
Introduction
Mast cells (MCs)1 and eosinophils2 are thought to play important roles in evoking allergic inflammation. Human MCs contribute to allergic inflammation by releasing a variety of mediators and cytokines.3-6 On the other hand, human eosinophils are considered to play a role in the bronchoconstriction of asthma patients through the release of leukotriene C47 and also to damage the bronchial epithelial cells through the release of granule proteins such as major basic protein (MBP).8 These 2 cell types also contribute to allergic inflammation by activating each other. Eosinophil MBP can induce MC degranulation,9 whereas human MCs can activate eosinophils through IgE-dependent production of cytokines such as interleukin-5 (IL-5).10
A draft reading of all human genome sequences has been completed.11,12 It is expected that in the near future, we will resolve previously unanswered questions such as the probability of development of various diseases by screening for single nucleotide polymorphisms over the whole genome sequence. Comprehension of the genome has also accelerated understanding of the transcriptome,13 all of the transcripts present in a cell, and the proteome,14 which controls all of the regulatory elements in a cell. Until recently, it required much time and labor to measure the expression levels of genes even for 100 transcripts. By using recently developed techniques, however, such as cDNA microarrays,15 oligonucleotide expression probe arrays,16 and serial analysis of gene expression,17 such systemic analysis of transcriptomes has become practical.
Among the newly developed techniques, high-density oligonucleotide expression probe array (Genechip; Affymetrix, Santa Clara, CA) is designed to measure the absolute levels of more than 10 000 transcripts regardless of the cell type by using the same set of inner standards on a 1.2-cm2 glass chip. The competition with another cell type required for cDNA microarray assay is not required with the Genechip.16,18-20 Thus, we can compare the expression levels of more than 10 000 transcripts even in different cell types by using the high-density oligonucleotide probe array. In the present study, we used the Genechip to measure and compare cultured human MCs and purified eosinophils for their expression levels of more than 10 000 transcripts. We found that MBP, which has previously been reported to be present only in eosinophils and basophils,21 was abundantly expressed in MCs. We also confirmed that MBP is expressed at both the transcript and protein levels in various types of MCs.
Materials and methods
Subjects
All human subjects in this study provided written informed consent, which was approved by the Ethical Review Board at their hospitals. Nonphagocytic mononuclear cells were separated from adult peripheral blood (PB) or umbilical cord blood (CB) samples by density-gradient centrifugation using Lymphocyte Separation Medium (Organon Teknika, Durham, NC) after depletion of phagocytes with silica (Immuno Biological Laboratories, Fujioka, Japan). The interface containing mononuclear cells was collected. Lineage-negative (Lin−) cells were negatively selected from the peripheral mononuclear cells using a magnetic separation column (MACS II; Miltenyi Biotec, Bergisch Gladbach, Germany) and a mixture of magnetic microbead-conjugated antibodies against CD4, CD8, CD11b, CD14, and CD19 (Miltenyi Biotec), according to the manufacturers' instructions. CD34+ cells were positively selected from CB-derived mononuclear cells using a CD34+ cell isolation kit (Miltenyi Biotec).
Cytokines and antibodies
Recombinant IL-3 was purchased from Intergen (Purchase, NY), and rIL-6 was kindly provided by Kirin Brewery (Maebashi, Japan). Bulk vials of recombinant human stem cell factor (SCF) were purchased from PeproTech EC (London, England). Recombinant IL-4 was purchased from R&D Systems (Minneapolis, MN). Antihuman tryptase monoclonal antibody (MoAb) was purchased from Chemicon (Temecula, CA). Two different anti-MBP MoAbs were purchased from Nichirei (BMK-13; Tokyo, Japan)22 and from Chemicon (AHE-2).23
Cell culture
The cells were suspended in Iscoves modified Dulbecco minimal essential medium (IMDM; Gibco BRL, Grand Island, NY) supplemented with 1% insulin-transferrin-selenium (Gibco BRL), 50 μM 2-ME (Gibco BRL), 1% penicillin+streptomycin (Gibco BRL), and 0.1% bovine serum albumin (BSA; Sigma, St Louis, MO) (complete IMDM). The Lin−106 PB cells were suspended in 0.3 mL of complete IMDM. The cells were mixed well with 2.7 mL serum-free Iscoves methylcellulose medium (Stem Cell Technologies, Vancouver, BC, Canada) supplemented with 200 ng/mL SCF, 50 ng/mL IL-6, and 1 ng/mL IL-3, as described previously.20 The cell suspension was inoculated at 0.3 mL per well to 24-well plates (Iwaki Glass, Tokyo, Japan) at 37°C in 5% CO2. Every 2 weeks, 0.3 mL fresh methylcellulose medium containing 100 ng/mL SCF and 50 ng/mL IL-6 was layered over the methylcellulose cultures. After 6 weeks, the whole cells were retrieved by dissolving the methylcellulose medium with phosphate-buffered saline (PBS). The cells were then suspended and cultured in complete IMDM supplemented with 100 g/mL SCF, 50 ng/mL IL-6, and 2% fetal calf serum (FCS; Cansera, Rexdale, ON, Canada) in 25-cm2flasks (Iwaki Glass). CB-derived CD34+ cells were cultured in the complete IMDM supplemented with 100 ng/mL SCF, 50 ng/mL IL-6, and 2% FCS (Cansera) in 25- or 75-cm2 flasks (Iwaki Glass), as described elsewhere.24
Purification of eosinophils, neutrophils, and mononuclear cells
Eosinophils, neutrophils, and mononuclear cells were separated from venous blood of normal volunteers. Eosinophils were isolated by using Percoll (1.090 g/mL; Pharmacia, Uppsala, Sweden) density centrifugation. The eosinophils were further purified by negative selection with anti-CD16–bound micromagnetic beads, as described previously.25 After this negative selection, the mean eosinophil purity was consistently greater than 99%. Neutrophils were isolated by 2-step density centrifugation. In brief, eosinophils were eliminated by Percoll density centrifugation as a first step. The buoyant fraction was collected and overlaid on Ficoll-Paque (1.077 g/mL; Pharmacia) to eliminate mononuclear cells. The mean neutrophil purity was consistently greater than 99%, and the viability was consistently greater than 95%. Mononuclear cells usually contained lymphocytes with 20% monocytes and 2% basophils.
Intracytoplasmic staining for MBP
Intracytoplasmic staining of eosinophils and CB- and PB-derived MCs was done by the method previously reported.26 Briefly, the cells were fixed with 4% paraformaldehyde, washed, and incubated with PBS-saponin containing 0.1% BSA for 1 hour at 37°C. These cells were then incubated with 50 mg/mL human IgG (ICN Biomedicals, Aurora, OH). Cells were then incubated with 3 μg/mL mouse anti-MBP MoAb (Nichirei) for 1 hour at 4°C, washed, and then incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti–mouse IgG (Becton Dickinson, San Jose, CA) for 30 minutes at 4°C. After staining, all cells were washed and resuspended in PBS containing 0.1% BSA. Control cells were stained with an irrelevant mouse IgG1 (Coulter Immunology, Hialeah, FL). Cell analysis was performed using FACScan and CellQuest software (BD Immunocytometry Systems, San Jose, CA). The distribution of histograms was evaluated as the coefficient of variation.
Confocal laser scanning microscopy
Cells treated with anti-MBP MoAbs (BMK-13) were stained with FITC-conjugated anti–mouse IgG. Confocal imaging was performed with a Fluoview FV300 confocal laser scanning unit (Olympus, Tokyo, Japan) mounted on the inverted-type fluorescence microscope (IX 70; Olympus) with a 60× oil-immersion lens. The images were collected as 1-mm-thick optical sections. In some experiments, differential interference images were also obtained. In one experiment, lung tissue was obtained at surgery (lobectomy) from a patient suffering from multiple lung cysts after informed consent was acquired. The lung tissue was prepared following the method described by Jaffe et al.5 In brief, tissue fragments were incubated in collagenase type II (2 μg/mL) at 37°C for 3 hours. Single-cell suspensions were obtained from the fragments. MCs were semipurified using the MACS system and anti–c-kit MoAb (Pharmingen). MCs were further purified to greater than 99.9% by culturing them in the serum-free IMDM supplemented with 100 ng/mL SCF for 4 weeks.
Immunohistochemical analysis for MBP and tryptase
Skin biopsy samples were obtained from patients with severe atopic dermatitis after informed consent as described above. Sections of 4 μm thick fixed with acetone at 4°C for 5 minutes were incubated with 10% normal goat serum, FITC-labeled anti-MBP MoAb (Nichirei), biotinylated antitryptase MoAb (Chemicon), and rhodamine-conjugated anti–mouse IgG. Negative controls were performed using isotype-matched unrelated antibodies. The results were examined under a fluorescence microscope (Carl Zeiss, Oberkochen, Germany, and Nikon, Tokyo, Japan).
Degranulation assay
MCs were sensitized with 1 μg/mL human myeloma IgE (generously provided by Dr Kimishige Ishizaka, La Jolla, CA) at 37°C for 48 hours in the presence of IL-4. After washing, cells were suspended in Tyrode solution (pH 7.4) containing 124 mM NaCl, 4 mM KCl, 0.64 mM NaH2PO4, 1 mM CaCl2, 0.6 mM MgCl2, 10 mM HEPES, and 0.03% human serum albumin. The cells were preincubated for 10 minutes and then challenged with either 1.5 μg/mL rabbit anti–human IgE (Dako, Glostrup, Denmark) or control Tyrode solution at 37°C for 30 minutes.
Genechip expression analysis
Gene expression was screened for with the Genechip Human Genome U95A probe array (Affymetrix, Santa Clara, CA), which contains the oligonucleotide probe set for approximately 12 000 genes, according to the manufacturer's protocol (Expression Analysis Technical Manual) and previous reports.16 18-20 Total RNA (3-10 μg) was extracted from approximately 107 cells. Double-stranded cDNA was synthesized by using a SuperScript Choice system (Life Technologies, Rockville, MD) and a T7-(dT)24 primer (Amersham Pharmacia Biotech, Buckinghamshire, England). The cDNA was subjected to in vitro transcription in the presence of biotinylated nucleoside triphosphates with the use of a BioArray HighYield RNA Transcript Labeling Kit (Enzo Diagnostics, Farmingdale, NY). The biotinylated cRNA was hybridized with a U95A probe array for 16 hours at 45°C. After washing, the hybridized biotinylated cRNA was stained with streptavidin-phycoerythrin (Molecular Probes, Eugene, OR) and then scanned with the HP Gene Array Scanner. The fluorescence intensity of each probe was quantified using a computer program, Genechip Analysis Suite 3.3 (Affymetrix). The expression level of a single mRNA was determined as the average fluorescence intensity among the intensities obtained by 16 to 20 paired (perfectly matched and single nucleotide–mismatched) primers consisting of 25-base oligonucleotides. If the intensities of mismatched primers are very high, gene expression is judged to be absent even if a high average fluorescence is obtained with the Genechip Analysis Suite 3.3 program.
Reverse transcriptase–polymerase chain reaction for the MBP transcript
Total RNA was isolated from the Isogen (Nippon Gene, Osaka, Japan) solution in which harvested MCs, peripheral eosinophils, or CB-derived immature eosinophils were dissolved. The immature eosinophils were obtained by culturing CB CD34+ cells in the presence of IL-5 plus IL-3, as described previously.27 28 The RNA was DNase-treated, converted to cDNA by reverse transcription (RT) using a kit from Invitrogen (San Diego, CA), and subjected to polymerase chain reaction (PCR) amplification. The primer sequences for MBP were as follows: 5′-GTG CTA AGA CGC TGC CTG AG-3′ for the 5′ primer and 5′-TTT CAG TGG GTT GAC GGC-3′ for the 3′ primer, spanning a fragment of 440 bp. The primer sequences for β-actin were as follows: 5′-TGA CGG GGT CAC CCA CAC TGT GCC-3′ for the 5′ primer and 5′-TAG AAG CAT TTG CGG TGGACG ATG-3′ for the 3′ primer, spanning a fragment of 661 bp (Continental Laboratory Product, San Diego, CA). PCR amplification was performed using a thermal cycler (Geneamp PCR System 9700; PE Biosystems) with an initial denaturation cycle for 1 minute at 94°C; 40 amplification cycles for 1 minute at 94°C, 1 minute at 55°C, and 2 minutes at 72°C; and a final extension phase consisting of one cycle of 10 minutes at 72°C. PCR products were visualized on 0.8% agarose gel (BRL Life Technologies) containing 0.05 μg/mL ethidium bromide (Sigma).
Statistical analysis
Statistical significance between paired groups was determined by the paired Student t test and considered significant forP < .05. Values are expressed as the mean ± SEM.
Results
Detection of MC-specific transcripts by Genechip
As shown in Table 1, we selected MC-specific transcripts by comparing the expression of 12 000 genes among PB-derived MCs, eosinophils, neutrophils, and mononuclear cells containing lymphocytes, monocytes, and basophils with the use of Genechip. Compensation between samples was done with housekeeping genes by referring to the inner standards (legend to Table 1). The MC-specific transcripts were selected by subtracting the compensated values for eosinophils, neutrophils, and mononuclear cells from those for PB-derived MCs. After elimination of redundant transcripts, 140 transcripts were found to be highly MC specific (the subtracted values were greater than 5% of the housekeeping genes).
Top 50 most-increased mast cell–specific transcripts
| Accession number* . | Name of transcript (protein) . | MCs (%AD†) . | Eo (%AD) . | MNC (%AD) . | Neu (%AD) . | Fold change‡ . |
|---|---|---|---|---|---|---|
| M33494 | Tryptase | 197.6 | 0.9 | 0.8 | 1.2 | 205.3 |
| M25915 | Clusterin | 134.0 | 3.1 | 9.4 | 1.2 | 29.5 |
| M16117 | Cathepsin G | 76.7 | 0.7 | 0.7 | < 0.1 | 167.0 |
| Z26248 | Eosinophil granule MBP | 67.9 | 0.5 | 1 | < 0.1 | 135.6 |
| M63138 | Cathepsin D | 107.4 | 9.1 | 20.5 | 12.9 | 7.6 |
| AF150241 | Prostaglandin D2 synthase | 59.7 | < 0.1 | < 0.1 | < 0.1 | 9324.9 |
| M73720 | Carboxypeptidase A | 57.5 | 0.5 | 0.9 | < 0.1 | 123.6 |
| X76534 | Glycoprotein (transmembrane) NMB | 47.4 | 0.1 | 0.1 | < 0.1 | 425.5 |
| AF002672 | Breast cancer suppressor candidate 1 | 45.1 | < 0.1 | 0.1 | < 0.1 | 1145.4 |
| M68891 | GATA-binding protein 2 | 56.7 | 4.8 | 8.5 | 0.8 | 12.0 |
| L76465 | 15 hydroxy-prostaglandin dehydrogenase | 38.4 | 0.6 | 0.9 | < 0.1 | 79.4 |
| D11139 | Tissue inhibitor of metalloproteinases | 52.3 | 3.3 | 10.4 | 3.3 | 9.2 |
| Y00451 | 5-aminolevulinate synthase | 40.4 | 1.6 | 2.5 | 2.2 | 19.1 |
| U18009 | Membrane protein of cholinergic synaptic vesicles | 43.4 | 2.6 | 6.7 | 1.31 | 12.4 |
| AB006780 | Galectin-3 | 49.4 | 3.9 | 13.6 | 0.7 | 8.2 |
| D16583 | L-histidine decarboxylase | 34.8 | 2.1 | 1.5 | < 0.1 | 28.6 |
| M89796 | FcMR I β chain | 29.4 | 0.2 | 0.5 | < 0.1 | 125.2 |
| M26683 | Interferon-γ treatment inducible | 34.4 | 4.6 | 0.5 | 1.0 | 16.8 |
| R92331 | Metallothionein 1E | 41.1 | 3.4 | 5.3 | 4.4 | 9.4 |
| L78833 | BRCA1, Rho7, and vatIgenes | 35.1 | 2.2 | 3.3 | 2.3 | 13.4 |
| X06182 | c-kitproto-oncogene | 27.0 | < 0.1 | 0.1 | < 0.1 | 483.1 |
| X56667 | Calretinin | 28.3 | 1.5 | 0.8 | 1.3 | 23.9 |
| AF022813 | NAG-2, transmembrane-4 superfamily | 29.5 | < 0.1 | 5.5 | < 0.1 | 16.0 |
| D86358 | Siglec6 | 23.3 | < 0.1 | 0.2 | < 0.1 | 411.6 |
| D87119 | GS3955 putative serine/threonine kinase | 38.6 | 1.5 | 13.2 | 0.9 | 7.4 |
| M94345 | Capping protein (actin filament) | 39.2 | 6.4 | 9.6 | 0.9 | 7.0 |
| X51956 | Phosphopyruvate hydratase | 24.3 | 0.2 | 1.9 | < 0.1 | 34.0 |
| AB000584 | TGF-beta superfamily | 22.7 | 0.3 | 0.2 | 0.3 | 85.7 |
| AF001294 | Tumor suppressing subtransferable candidate 3 | 22.9 | 1.0 | 0.3 | < 0.1 | 48.9 |
| J04988 | 90-kd heat shock protein | 50.8 | 2.9 | 26.1 | 0.8 | 5.1 |
| AL050224 | RNA polymerase I | 22.7 | < 0.1 | 1.0 | 1.2 | 29.4 |
| M28225 | Monocyte chemoattractant peptide-1 | 23.2 | 2.1 | 0.3 | 0.6 | 23.5 |
| L35594 | Dynactin subunit | 20.5 | 0.2 | 0.1 | < 0.1 | 209.4 |
| H68340 | RNA helicase-related protein | 25.2 | 1.7 | 3.5 | < 0.1 | 14.4 |
| M69136 | Chymase | 22.1 | 0.4 | 0.6 | 1.3 | 29.4 |
| M15518 | Tissue-type plasminogen activator | 19.9 | < 0.1 | < 0.1 | 0.1 | 242.5 |
| U50136 | Leukotriene C4synthase | 39.9 | 17.2 | 1.8 | 2.1 | 5.7 |
| X64559 | Tetranectin (plasminogen-binding protein) | 21.9 | 1.5 | 1.6 | < 0.1 | 21.2 |
| R93527 | Metallothionein 1H | 31.6 | 3.8 | 5.6 | 3.6 | 7.3 |
| X75252 | Phosphatidylethanolamine binding protein | 25.3 | 0.6 | 7.7 | 0.3 | 8.8 |
| X67951 | Peroxiredoxin 1 | 35.0 | 3.4 | 15.1 | < 0.1 | 5.6 |
| M93311 | Metallothionein 3 | 30.0 | 2.7 | 8.4 | 2.6 | 6.6 |
| AF038844 | MKP-1–like protein tyrosine phosphatase | 16.4 | < 0.1 | 0.6 | < 0.1 | 80.0 |
| U03688 | Dioxin-inducible cytochrome P450 (CYP1B1) | 23.3 | 2.0 | 5.6 | 0.3 | 8.8 |
| U78027 | Bruton's tyrosine kinase | 26.9 | 6.2 | 4.0 | 1.3 | 7.0 |
| K01383 | Metallothionein 1A | 19.0 | < 0.1 | 3.5 | 0.4 | 14.7 |
| M19481 | Follistatin | 24.4 | 3.1 | 1.7 | 4.4 | 7.9 |
| M69177 | Monoamine oxidase B | 15.0 | < 0.1 | 0.2 | 0.2 | 125.0 |
| X06948 | FcεRI α chain | 27.6 | 2.2 | 11.3 | < 0.1 | 6.1 |
| AF045229 | Regulator of G-protein signaling 10 | 24.8 | 0.4 | 10.7 | 0.2 | 6.6 |
| P1 cre recombinase protein (inner standard 1)1-153 | 81.5 | 131.5 | 106.2 | 97.4 | ||
| P1 cre recombinase protein (inner standard 2) | 62.4 | 109.2 | 79.9 | 56.3 | ||
| Mean of the 6 AD† levels of housekeeping genes | 12 071 | 7759 | 16 498 | 9779 |
| Accession number* . | Name of transcript (protein) . | MCs (%AD†) . | Eo (%AD) . | MNC (%AD) . | Neu (%AD) . | Fold change‡ . |
|---|---|---|---|---|---|---|
| M33494 | Tryptase | 197.6 | 0.9 | 0.8 | 1.2 | 205.3 |
| M25915 | Clusterin | 134.0 | 3.1 | 9.4 | 1.2 | 29.5 |
| M16117 | Cathepsin G | 76.7 | 0.7 | 0.7 | < 0.1 | 167.0 |
| Z26248 | Eosinophil granule MBP | 67.9 | 0.5 | 1 | < 0.1 | 135.6 |
| M63138 | Cathepsin D | 107.4 | 9.1 | 20.5 | 12.9 | 7.6 |
| AF150241 | Prostaglandin D2 synthase | 59.7 | < 0.1 | < 0.1 | < 0.1 | 9324.9 |
| M73720 | Carboxypeptidase A | 57.5 | 0.5 | 0.9 | < 0.1 | 123.6 |
| X76534 | Glycoprotein (transmembrane) NMB | 47.4 | 0.1 | 0.1 | < 0.1 | 425.5 |
| AF002672 | Breast cancer suppressor candidate 1 | 45.1 | < 0.1 | 0.1 | < 0.1 | 1145.4 |
| M68891 | GATA-binding protein 2 | 56.7 | 4.8 | 8.5 | 0.8 | 12.0 |
| L76465 | 15 hydroxy-prostaglandin dehydrogenase | 38.4 | 0.6 | 0.9 | < 0.1 | 79.4 |
| D11139 | Tissue inhibitor of metalloproteinases | 52.3 | 3.3 | 10.4 | 3.3 | 9.2 |
| Y00451 | 5-aminolevulinate synthase | 40.4 | 1.6 | 2.5 | 2.2 | 19.1 |
| U18009 | Membrane protein of cholinergic synaptic vesicles | 43.4 | 2.6 | 6.7 | 1.31 | 12.4 |
| AB006780 | Galectin-3 | 49.4 | 3.9 | 13.6 | 0.7 | 8.2 |
| D16583 | L-histidine decarboxylase | 34.8 | 2.1 | 1.5 | < 0.1 | 28.6 |
| M89796 | FcMR I β chain | 29.4 | 0.2 | 0.5 | < 0.1 | 125.2 |
| M26683 | Interferon-γ treatment inducible | 34.4 | 4.6 | 0.5 | 1.0 | 16.8 |
| R92331 | Metallothionein 1E | 41.1 | 3.4 | 5.3 | 4.4 | 9.4 |
| L78833 | BRCA1, Rho7, and vatIgenes | 35.1 | 2.2 | 3.3 | 2.3 | 13.4 |
| X06182 | c-kitproto-oncogene | 27.0 | < 0.1 | 0.1 | < 0.1 | 483.1 |
| X56667 | Calretinin | 28.3 | 1.5 | 0.8 | 1.3 | 23.9 |
| AF022813 | NAG-2, transmembrane-4 superfamily | 29.5 | < 0.1 | 5.5 | < 0.1 | 16.0 |
| D86358 | Siglec6 | 23.3 | < 0.1 | 0.2 | < 0.1 | 411.6 |
| D87119 | GS3955 putative serine/threonine kinase | 38.6 | 1.5 | 13.2 | 0.9 | 7.4 |
| M94345 | Capping protein (actin filament) | 39.2 | 6.4 | 9.6 | 0.9 | 7.0 |
| X51956 | Phosphopyruvate hydratase | 24.3 | 0.2 | 1.9 | < 0.1 | 34.0 |
| AB000584 | TGF-beta superfamily | 22.7 | 0.3 | 0.2 | 0.3 | 85.7 |
| AF001294 | Tumor suppressing subtransferable candidate 3 | 22.9 | 1.0 | 0.3 | < 0.1 | 48.9 |
| J04988 | 90-kd heat shock protein | 50.8 | 2.9 | 26.1 | 0.8 | 5.1 |
| AL050224 | RNA polymerase I | 22.7 | < 0.1 | 1.0 | 1.2 | 29.4 |
| M28225 | Monocyte chemoattractant peptide-1 | 23.2 | 2.1 | 0.3 | 0.6 | 23.5 |
| L35594 | Dynactin subunit | 20.5 | 0.2 | 0.1 | < 0.1 | 209.4 |
| H68340 | RNA helicase-related protein | 25.2 | 1.7 | 3.5 | < 0.1 | 14.4 |
| M69136 | Chymase | 22.1 | 0.4 | 0.6 | 1.3 | 29.4 |
| M15518 | Tissue-type plasminogen activator | 19.9 | < 0.1 | < 0.1 | 0.1 | 242.5 |
| U50136 | Leukotriene C4synthase | 39.9 | 17.2 | 1.8 | 2.1 | 5.7 |
| X64559 | Tetranectin (plasminogen-binding protein) | 21.9 | 1.5 | 1.6 | < 0.1 | 21.2 |
| R93527 | Metallothionein 1H | 31.6 | 3.8 | 5.6 | 3.6 | 7.3 |
| X75252 | Phosphatidylethanolamine binding protein | 25.3 | 0.6 | 7.7 | 0.3 | 8.8 |
| X67951 | Peroxiredoxin 1 | 35.0 | 3.4 | 15.1 | < 0.1 | 5.6 |
| M93311 | Metallothionein 3 | 30.0 | 2.7 | 8.4 | 2.6 | 6.6 |
| AF038844 | MKP-1–like protein tyrosine phosphatase | 16.4 | < 0.1 | 0.6 | < 0.1 | 80.0 |
| U03688 | Dioxin-inducible cytochrome P450 (CYP1B1) | 23.3 | 2.0 | 5.6 | 0.3 | 8.8 |
| U78027 | Bruton's tyrosine kinase | 26.9 | 6.2 | 4.0 | 1.3 | 7.0 |
| K01383 | Metallothionein 1A | 19.0 | < 0.1 | 3.5 | 0.4 | 14.7 |
| M19481 | Follistatin | 24.4 | 3.1 | 1.7 | 4.4 | 7.9 |
| M69177 | Monoamine oxidase B | 15.0 | < 0.1 | 0.2 | 0.2 | 125.0 |
| X06948 | FcεRI α chain | 27.6 | 2.2 | 11.3 | < 0.1 | 6.1 |
| AF045229 | Regulator of G-protein signaling 10 | 24.8 | 0.4 | 10.7 | 0.2 | 6.6 |
| P1 cre recombinase protein (inner standard 1)1-153 | 81.5 | 131.5 | 106.2 | 97.4 | ||
| P1 cre recombinase protein (inner standard 2) | 62.4 | 109.2 | 79.9 | 56.3 | ||
| Mean of the 6 AD† levels of housekeeping genes | 12 071 | 7759 | 16 498 | 9779 |
MCs indicates mast cells; Eo, eosinophil; MNC, mononuclear cell; Neu, neutrophil; MBP, major basic protein; TGF, transforming growth factor; MKP, mitogen-activated protein kinase phosphatase; AD, average difference.
The GenBank accession number (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Nucleotide) of each transcript is shown.
The level of gene expression was obtained as the AD using Genechip software. The percentages of the specific AD level versus the mean AD level of 6 probe sets for housekeeping genes (β-actin and glyceraldehyde-3-phosphate dehydrogenase) were then calculated. The mean percent AD levels obtained from PB-derived MCs (n = 6), eosinophils (Eo, n = 2), mononuclear cells (MNC, n = 4), and neutrophils (Neu, n = 1) are shown.
The fold change was obtained by dividing the MC (mast cell) value by the mean of the 3 other transcript levels (Eo, MNC, and Neu). The transcripts showing at least a 5-fold change are listed in the table.
Inner standards such as PI cre recombinase protein were added to cRNA of the target sample just before the hybridization (see “Materials and methods”) to evaluate the difference between samples. The inner standards were found at similar levels (< 2-fold change) after compensation with the housekeeping genes.
It is interesting that the top 50 PB-derived MC-specific transcripts shown in Table 1 include several genes related to remodeling of tissues, such as tissue-type plasminogen activator (the 36th most-increased transcript in Table 1)29 and tissue inhibitor of metalloproteinases (the 12th most-increased transcript in Table 1),30 as reported previously. Clusterin, an anti-inflammatory multifunctional protein, also called apolipoprotein J,31 was ranked as the second most-increased transcript in Table 1. Clusterin present in MCs is considered to play a role in regression of hemangioma, the most frequent tumor of infancy.32
Detection of eosinophil-specific transcripts by Genechip
By employing the same protocol as for selecting MC-specific transcripts, we judged 34 transcripts to be highly eosinophil specific (we compared eosinophils versus MCs, neutrophils, and mononuclear cells). The top 30 transcripts are shown in Table2. CRTH2, a chemoattractant receptor specifically expressed on type-2 helper T cells and eosinophils,33 was judged to be eosinophil specific. Many increased transcripts in eosinophils overlapped with those in neutrophils and mononuclear cells containing basophils, whereas MCs tended to have unique transcripts compared with neutrophils, mononuclear cells, and eosinophils.
Top 30 increased eosinophil-specific transcripts
| Accession number* . | Name of transcript (protein) . | Eo (%AD†) . | MCs (%AD) . | MNC (%AD) . | Neu (%AD) . | Fold change‡ . |
|---|---|---|---|---|---|---|
| X55988 | Eosinophil-derived neurotoxin | 86.8 | 0.2 | 7.3 | 0.5 | 32.2 |
| L01664 | Charcot-Leyden crystal protein | 78.0 | < 0.1 | 9.2 | 0.2 | 24.5 |
| X81479 | Glycoprotein EMR1 hormone receptor | 59.1 | 1.7 | 5.2 | 2.8 | 18.4 |
| M84526 | Complement factor D | 45.9 | 0.9 | 10.9 | 4.8 | 8.3 |
| J04130 | Macrophage inflammatory protein-1 β | 36.8 | < 0.1 | 6.0 | 1.9 | 14.0 |
| D90144 | Macrophage inflammatory protein-1 α | 41.5 | 0.2 | 4.7 | 8.2 | 9.5 |
| AJ004832 | Neuropathy target esterase | 36.0 | 3.2 | 8.7 | 3.0 | 7.2 |
| U09937 | Urokinase-type plasminogen receptor | 43.6 | 9.3 | 4.6 | 8.6 | 5.8 |
| X70326 | MARCKS-related protein | 42.1 | 2.7 | 10.0 | 10.9 | 5.4 |
| U81375 | Solute carrier family 29 | 29.4 | 10.7 | 0.5 | < 0.1 | 7.9 |
| AB008535 | CRTH2 | 18.2 | 0.6 | 0.4 | < 0.1 | 56.6 |
| M23892 | Arachidonate 15-lipoxygenase | 17.9 | 0.3 | 0.5 | 0.3 | 46.9 |
| AA131149 | S100 calcium-binding protein P | 18.5 | 0.2 | 0.9 | 1.5 | 21.1 |
| AB018339 | Synaptic nuclei expressed gene 1β | 24.3 | 0.8 | 7.3 | 0.4 | 8.5 |
| AF079167 | Lectin-like oxidized LDL receptor | 13.5 | < 0.1 | < 0.1 | 0.2 | 128.9 |
| D13626 | G-protein–coupled receptor for UDP-glucose | 14.2 | 1.1 | 1.0 | 0.7 | 15.0 |
| M34455 | Indoleamine 2,3-dioxygenase | 13.5 | 0.8 | 1.1 | 0.9 | 14.1 |
| X74328 | Cannabinoid receptor 2 | 13.0 | 0.3 | 0.9 | 1.7 | 13.7 |
| L77730 | A3 adenosine receptor | 10.2 | 0.4 | < 0.1 | < 0.1 | 55.5 |
| S59049 | Regulator of G-protein signaling 1 | 12.0 | 2.3 | 1.0 | 0.1 | 10.2 |
| L41816 | CAM kinase I | 10.2 | 1.7 | 1.1 | < 0.1 | 10.7 |
| U21931 | Fructose-1, 6-bisphosphatase 1 | 15.9 | 1.0 | 6.6 | 1.2 | 5.4 |
| U28694 | CCR3 | 16.1 | 0.3 | 1.0 | 8.0 | 5.2 |
| M75914 | IL-5 receptor α chain | 8.8 | 0.7 | 0.7 | 1.1 | 10.5 |
| AF010193 | SMAD7 | 10.9 | 2.2 | 2.4 | 0.2 | 6.7 |
| AF000545 | Purinergic receptor P2Y10 | 14.1 | 0.3 | 3.7 | 4.4 | 5.1 |
| U48250 | Protein kinase C binding protein 2 | 6.2 | 0.1 | 0.1 | 0.4 | 30.4 |
| S68134 | Cyclic AMP-responsive element modulator, β unit | 13.2 | 5.6 | 1.6 | 0.4 | 5.2 |
| L19314 | Phosphorylase kinase, β unit | 7.3 | 1.5 | 0.1 | < 0.1 | 12.7 |
| L08044 | Trefoil factor 3 (intestinal) | 5.6 | < 0.1 | < 0.1 | < 0.1 | 132.9 |
| P1 cre recombinase protein (inner standard 1)2-153 | 131.5 | 81.5 | 106.2 | 97.4 | ||
| P1 cre recombinase protein (inner standard 2) | 109.2 | 62.4 | 79.9 | 56.3 | ||
| Mean of the 6 AD†values | 7759 | 12 071 | 16 498 | 9779 |
| Accession number* . | Name of transcript (protein) . | Eo (%AD†) . | MCs (%AD) . | MNC (%AD) . | Neu (%AD) . | Fold change‡ . |
|---|---|---|---|---|---|---|
| X55988 | Eosinophil-derived neurotoxin | 86.8 | 0.2 | 7.3 | 0.5 | 32.2 |
| L01664 | Charcot-Leyden crystal protein | 78.0 | < 0.1 | 9.2 | 0.2 | 24.5 |
| X81479 | Glycoprotein EMR1 hormone receptor | 59.1 | 1.7 | 5.2 | 2.8 | 18.4 |
| M84526 | Complement factor D | 45.9 | 0.9 | 10.9 | 4.8 | 8.3 |
| J04130 | Macrophage inflammatory protein-1 β | 36.8 | < 0.1 | 6.0 | 1.9 | 14.0 |
| D90144 | Macrophage inflammatory protein-1 α | 41.5 | 0.2 | 4.7 | 8.2 | 9.5 |
| AJ004832 | Neuropathy target esterase | 36.0 | 3.2 | 8.7 | 3.0 | 7.2 |
| U09937 | Urokinase-type plasminogen receptor | 43.6 | 9.3 | 4.6 | 8.6 | 5.8 |
| X70326 | MARCKS-related protein | 42.1 | 2.7 | 10.0 | 10.9 | 5.4 |
| U81375 | Solute carrier family 29 | 29.4 | 10.7 | 0.5 | < 0.1 | 7.9 |
| AB008535 | CRTH2 | 18.2 | 0.6 | 0.4 | < 0.1 | 56.6 |
| M23892 | Arachidonate 15-lipoxygenase | 17.9 | 0.3 | 0.5 | 0.3 | 46.9 |
| AA131149 | S100 calcium-binding protein P | 18.5 | 0.2 | 0.9 | 1.5 | 21.1 |
| AB018339 | Synaptic nuclei expressed gene 1β | 24.3 | 0.8 | 7.3 | 0.4 | 8.5 |
| AF079167 | Lectin-like oxidized LDL receptor | 13.5 | < 0.1 | < 0.1 | 0.2 | 128.9 |
| D13626 | G-protein–coupled receptor for UDP-glucose | 14.2 | 1.1 | 1.0 | 0.7 | 15.0 |
| M34455 | Indoleamine 2,3-dioxygenase | 13.5 | 0.8 | 1.1 | 0.9 | 14.1 |
| X74328 | Cannabinoid receptor 2 | 13.0 | 0.3 | 0.9 | 1.7 | 13.7 |
| L77730 | A3 adenosine receptor | 10.2 | 0.4 | < 0.1 | < 0.1 | 55.5 |
| S59049 | Regulator of G-protein signaling 1 | 12.0 | 2.3 | 1.0 | 0.1 | 10.2 |
| L41816 | CAM kinase I | 10.2 | 1.7 | 1.1 | < 0.1 | 10.7 |
| U21931 | Fructose-1, 6-bisphosphatase 1 | 15.9 | 1.0 | 6.6 | 1.2 | 5.4 |
| U28694 | CCR3 | 16.1 | 0.3 | 1.0 | 8.0 | 5.2 |
| M75914 | IL-5 receptor α chain | 8.8 | 0.7 | 0.7 | 1.1 | 10.5 |
| AF010193 | SMAD7 | 10.9 | 2.2 | 2.4 | 0.2 | 6.7 |
| AF000545 | Purinergic receptor P2Y10 | 14.1 | 0.3 | 3.7 | 4.4 | 5.1 |
| U48250 | Protein kinase C binding protein 2 | 6.2 | 0.1 | 0.1 | 0.4 | 30.4 |
| S68134 | Cyclic AMP-responsive element modulator, β unit | 13.2 | 5.6 | 1.6 | 0.4 | 5.2 |
| L19314 | Phosphorylase kinase, β unit | 7.3 | 1.5 | 0.1 | < 0.1 | 12.7 |
| L08044 | Trefoil factor 3 (intestinal) | 5.6 | < 0.1 | < 0.1 | < 0.1 | 132.9 |
| P1 cre recombinase protein (inner standard 1)2-153 | 131.5 | 81.5 | 106.2 | 97.4 | ||
| P1 cre recombinase protein (inner standard 2) | 109.2 | 62.4 | 79.9 | 56.3 | ||
| Mean of the 6 AD†values | 7759 | 12 071 | 16 498 | 9779 |
MARCKS indicates myristoylated alanine-rich C kinase substrate; LDL, low-density lipoprotein; UDP, uridine 5′-diphosphate; CAM kinase, calmodulin-dependent protein kinase.
The GenBank accession number (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Nucleotide) of each transcript is shown.
The level of gene expression was obtained as the AD using Genechip software. The percentages of the specific AD level versus the mean AD level of 6 probe sets for housekeeping genes (β-actin and glyceraldehyde-3-phosphate dehydrogenase) were then calculated. The mean percent AD levels obtained from cultured eosinophils (Eo, n = 2), PB-derived MCs (n = 6), mononuclear cells (MNC, n = 4), and neutrophils (Neu, n = 1) are shown.
The fold change was obtained by dividing the eosinophil value (Eo) by the mean of the 3 other transcript levels (MCs, MNC, and Neu). The transcripts showing at least a 5-fold change are listed in the table.
Inner standards such as P1 cre recombinase protein were added to cRNA of the target sample just before the hybridization (see “Materials and methods”) to evaluate the difference between samples. The inner standards were found at similar levels (< 2-fold change) after compensation with the housekeeping genes.
Abundant expression of MBP transcripts in MCs but not in eosinophils
Of particular interest is that MBP ranked as the fourth most-increased transcript in MCs but not in eosinophils. Indeed, it has been reported by others34 and by us28 that the level of MBP transcripts in CB-derived developing eosinophils decreases after complete maturation. We also used Genechip to measure the transcripts of major granule proteins in eosinophils derived from 6 patients with atopic dermatitis. The percentage expression levels (per housekeeping gene) of Charcot-Leyden crystal protein, eosinophil-derived neurotoxin, eosinophil cationic protein, and MBP in these eosinophils were, respectively, 150.8% ± 3.4%, 110.1% ± 11.0%, 51.4% ± 21.2%, and 1.6% ± 0.7%, which are relatively higher than the values shown in Table 2.
CB-derived MCs showed lower levels of α- and β-chains of high-affinity receptors for IgE (FcεRI)20 and chymase26 transcripts than PB-derived MCs. However, they had a similar level of MBP (49.9% ± 2.8%, n = 3) compared with adult-type cultured cells (67.9% ± 31.6%, n = 3). Three other transcripts of eosinophil proteins (ie, Charcot-Leyden crystal protein, eosinophil-derived neurotoxin, and eosinophil cationic protein) were judged to be absent in MCs (data not shown).
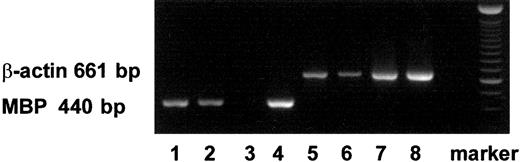
Using RT-PCR, we also detected mRNA expression of MBP in PB- and CB-derived MCs and in immature eosinophils (Figure1).
Expression of transcripts for MBP in MCs and eosinophils with RT-PCR.
The transcripts for MBP (440 bp) and those for β-actin (661 bp) are shown in lanes 1 to 4 and lanes 5 to 8, respectively. The transcripts were obtained from PB-derived MCs (lanes 1, 5), CB-derived MCs (lanes 2, 6), PB-derived primary eosinophils (lanes 3, 7), and CB-derived immature cultured eosinophils (lanes 4, 8).26 27Similar results were obtained with 2 other experiments.
Expression of transcripts for MBP in MCs and eosinophils with RT-PCR.
The transcripts for MBP (440 bp) and those for β-actin (661 bp) are shown in lanes 1 to 4 and lanes 5 to 8, respectively. The transcripts were obtained from PB-derived MCs (lanes 1, 5), CB-derived MCs (lanes 2, 6), PB-derived primary eosinophils (lanes 3, 7), and CB-derived immature cultured eosinophils (lanes 4, 8).26 27Similar results were obtained with 2 other experiments.
The presence of immunoreactive MBP in MC granules
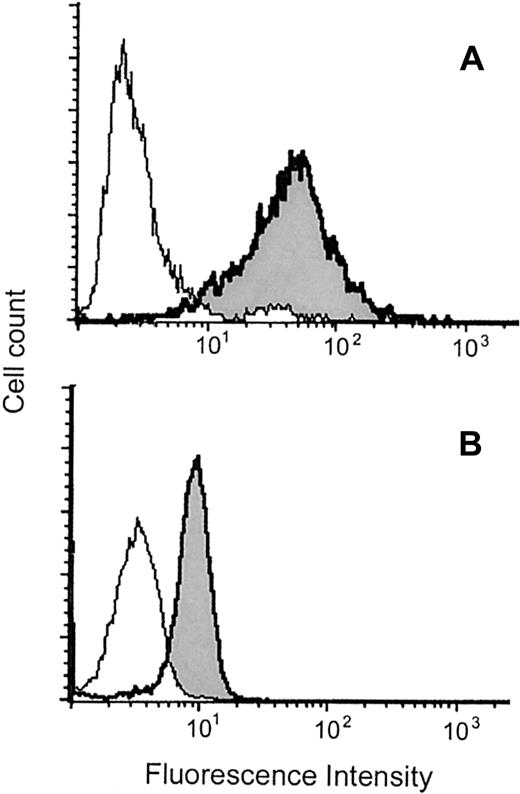
We asked whether MBP is also detected as a protein in MCs. As shown in Figure 2, MBP was detected in saponin-permeabilized MCs by flow cytometry.
Flow cytometric analysis of intracytoplasmic MBP in MCs and eosinophils.
Representative results obtained with permeabilized CB-derived MCs (A) and eosinophils (B) are shown. The histograms indicated as shaded areas with bold lines were obtained using the anti-MBP MoAb (BMK-13), whereas those indicated as open areas with thin lines were obtained using control antibody. The experiments using another anti-MBP MoAb (AHE-2) derived from a different hybridoma gave similar histograms. The mean fluorescence intensity ratios of MBP to control in PB- and CB-derived MCs and eosinophils were, respectively, 14.3 ± 2.1 (n = 10) and 2.4 ± 0.5 (n = 3). The levels of coefficient of variation for MCs and eosinophils were, respectively, 86.2% ± 6.4% and 37.4% ± 5.3% (n = 10 and n = 3), indicating that the intensities of MBP fluorescence in MCs varied widely compared with those in eosinophils.
Flow cytometric analysis of intracytoplasmic MBP in MCs and eosinophils.
Representative results obtained with permeabilized CB-derived MCs (A) and eosinophils (B) are shown. The histograms indicated as shaded areas with bold lines were obtained using the anti-MBP MoAb (BMK-13), whereas those indicated as open areas with thin lines were obtained using control antibody. The experiments using another anti-MBP MoAb (AHE-2) derived from a different hybridoma gave similar histograms. The mean fluorescence intensity ratios of MBP to control in PB- and CB-derived MCs and eosinophils were, respectively, 14.3 ± 2.1 (n = 10) and 2.4 ± 0.5 (n = 3). The levels of coefficient of variation for MCs and eosinophils were, respectively, 86.2% ± 6.4% and 37.4% ± 5.3% (n = 10 and n = 3), indicating that the intensities of MBP fluorescence in MCs varied widely compared with those in eosinophils.
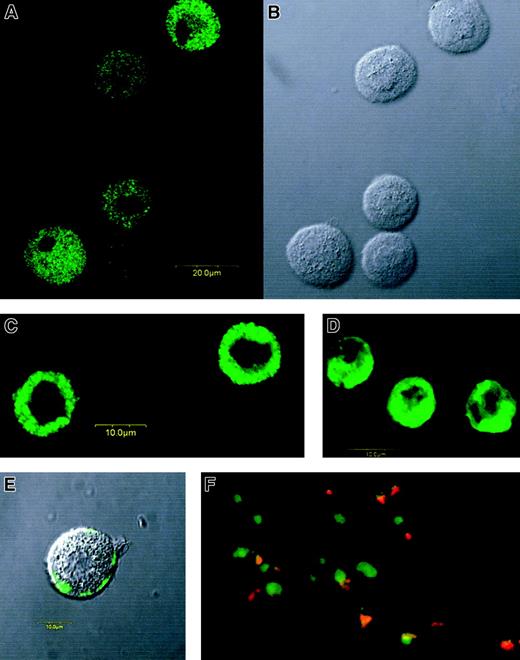
Using a confocal laser scanning microscope, we found MBP not only in purified eosinophils (Figure 3D), but also in CB-derived MCs (Figure 3A,B) and in lung-derived purified MCs (Figure 3C) under the condition that more than 99% of the cells were tryptase positive.
Immunofluorescent staining of MBP in MCs and eosinophils.
Confocal fluorescence image with anti-MBP MoAb (A) and differential interference contrast image (B) of permeabilized CB-derived cultured MCs (1-μm section). Confocal fluorescence images with anti-MBP MoAb of permeabilized lung-derived purified MCs (C) and permeabilized PB-derived purified eosinophils (D). MBP was also detected even in unpermeabilized CB-derived cultured MCs, but only after anti-IgE challenge. (E) Confocal fluorescence and differential interference contrast images were synthesized in the figure. (F) Double staining for MBP (FITC) and tryptase (rhodamine) of a skin biopsy sample obtained from a patient with severe atopic dermatitis. Green images represent MBP-only positive cells, whereas orange-yellow images show MBP and tryptase double-positive cells. Tryptase-only positive cells (red images) were not found in this study, including samples from 2 patients with atopic dermatitis and 2 normal control subjects. Use of a control antibody confirmed all of the positive staining findings to be specific for anti-MBP and antitryptase antibodies.
Immunofluorescent staining of MBP in MCs and eosinophils.
Confocal fluorescence image with anti-MBP MoAb (A) and differential interference contrast image (B) of permeabilized CB-derived cultured MCs (1-μm section). Confocal fluorescence images with anti-MBP MoAb of permeabilized lung-derived purified MCs (C) and permeabilized PB-derived purified eosinophils (D). MBP was also detected even in unpermeabilized CB-derived cultured MCs, but only after anti-IgE challenge. (E) Confocal fluorescence and differential interference contrast images were synthesized in the figure. (F) Double staining for MBP (FITC) and tryptase (rhodamine) of a skin biopsy sample obtained from a patient with severe atopic dermatitis. Green images represent MBP-only positive cells, whereas orange-yellow images show MBP and tryptase double-positive cells. Tryptase-only positive cells (red images) were not found in this study, including samples from 2 patients with atopic dermatitis and 2 normal control subjects. Use of a control antibody confirmed all of the positive staining findings to be specific for anti-MBP and antitryptase antibodies.
We also tested whether MBP in granules of IgE-sensitized MCs is released by anti-IgE stimulation. We could not detect, using flow cytometry, a significant decrease of MBP fluorescence in permeabilized PB- and CB-derived cultured MCs after degranulation. Yet, using confocal laser scanning microscopy, we found MBP on the outside of these unpermeabilized MC membranes after anti-IgE stimulation (Figure3E), suggesting that MBP is released from granules to at least the outside of the membrane when the cells are suspended without aggregation, and that this immunoreactive MBP is quite an adhesive protein. Thirty-three percent to 43% of the histamine content was specifically released from these MCs by anti-IgE stimulation. We never detected MBP in unpermeabilized MCs when anti-IgE stimulation was not performed.
Double staining for tryptase and MBP revealed that most of the cells at the site of allergic inflammation in skin were double positive (Figure3F).
Discussion
To evaluate the significance of proteins present in MCs and eosinophils, we examined genes selectively transcribed in these cells by using high-density oligonucleotide probe arrays that allow measurement of approximately 12 000 kinds of transcripts at once. We found 34 eosinophil-specific transcripts, whereas PB-derived MCs expressed 140 cell-type–specific transcripts under the same criteria of selection. This difference may be due to the fact that the MC lineage is somewhat different from the myeloid lineage including eosinophils and basophils.35 Four isotypes of metallothioneins,36 antiapoptotic proteins, were ranked as the 19th, 39th, 42nd, and 46th most-increased transcripts among the MC-specific genes (Table 1). The expression of these proteins has been reported to be IL-6 dependent,37 suggesting that these proteins might have been up-regulated by IL-6 in the MC culture. Thus, one may claim that increased transcripts in MCs simply reflect the effect of the culture condition. Indeed, when eosinophils cultured with IL-5 for 6 hours were substituted for the unprimed eosinophils shown in Tables 1 and 2, eosinophil-specific transcripts increased to 57 and MC-specific transcripts decreased to 132. However, MC-specific transcripts were still more than double the eosinophil-specific transcripts (data not shown).
The transcript for MBP is almost absent in purified PB eosinophils, whereas transcripts for 4 other major eosinophil granular proteins were expressed at higher levels. Because immature CB-derived eosinophils express MBP mRNA but lose it after maturation,28,34 PB eosinophils seem to be mature enough to produce more MBP. A similar dissociation between protein and mRNA after maturation is also found for histidine decarboxylase38 and chymase26in human MCs.
MBP was ranked as the fourth most-increased transcript in human MCs when approximately 12 000 genes were examined. The average expression level (average difference in fluorescence intensities between 20 perfectly matched and 20 mismatched probes) of MBP transcripts was 59% (average of 6 experiments) of that of the housekeeping genes. As reported previously,20 the expression levels of more than 10% housekeeping genes were highly reproducible using this method. In addition, using RT-PCR, we detected mRNA of MBP in cultured MCs.
In an earlier immunohistochemical study using a polyclonal antibody against MBP, it was reported that the protein is detected only in eosinophils and, in a lesser amount, in basophils, but not in MCs.21 Later, the same group found that MBP is deposited in tissues without eosinophil infiltration in the IgE-mediated immediate-type reaction.39 However, they explained the results as occurring because eosinophils became invisible after degranulation.39 In another report, they also indicated that MBP is detected in some MC types, although they speculated that MCs incorporated free MBP.40 We found that both immunoreactive protein and mRNA of MBP are abundant in MC granules. MBP was always detected in all MC types in this study, including CB-derived, PB-derived, and lung and skin MCs using the 2 MoAbs derived from different hybridomas. The MBP levels varied between individual MCs, whereas the protein levels in eosinophils were relatively constant, as shown in Figure 1. Some MCs might be judged to be negative for MBP if tested by the less sensitive methods employed in the earlier studies.
MBP has been thought to play a key role in allergic inflammation because this highly basic protein is deposited on the damaged lung epithelium of patients with asthma and in the skin of patients with atopic dermatitis.41-43 We could show that MBP present in MC granules was released at least to the outside of the membrane along with histamine release. There are 2 types of immunoreactive MBP, 14-kd MBP and 24-kd proform MBP,44 and the antibody that can differentiate these specific domains is not commercially available. Proform MBP is not cytotoxic because it contains a highly acidic domain in addition to a highly basic MBP domain. We did not have a suitable antibody applicable to immunoblotting for differentiating the 2 MBP proteins. Thus, it is uncertain whether the immunoreactive protein found in MC granules is MBP or proform MBP. In any case, because MBP is assumed to conjugate with highly sulfated heparin proteoglycan of MCs in the event of degranulation, the biologic significance of MBP present in MCs remains to be determined. Moreover, the biologic significance of MBP present in eosinophils remains controversial because, in a murine model of asthma, MBP was recently reported not to contribute to allergic pathogenesis.45 Yet, the present finding that MCs can produce abundant immunoreactive MBP is crucial because many reports regarding allergic pathogenesis have been based on earlier findings that MBP is almost unique to eosinophils and not produced by MCs.21,39 40
In conclusion, we found MBP abundantly expressed in human MCs while we were screening for gene expression among the transcriptomes. This result was unexpected and was somewhat different from our aim. Such unexpected findings may result in changing our understanding of cell biology as well as experimental procedures.
We would like to thank Dr Kiyoshi Kawashima, Dr Shigenobu Shoda, and the staff of the Department of Obstetrics, Gyoda Chuo Hospital, for their continuous support by generously providing umbilical cord blood. We thank Dr Harumi Mukai and Dr Haruhiko Ninomiya at the University of Tsukuba and Dr Kentaro Yoshimatsu at Tsukuba Research Laboratory, Eisai Company Limited, for valuable discussion; and Ms Noriko Hashimoto at National Children's Medical Research Center for her skillful technical assistance.
Supported in part by a grant from the Organization for Pharmaceutical Safety and Research and the Ministry of Health, Labour, and Welfare (the Millennium Genome Project, MPJ-5).
T.N. and K.M. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hirohisa Saito, Department of Allergy & Immunology, National Children's Medical Research Center, 3-35-31 Taishido, Setagaya-ku, Tokyo 154-8509, Japan; e-mail:hsaito@nch.go.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal