The transmembrane 4 superfamily (TM4SF) has come into prominence for its association with a wide range of cell surface molecules, especially integrins. We report that TM4SF molecules CD9, CD63, and CD81 are physically associated with c-kit receptor tyrosine kinase in the human factor–dependent myeloid cell line, MO7e. We characterized this complex using coimmunoprecipitation and colocalization methods. The c-kit coimmunoprecipitated with anti-TM4SF antibodies showed several distinct phenotypes compared to the total c-kit immunoprecipitated with anti–c-kit antibody. These included: (1) higher basal level of tyrosine phosphorylation without elevated kinase activity in the absence of Steel factor (SLF), (2) deficient enhancement of tyrosine phosphorylation and kinase activity in response to SLF, (3) elevated binding rate of SLF shown in chemical cross-linking studies, and (4) little internalization and degradation after SLF treatment. Cocapping studies in living cells showed that c-kit colocalized with TM4SF molecules after SLF stimulation, suggesting confirmation of the biochemical data obtained by the coimmunoprecipitation studies. Colocalization of c-kit with CD81 by SLF was also observed in cord blood CD34+ cells, suggesting the existence of functional units of c-kit in TM4SF complexes in primary hematopoietic cells. This suggests that some TM4SF members may negatively modulate function of c-kit receptor tyrosine kinase and thus regulate receptor sensitivity to SLF in hematopoietic progenitors.
Introduction
Steel factor (SLF) and c-kit interaction is implicated in maintenance of hematopoiesis. SLF is a potent costimulating cytokine that synergizes with other cytokines to stimulate growth of hematopoietic progenitors.1 Both soluble and membrane-bound forms are generated by differential splicing and proteolytic cleavage. Biologically differential effects of these 2 forms have been reported.2,3 c-kit is a member of the type III receptor tyrosine kinase family, which includes receptors for platelet-derived growth factor, macrophage colony-stimulating factor, and Flt3 ligand.4 Interaction of c-kit receptor with its ligand SLF elicits receptor dimerization followed by activation of intrinsic tyrosine kinase and receptor transphosphorylation.5 The tyrosine-phosphorylated c-kit binds an array of specific SH2-containing molecules, which are in turn phosphorylated by the c-kit tyrosine kinase.6 7
Transmembrane 4 superfamily (TM4SF) proteins are cell surface molecules characterized by the presence of 4 highly conserved hydrophobic transmembrane domains, forming 2 unequal sized extracellular loops with intracellular short amino and carboxyl tails. Although their precise functional role is still unclear, involvement in various aspects of cellular function has been suggested.8 A conspicuous feature of TM4SF proteins is their association with each other or other cell surface molecules, where they form a large molecular complex on the cell surface. A number of recent studies have focused on their association with integrin family proteins, suggesting a regulatory role for them in cell adhesive properties.9-13 In particular, the importance of this association for cell migration has been indicated.14-18 In this context, expression of several members of the TM4SF is correlated with metastatic potential of cancer cells.19-22
Hematopoiesis is collaboratively maintained by hematopoietic growth factors and interaction with extracellular components through adhesion receptors such as integrins.23,24 A well-known effect of hematopoietic growth factors includes modulation of integrin function through mechanisms that have not been fully elucidated.25-28 In addition, evidence has been presented that cross-talk and functional overlap exist between cytokine receptors and integrins.29 In a search for a molecular link between 2 types of receptors, we found that c-kit forms a complex with the integrin-associated TM4SF proteins CD9, CD63, and CD81 in factor-dependent cells. The unique characteristics of c-kit in the complex may suggest a novel regulatory mechanism of c-kit function by TM4SF proteins.
Materials and methods
Cytokines and antibodies
Recombinant human SLF was purchased from Biosource International (Camarillo, CA). Recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF) was kindly provided by Immunex (Seattle, WA). Anti-CD9 monoclonal antibody (mAb; clone MM2/57) was obtained from Biodesign International (Kennebuck, MA). Anti-CD31 mAb (clone 5.6E), anti-CD43 mAb (clone DF-T1), anti-CD50 mAb (clone HP2/19), anti-CD81 mAb (clone JS64), fluorescein isothiocyanate (FITC)–conjugated anti-CD63 mAb (clone CLBGran/12) and biotin-conjugated anti-c-kit mAb (clone 17F11) were from Immunotech (Westbrook, ME). Anti-CD63 mAb(1786) was from Chemicon (Temecula, CA). Anti-CD29 mAb (clone 13) was from Becton Dickinson (Bedford MA). Anti-c-kit mAb (cloneYB5.B8) was from Pharmingen (San Diego, CA). Anti–c-kit polyclonal antibody raised against amino acids 958-976 of c-kit of human origin, antigranulocyte-macrophage colony-stimulating factor receptor α (anti–GM-CSFRα) mAb (S-50) and FITC-conjugated anti-CD81 mAb (clone 1.3.3.22) were from Santa Cruz Biotechnology (Santa Cruz, CA). FITC-conjugated anti-CD50 (clone TP1/25.1) and biotin-conjugated anti-CD31 (clone MBC78.2) were from Caltag Laboratories (Burlingame, CA). Antiphosphotyrosine (anti-pTyr) mAb 4G10 and anti–GM-CSFRβ rabbit serum (06-405) were from Upstate Biotechnology (Lake Placid, NY).
Cell lines and cord blood CD34+ cells
The human growth factor–dependent myeloid cell line, MO7e, was cultured in RPMI 1640 supplemented with 20% fetal bovine serum (FBS) and 100 U/mL GM-CSF. The biologic characteristics of this cell line were previously described.30,31 Before SLF stimulation, cells were washed twice, resuspended in RPMI 1640 supplemented with 1% bovine serum albumin (BSA), and incubated for 18 hours at 37°C. Other c-kit+ leukemia cell lines used were HEL and Kasumi-1.32 Cord blood CD34+ cells were obtained as previously described.33 Mononuclear cells were isolated from cord blood by Ficoll-Hypaque (Pharmacia, Piscataway, NJ) gradient centrifugation and were enriched for CD34+ cells (≥ 95% pure) using a magnetic cell sorting (MACS) system (Miltenyi Biotec, Auburn, CA).
Immunoprecipitation and immunoblotting
Cells were lysed with lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 μg/mL aprotinin, 10 μg/mL leupeptin, 100 mM NaF, and 1 mM sodium orthovanadate) containing 1% CHAPS (Calbiochem, San Diego, CA) on ice for 20 minutes, except where otherwise indicated. Insoluble fractions were removed by centrifugation and the supernatants were precleared with protein A-Sepharose (Pharmacia) or protein G-agarose (Santa Cruz) beads coated with normal IgG of the same species as the antibody used for immunoprecipitation. The lysates were then incubated with protein A-Sepharose or protein G-agarose beads precoated with each antibody. Immunoprecipitates were washed with lysis buffer extensively and separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA). For detection of TM4SF proteins, nonreducing conditions were used. Membranes were blocked in Tris-buffered saline containing 0.5% Tween 20 and 1% BSA for 1 hour at room temperature and incubated with appropriate primary antibodies for 2 hours. Blots were visualized using horseradish peroxidase–conjugated secondary antibody and an enhanced chemiluminescence system (Amersham, Piscataway, NJ). To reprobe with another first antibody, membranes were incubated in striping buffer (62.5 mM Tris-HCl, [pH 6], 100 mM 2-mercaptoethanol [2-ME], and 2% SDS) for 30 minutes at 50°C, washed, and then used for further study. For reprecipitation experiments, immunoprecipitated proteins with anti-TM4SF antibodies from CHAPS cell lysate were eluted with NP-40 lysis buffer. Then, eluted proteins were subjected to additional immunoprecipitation with anti–c-kit antibody.
Immune complex kinase assay
Immune complex kinase assay was performed as previously described.2 The immunoprecipitates were washed 3 times with lysis buffer and 3 times with kinase buffer (50 mM Tris-HCl, pH 7.4, 25 mM β-glycerophosphate, 2 mM sodium orthovanadate, 1 mmol/L dithiothreitol, 10 mmol/L MgCl2). Ten microcuries of [32P-γ-adenosine triphosphate was added and incubated for 10 minutes at room temperature. The reaction was terminated by addition of SDS-PAGE sample buffer containing 2-ME and boiled for 5 minutes.
Surface cross-linking
Steel factor was labeled with 125I (Amersham) using IODO-BEADS (Pierce, Rockford, IL) according to the manufacturer's instruction. MO7e cells were stimulated with 125I-labeled SLF at 37°C for indicated time periods and then cross-linked with 2 mmol/mL bis (sulfosuccinimidyl) suberate (BS3, Pierce) for 2 hours at 4°C. After washing with phosphate-buffered saline (PBS), the reaction was terminated with 20 mM Tris-HCl.
Internalization of 125I-SLF
Internalization was assessed as previously described.34 After stimulation with125I-labeled SLF, MO7e cells were resuspended for 5 minutes on ice-cold PBS containing 1 mg/mL BSA or on the same buffer adjusted to pH 3.7 with acetic acid. Then the cells were washed with binding buffer at pH 7.4, and the amount of total and internalized (acid nonreleasable) cell-associated radioactivity was quantitated with a 5500B Beckman γ counter (Beckman Instruments, Fullerton, CA).
Metabolic labeling
The method of metabolic labeling was described elsewhere.34 Exponentially growing MO7e cells were washed and resuspended in methionine- and cysteine-free RPMI 1640 containing 5% FBS and 100 U/mL GM-CSF for 1 hour, then 100 μCi/mL (3.7 MBq/mL) Tran35S-Label (ICN Pharmaceuticals, Costa Mesa, CA) was added and culture was continued for 3 hours. After in vivo labeling, cells were washed 3 times and resuspended RPMI 1640 containing 5% FBS with a 10-fold molar excess of unlabeled methionine and incubated for 1 hour. The cells were then treated with 50 ng/mL SLF for indicated time periods at 37°C.
Surface biotinylation
MO7e cells were treated with or without 50 ng/mL SLF for 90 minutes at 37°C. After washing 3 times with PBS, cell surface proteins were labeled with Sulfo-NHS-LC-biotin (Pierce) according to the manufacturer's protocol.
Analysis by a confocal laser scanning microscopy
Colocalization assay was performed as previously described with modifications.35 Cells were first incubated with mAb at 4°C for 30 minutes and then labeled with FITC-conjugated F(ab′)2 goat antimouse IgG. After washing, the cells were incubated at 37°C for 30 minutes to induce capping. For cord blood cells, we used FITC-conjugated first antibodies and purified F(ab′)2 sheep antimouse IgG (Sigma, St Louis, MO) for capping induction. Then, the cells were blocked with mouse immunoglobulins and labeled with biotin-conjugated second mAb and tetrarhodamine isothiocyanate (TRITC)–conjugated ExtraAvidin (Sigma) at 4°C. After washing, the cells were fixed with 2% formalin in PBS, and intensities of fluorescence from FITC and TRITC were visualized separately by confocal laser scanning microscopy (CSU10; Yokokawa Electric, Tokyo, Japan) using a krypton/argon laser.
Results
c-kit forms a complex with the TM4SF proteins CD9, CD63, and CD81 in MO7e cells
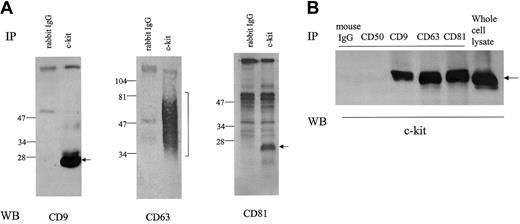
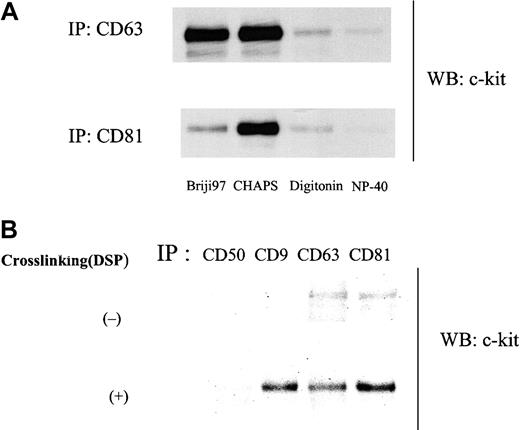
As shown in Figure 1A, anti–c-kit antibody coimmunoprecipitated TM4SF proteins CD9, CD63, and CD81 from the cell lysate of MO7e in the presence of the mild detergent CHAPS. In reciprocal experiments, we could detect c-kit protein in CD9, CD63, and CD81 immunoprecipitates (Figure 1B). To investigate the potential association of other cell surface molecules with CD9 or CD81, we probed for coimmunoprecipitation using antibodies against CD31, CD43, CD50, and GM-CSFRα and β chains. We did not detect CD9 or CD81 in the immunoprecipitates with these mouse or rabbit antibodies by this method (data not shown), suggesting specificity of the association of c-kit with TM4SF proteins in MO7e cells. As for integrin β1, its association with CD151, a member of TM4SF, has been reported by others in MO7e cells.13 We also detected the association of integrin β1 with CD9 and CD81 (data not shown). Extensive immunodepletion with anti–c-kit antibody had little effect on CD9 and CD81 amounts coimmunoprecipitated with anti-integrin antibody and vice versa (data not shown), suggesting that at least part of c-kit/TM4SF complexes are distinct from integrin/TM4SF complexes. As a rule, TM4SF associations reported so far are detergent dependent. Thus, we examined the influence of solubilizing detergents on c-kit/TM4SF association. Amounts of c-kit associated with both CD63 and CD81 were substantially diminished in digitonin and NP-40 lysates compared with CHAPS lysates. Briji97 reduced the stability of c-kit/CD81 but not c-kit/CD63 association (Figure2A). In the case of CD9, a similar pattern was observed as with CD81 (data not shown). Even under stringent detergent conditions (1% NP40 + 0.1% SDS + 0.5% sodium deoxycholate), we could detect low levels of c-kit association with CD63 or CD81 but not with CD9 (Figure 2B). With chemical cross-linking with dithiobis (succinimiddyl proprionate) (DSP) before lysis and immunoprecipitation, c-kit amounts in CD9 immunoprecipitate were almost equal to those in CD63 or CD81 immunoprecipitates. In reciprocal experiments with biotin-labeled MO7e cells under stringent detergent conditions (1% NP40 + 0.1% SDS + 0.5% sodium deoxycholate), precipitation with c-kit antibody coprecipitated a band of the molecular size of CD81. However, we could not confirm that the band was CD81, perhaps because the actual CD81 protein levels were below the limit of detection with Western blotting. Despite the similar levels of expression of CD9, CD63, CD81, and c-kit in HEL cells compared with those in MO7e cells, we could not detect the TM4SF molecules in the c-kit immunoprecipitates from HEL cell lysate. However, small amounts of CD81 were coimmunoprecipitated with c-kit in Kasumi-1 cells, which expressed undetectable levels of CD9 and CD63 by FACS analysis (data not shown). These data suggest that association of TM4SF proteins with c-kit may be cell type specific.
Association of c-kit with TM4SF molecules in MO7e cells.
Cells were solubilized with lysis buffer containing 1% CHAPS. (A) Cell lysates were immunoprecipitated with normal rabbit IgG or anti–c-kit polyclonal antibody. After being separated by SDS-PAGE under nonreducing conditions, proteins were immunoblotted with anti-CD9, anti-CD63, or anti-CD81 mAb. (B) The cell lysates were immunoprecipitated with indicated antibodies. Anti-CD50 (ICAM-3) mAb was used as negative control. After being separated by SDS-PAGE, proteins were immunoblotted with anti–c-kit polyclonal antibody. These are the representative results from 3 separate experiments.
Association of c-kit with TM4SF molecules in MO7e cells.
Cells were solubilized with lysis buffer containing 1% CHAPS. (A) Cell lysates were immunoprecipitated with normal rabbit IgG or anti–c-kit polyclonal antibody. After being separated by SDS-PAGE under nonreducing conditions, proteins were immunoblotted with anti-CD9, anti-CD63, or anti-CD81 mAb. (B) The cell lysates were immunoprecipitated with indicated antibodies. Anti-CD50 (ICAM-3) mAb was used as negative control. After being separated by SDS-PAGE, proteins were immunoblotted with anti–c-kit polyclonal antibody. These are the representative results from 3 separate experiments.
Influence of solubilizing detergents on c-kit/TM4SF association.
(A) Cells were solubilized with lysis buffer containing 1% indicated detergents. Cell lysates were immunoprecipitated with anti-CD63 or anti-CD81 mAb. Immunoprecipitated proteins were separated by SDS-PAGE and immunoblotted with anti–c-kit polyclonal antibody. (B) Cells were solubilized with lysis buffer containing 1% NP40 plus 0.1% SDS plus 0.5% sodium deoxycholate with or without pretreatment with 0.5 mM DSP and submitted to immunoprecipitation with indicated antibodies. After being separated by SDS-PAGE, proteins were immunoblotted with anti–c-kit polyclonal antibody. These are both the representative results from 3 separate experiments.
Influence of solubilizing detergents on c-kit/TM4SF association.
(A) Cells were solubilized with lysis buffer containing 1% indicated detergents. Cell lysates were immunoprecipitated with anti-CD63 or anti-CD81 mAb. Immunoprecipitated proteins were separated by SDS-PAGE and immunoblotted with anti–c-kit polyclonal antibody. (B) Cells were solubilized with lysis buffer containing 1% NP40 plus 0.1% SDS plus 0.5% sodium deoxycholate with or without pretreatment with 0.5 mM DSP and submitted to immunoprecipitation with indicated antibodies. After being separated by SDS-PAGE, proteins were immunoblotted with anti–c-kit polyclonal antibody. These are both the representative results from 3 separate experiments.
Tyrosine kinase of c-kit is functionally inactive in the TM4SF complex
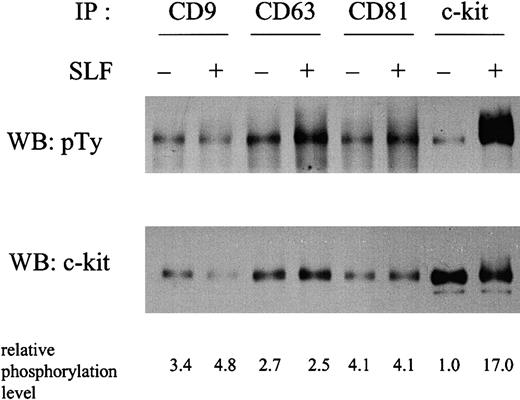
To investigate the possible functional relevance of the c-kit–TM4SF association, we examined the tyrosine phosphorylation status of the c-kit associated with TM4SF molecules in the absence or presence of SLF stimulation in MO7e cells (Figure3). Compared with the total c-kit immunoprecipitated with anti–c-kit antibody, the c-kit associated with TM4SF molecules showed a higher basal level of tyrosine phosphorylation without SLF stimulation. Taking into account the amount of c-kit protein in each lane, an approximately 3- to 4-fold increase of phosphorylation was observed in the fraction of c-kit associated with TM4SF molecules. Relative phosphorylation levels after normalization by protein amount are shown in the bottom of Figure 3. To rule out the possibility of other tyrosine phosphorylated bands overlapping the c-kit band, we used reimmunoprecipitation methods. After second immunoprecipitation with anti–c-kit antibody, we observed essentially the same tyrosine phosphorylation level in the c-kit associated with TM4SF proteins (data not shown). To examine the potential contribution of endogenous SLF to the phosphorylation of the c-kit in the TM4SF complex, we performed reverse transcription–polymerase chain reaction analysis as previously described.36 We could not detect SLF expression in MO7e cells (data not shown). SLF induced a 17-fold enhancement of tyrosine phosphorylation in the total c-kit fraction. However, such enhancement was almost completely suppressed in the c-kit fraction associated with TM4SF molecules. We consistently observed less than 2-fold SLF enhancement of phosphorylation of the c-kit associated with TM4SF molecules. No significant changes in the tyrosine phosphorylation levels of the c-kit associated with TM4SF molecules were observed at 2, 5, 10, 30, and 120 minutes after SLF stimulation (data not shown). These results led us to investigate the kinase activity of c-kit by immune complex kinase assay. Because TM4SF proteins possibly associate with other kinase activities,11 we took advantage of reimmunoprecipitation methods to eliminate such activities. After a second immunoprecipitation with anti–c-kit mAb YB5.B8, the immunoprecipitates were subjected to immune complex kinase assay. We found that the higher basal phosphorylation level of c-kit in the TM4SF complex in the absence of SLF led to no elevated kinase activity (Figure4). Furthermore, in contrast to the total c-kit fraction, no enhancement of c-kit kinase activity by SLF was observed in any of the TM4SF complexes (Figure 4). These results suggest that the tyrosine kinase activity of c-kit is functionally inactive in the TM4SF complex.
Tyrosine phosphorylation status of TM4SF-associated and total c-kit fractions in the absence or the presence of SLF.
MO7e cells were stimulated with or without 50 ng/mL SLF at 37°C for 5 minutes. Cells were solubilized with lysis buffer containing 1% CHAPS. Cell lysates were immunoprecipitated with indicated antibodies. After being separated by SDS-PAGE, proteins were immunoblotted with antiphosphotyrosine antibody (upper panel). The same membrane was reprobed with anti–c-kit polyclonal antibody (lower panel). The relative tyrosine phosphorylation levels normalized by c-kit protein amounts shown in lower panel were indicated at the bottom. These are the representative results from 3 separate experiments.
Tyrosine phosphorylation status of TM4SF-associated and total c-kit fractions in the absence or the presence of SLF.
MO7e cells were stimulated with or without 50 ng/mL SLF at 37°C for 5 minutes. Cells were solubilized with lysis buffer containing 1% CHAPS. Cell lysates were immunoprecipitated with indicated antibodies. After being separated by SDS-PAGE, proteins were immunoblotted with antiphosphotyrosine antibody (upper panel). The same membrane was reprobed with anti–c-kit polyclonal antibody (lower panel). The relative tyrosine phosphorylation levels normalized by c-kit protein amounts shown in lower panel were indicated at the bottom. These are the representative results from 3 separate experiments.
Immune complex kinase activity of TM4SF-associated and total c-kit fractions in the absence or the presence of SLF.
MO7e cells were stimulated with or without 50 ng/mL SLF at 37°C for 10 minutes. Cells were solubilized with lysis buffer containing 1% CHAPS. The cell lysates were immunoprecipitated with indicated first antibodies. Immunoprecipitated materials were eluted with 1% NP-40 lysis buffer and supernatants were immunoprecipitated with anti–c-kit mAb (cloneYB5.B8). In the case of anti–c-kit mAb as a first immunoprecipitation antibody, immunoprecipitated proteins were treated with NP-40 in the same way. The immune complexes were incubated with 10 μCi (0.37 MBq) [32P-γ] adenosine triphosphate for 10 minutes at room temperature. Immune complexes were separated by SDS-PAGE and transferred onto Immobilon-P membrane. [32P] incorporation into c-kit protein was visualized by autoradiography (upper panel). The same membrane was immunoblotted with anti–c-kit polyclonal antibody (lower panel). These are the representative results from 2 separate experiments.
Immune complex kinase activity of TM4SF-associated and total c-kit fractions in the absence or the presence of SLF.
MO7e cells were stimulated with or without 50 ng/mL SLF at 37°C for 10 minutes. Cells were solubilized with lysis buffer containing 1% CHAPS. The cell lysates were immunoprecipitated with indicated first antibodies. Immunoprecipitated materials were eluted with 1% NP-40 lysis buffer and supernatants were immunoprecipitated with anti–c-kit mAb (cloneYB5.B8). In the case of anti–c-kit mAb as a first immunoprecipitation antibody, immunoprecipitated proteins were treated with NP-40 in the same way. The immune complexes were incubated with 10 μCi (0.37 MBq) [32P-γ] adenosine triphosphate for 10 minutes at room temperature. Immune complexes were separated by SDS-PAGE and transferred onto Immobilon-P membrane. [32P] incorporation into c-kit protein was visualized by autoradiography (upper panel). The same membrane was immunoblotted with anti–c-kit polyclonal antibody (lower panel). These are the representative results from 2 separate experiments.
Binding of SLF and subsequent dimerization occurs effectively in the c-kit associated with TM4SF proteins
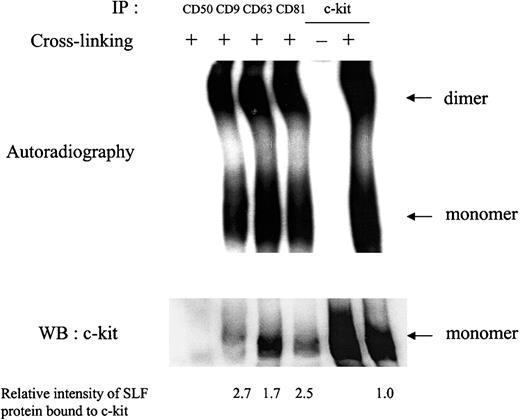
To assess whether the impairment in c-kit response to SLF when c-kit is associated with TM4SF proteins was a result of deficiency of SLF binding or dimerization, we performed chemical cross-linking studies. After treatment with 125I-labeled SLF, cell surface proteins were cross-linked with a membrane impermeable cross-linker BS3. Anti-TM4SF antibodies coimmunoprecipitated SLF-bound monomeric and dimeric forms of c-kit (Figure 5), suggesting that binding of SLF and subsequent dimerization occurred effectively in the complex. The relative intensity of radiolabeled SLF bound to c-kit was approximately 2- or 3-fold higher in the c-kit associated with TM4SF compared with that of the total c-kit after normalization by c-kit protein amount shown in Western blotting (Figure 5). In this experiment, we used saturating concentrations of SLF.
Ligand binding and dimerization after SLF treatment.
MO7e cells were treated with 125I-labeled SLF at 37°C for 5 minutes. Then, cell surface proteins were cross-linked with 2 mM BS3. Cells were solubilized with lysis buffer containing 1% CHAPS and processed for immunoprecipitation with indicated antibodies. After being separated by SDS-PAGE, c-kit bound to radiolabeled SLF was detected by autoradiography (upper panel). The same membrane was immunoblotted with anti–c-kit polyclonal antibody (lower panel). Relative SLF binding rates normalized by the protein amounts in lower panel were shown at the bottom. These are the representative results from 2 separate experiments.
Ligand binding and dimerization after SLF treatment.
MO7e cells were treated with 125I-labeled SLF at 37°C for 5 minutes. Then, cell surface proteins were cross-linked with 2 mM BS3. Cells were solubilized with lysis buffer containing 1% CHAPS and processed for immunoprecipitation with indicated antibodies. After being separated by SDS-PAGE, c-kit bound to radiolabeled SLF was detected by autoradiography (upper panel). The same membrane was immunoblotted with anti–c-kit polyclonal antibody (lower panel). Relative SLF binding rates normalized by the protein amounts in lower panel were shown at the bottom. These are the representative results from 2 separate experiments.
The c-kit in the TM4SF complex is neither internalized nor degraded after SLF treatment
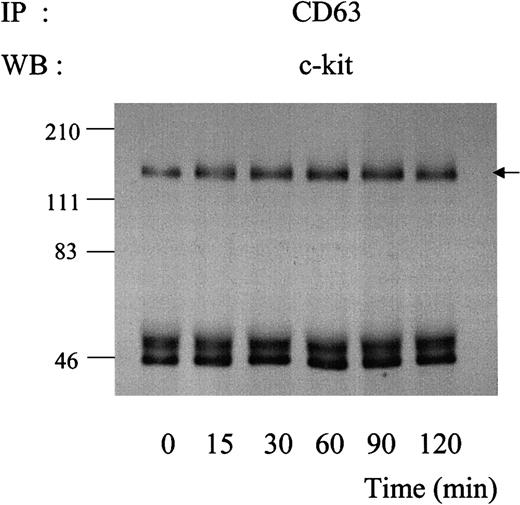
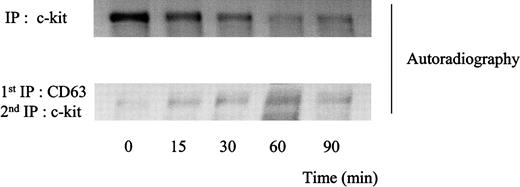
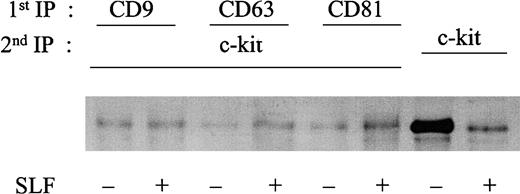
Binding of SLF is known to result in rapid kinase activity-dependent c-kit internalization and degradation.34,37 We investigated the kinetics of association of c-kit/TM4SF proteins after SLF stimulation. As shown in Figure 6, amounts of c-kit associated with CD63 were not changed up to 2 hours after treatment with SLF. Similar results were obtained in the cases of CD9 or CD81 (data not shown). Using 35S-methionine-cysteine metabolic labeling, we examined whether the stable association indicated that the c-kit associated with TM4SF was not degraded after SLF treatment. Consistent with our previous finding,34 the density of the total c-kit protein rapidly decreased during exposure to SLF. In contrast, the density of the c-kit associated with CD63 was almost constant, suggesting that little degradation occurred in the TM4SF complex after stimulation with SLF (Figure 7). Next, we examined whether the c-kit in the complex was internalized. We used surface biotinylation as an indicator for cell-surface c-kit protein expression. SLF caused no reduction of the biotinylation level of the c-kit in the TM4SF complex, whereas that of the total c-kit was greatly reduced (Figure 8). These results indicated that the c-kit associated with TM4SF molecules was not internalized but was still on the cell surface even after SLF treatment.
Kinetics of association of c-kit with CD63 during exposure to SLF.
MO7e cells were treated with 50 ng/mL SLF at 37°C for indicated time periods. Cell lysates were immunoprecipitated with anti-CD63 antibody. After being separated by SDS-PAGE, immunoprecipitated proteins were blotted with anti–c-kit polyclonal antibody. These are the representative results from 2 separate experiments.
Kinetics of association of c-kit with CD63 during exposure to SLF.
MO7e cells were treated with 50 ng/mL SLF at 37°C for indicated time periods. Cell lysates were immunoprecipitated with anti-CD63 antibody. After being separated by SDS-PAGE, immunoprecipitated proteins were blotted with anti–c-kit polyclonal antibody. These are the representative results from 2 separate experiments.
Degradation patterns of c-kit protein after stimulation of MO7e cells with SLF.
Cellular proteins were labeled with35S-methionine-cysteine. Thereafter, the cells were treated with 50 ng/mL SLF at 37°C for indicated time periods. The cells were solubilized with lysis buffer containing 1% CHAPS and processed for immunoprecipitation with anti-CD63 mAb or anti–c-kit polyclonal antibody. CD63 immunoprecipitates were eluted with 1% NP-40 lysis buffer and supernatants were immunoprecipitated with anti–c-kit polyclonal antibody. Immunoprecipitated proteins were separated by SDS-PAGE and detected by autoradiography. These are the representative results from 2 separate experiments.
Degradation patterns of c-kit protein after stimulation of MO7e cells with SLF.
Cellular proteins were labeled with35S-methionine-cysteine. Thereafter, the cells were treated with 50 ng/mL SLF at 37°C for indicated time periods. The cells were solubilized with lysis buffer containing 1% CHAPS and processed for immunoprecipitation with anti-CD63 mAb or anti–c-kit polyclonal antibody. CD63 immunoprecipitates were eluted with 1% NP-40 lysis buffer and supernatants were immunoprecipitated with anti–c-kit polyclonal antibody. Immunoprecipitated proteins were separated by SDS-PAGE and detected by autoradiography. These are the representative results from 2 separate experiments.
Biotinylation of c-kit protein in the absence or presence of SLF.
MO7e cells were treated with or without 50 ng/mL SLF for 90 minutes at 37°C. Then, cell surface proteins were labeled with biotin. The cells were solubilized with lysis buffer containing 1% CHAPS. Cell lysates were immunoprecipitated with indicated first antibodies. Immunoprecipitated materials were eluted with 1% NP-40 lysis buffer and supernatants were immunoprecipitated with anti–c-kit polyclonal antibody. Immunoprecipitated proteins were separated by SDS-PAGE and transferred onto Immobilon-P membrane. Biotin-labeled proteins were revealed with streptavidin-biotinylated horseradish peroxidase complex. These are the representative results from 2 separate experiments.
Biotinylation of c-kit protein in the absence or presence of SLF.
MO7e cells were treated with or without 50 ng/mL SLF for 90 minutes at 37°C. Then, cell surface proteins were labeled with biotin. The cells were solubilized with lysis buffer containing 1% CHAPS. Cell lysates were immunoprecipitated with indicated first antibodies. Immunoprecipitated materials were eluted with 1% NP-40 lysis buffer and supernatants were immunoprecipitated with anti–c-kit polyclonal antibody. Immunoprecipitated proteins were separated by SDS-PAGE and transferred onto Immobilon-P membrane. Biotin-labeled proteins were revealed with streptavidin-biotinylated horseradish peroxidase complex. These are the representative results from 2 separate experiments.
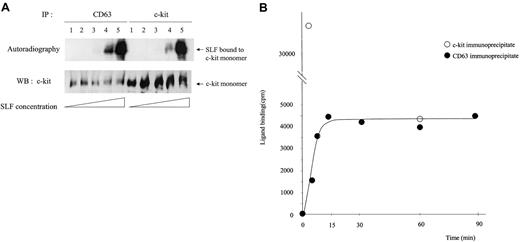
Binding efficiency of SLF is altered in the fraction of c-kit that is associated with CD63
We treated MO7e cells with different concentrations of125I-SLF for 5 minutes at 37°C and then compared the binding efficiency to the total c-kit and to the CD63-associated c-kit (Figure 9A). Because the usual dimer level shown in Western blotting was not high and the dimer-to-monomer ratio was the same in both the TM4SF-associated c-kit and nonassociated fractions, only SLF bound to monomer is shown in Figure 9A. The maximum concentration of SLF was approximately 20 ng/mL as judged by the induced phosphorylation level of c-kit in MO7e cells. We observed up to approximately 10-fold increase of SLF binding (lane 4) to the c-kit associated with CD63 compared with the total c-kit. As the concentration of SLF increased, the difference between125I-SLF bound to CD63-associated c-kit and that bound to total c-kit always decreased. Because the c-kit in the TM4SF complex is not internalized, the binding rate difference may reflect different internalization behavior. To exclude this possibility, we measured the internalization rate of 125I-SLF for 5 minutes at 37°C at 2 different concentrations of 125I-SLF. One was a concentration close to saturation and another was one fifth of that. Internalization percentages (average ± SD) were 55.5% ± 4.0% and 37.7% ± 10.5%, respectively. Thus even considering differences of internalization, it is highly possible that binding of SLF to c-kit is enhanced in the TM4SF complex, especially at low concentrations of SLF. We further examined the kinetics of binding of125I-SLF to the CD63-associated c-kit. After chemical cross-linking and subsequent immunoprecipitation, the radioactivities of CD63 immunoprecipitates were measured. Figure 9B shows that the125I-SLF binding to the c-kit/CD63 complex reached a plateau by 15 minutes. In contrast, the radioactivity of total c-kit immunoprecipitates at 60 minutes was 12% of that of c-kit immunoprecipitates at 5 minutes. The radioactivity of CD50 immunoprecipitates was less than 5% of that of the CD63 immunoprecipitates, suggesting that radioactivity in the CD63 immunoprecipitates was due specifically to 125I-SLF bound to the c-kit/CD63 complex. These results suggest that reaction of SLF and the c-kit/CD63 complex reaches equilibrium, whereas SLF binding to total c-kit decreased dramatically due to internalization.
Characteristics of SLF binding to the c-kit associated with CD63.
(A) MO7e cells were treated with 5 times increasing concentrations of125I-labeled SLF at 37°C for 5 minutes. After cross-linking with BS3, cells were solubilized with lysis buffer containing 1% CHAPS and processed for immunoprecipitation with anti-CD63 or anti–c-kit antibody. After being separated by SDS-PAGE, c-kit bound to radiolabeled SLF was detected by autoradiography (upper panel). The same membrane was immunoblotted with anti–c-kit polyclonal antibody (lower panel). These are the representative results from 4 separate experiments. (B) MO7e cells were treated with125I-labeled SLF (106 cpm/107cells/mL) for indicated time periods at 37°C. After cross-linking and subsequent immunoprecipitation with CD63, radioactivities of immunoprecipitates were measured with a γ counter. These are the representative results from 2 separate experiments.
Characteristics of SLF binding to the c-kit associated with CD63.
(A) MO7e cells were treated with 5 times increasing concentrations of125I-labeled SLF at 37°C for 5 minutes. After cross-linking with BS3, cells were solubilized with lysis buffer containing 1% CHAPS and processed for immunoprecipitation with anti-CD63 or anti–c-kit antibody. After being separated by SDS-PAGE, c-kit bound to radiolabeled SLF was detected by autoradiography (upper panel). The same membrane was immunoblotted with anti–c-kit polyclonal antibody (lower panel). These are the representative results from 4 separate experiments. (B) MO7e cells were treated with125I-labeled SLF (106 cpm/107cells/mL) for indicated time periods at 37°C. After cross-linking and subsequent immunoprecipitation with CD63, radioactivities of immunoprecipitates were measured with a γ counter. These are the representative results from 2 separate experiments.
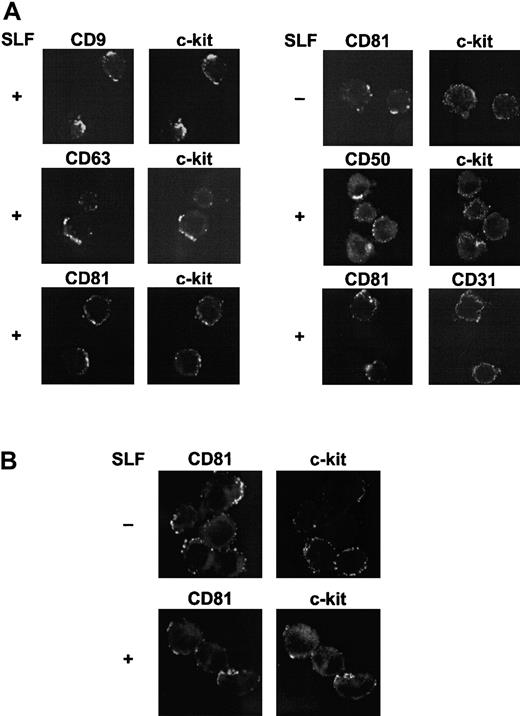
Cocapping of c-kit with TM4SF proteins after stimulation with SLF
The results of the biochemical data suggested that major portions of c-kit were degraded and only the TM4SF-associated c-kit might remain on the cell surface after SLF stimulation. To confirm this interpretation, the molecular association was examined by cocapping experiments using confocal microscopy. These experiments demonstrated that c-kit strongly cocapped with TM4SF molecules CD9, CD63, and CD81 after SLF stimulation in MO7e cells. As controls, capping of CD50 had no effect on c-kit distribution and CD31 did not cocap with CD81. High levels of expressions of CD31 and CD50, comparable to those of c-kit and TM4SF molecules, were confirmed by FACS analysis (data not shown). Without SLF stimulation, cocapping of c-kit with CD81 was ambiguous (Figure 10A). To investigate the possibility that the regulatory mechanism of c-kit function in the TM4SF complex is present in primary hematopoietic progenitors, we used purified cord blood CD34+ cells (> 95% pure). Because levels of c-kit expression in cord blood CD34+ cells are not high, we treated the cells with thrombopoietin (TPO), Flt3 ligand, and interleukin 6 (IL-6) for 3 days. More than 90% of cells were highly positive both for c-kit and for CD34 after stimulation with these cytokines as assessed by FACS analysis (data not shown). Because of low expression level of CD9 in these cell populations, CD9 was excluded from the cocapping experiments. Colocalization of c-kit with CD81 was manifested with SLF stimulation (Figure 10B). However, little colocalization of c-kit with CD63 as well as CD50 was observed (data not shown). Although the level of c-kit expression was low in freshly isolated cord blood CD34+ cells, these purified cells showed a pattern of cocapping similar to that of cells pretreated with TPO, Flt3 ligand, and IL-6 for 3 days (data not shown).
Colocalization of TM4SF molecules with c-kit.
(A) MO7e cells were treated with or without 50 ng/mL SLF for 60 minutes at 37°C. Cell surface TM4SF molecules or CD50 were cross-linked first with primary mAbs and then with FITC-conjugated antimouse antibody and allowed to cap at 37°C. The cells were then stained with biotin-labeled anti–c-kit or -CD31 mAbs and TRITC-conjugated streptavidin. Fluorescence intensities from FITC (left column) and TRITC (right column) were separately determined by a confocal laser scanning microscopy. These are the representative results from 3 separate experiments. (B) Cord blood CD34+ cells were incubated with IL-6, TPO, and Flt3 ligand for 3 days. The cells were treated with or without 50 ng/mL SLF for 60 minutes at 37°C. After capping induction with FITC-conjugated anti-CD81 mAb and purified antimouse antibody, the cells were stained with biotin-labeled anti–c-kit mAb and TRITC-conjugated streptavidin, and visualized by confocal laser scanning microscopy. These are the representative results from 2 separate experiments.
Colocalization of TM4SF molecules with c-kit.
(A) MO7e cells were treated with or without 50 ng/mL SLF for 60 minutes at 37°C. Cell surface TM4SF molecules or CD50 were cross-linked first with primary mAbs and then with FITC-conjugated antimouse antibody and allowed to cap at 37°C. The cells were then stained with biotin-labeled anti–c-kit or -CD31 mAbs and TRITC-conjugated streptavidin. Fluorescence intensities from FITC (left column) and TRITC (right column) were separately determined by a confocal laser scanning microscopy. These are the representative results from 3 separate experiments. (B) Cord blood CD34+ cells were incubated with IL-6, TPO, and Flt3 ligand for 3 days. The cells were treated with or without 50 ng/mL SLF for 60 minutes at 37°C. After capping induction with FITC-conjugated anti-CD81 mAb and purified antimouse antibody, the cells were stained with biotin-labeled anti–c-kit mAb and TRITC-conjugated streptavidin, and visualized by confocal laser scanning microscopy. These are the representative results from 2 separate experiments.
Discussion
The TM4SF proteins form multimolecular complexes containing several types of cell surface molecules. For example, TM4SF proteins are components of the CD19/CD21/Leu13 signaling complex in B cells.38 CD81 and CD82 are reported to be associated with CD4 or CD8 in T cells.35 Association of CD9 with membrane-bound growth factors of the epidermal growth factor (EGF) family has been reported.39-41 Recently the integrin/TM4SF complex has been studied intensively in a number of cell systems.9-18 Here, we provide evidence that TM4SF proteins can also associate with c-kit receptor tyrosine kinase.
The c-kit in the TM4SF complex exhibited several distinct phenotypes compared with that in the total c-kit fraction. The results of biochemical characterization of this complex indicate that the c-kit in the TM4SF complex may be functionally different from the c-kit not associated with TM4SF proteins. The concept that TM4SF molecules directly alter the function of associated partners has come from a few studies. For example, membrane-anchored heparin-binding EGF-like growth factor has the property of functioning as diphtheria toxin receptor. CD9 association up-regulated the juxtacrine mitogeneic activity of heparin-binding EGF-like growth factor as well as the sensitivity to diphtheria toxin.39,40 Recently CD9 has been reported to increase the diphtheria toxin receptor affinity.42 CD19 mutants lacking the capacity of association with CD81 could not deliver the signal to cell aggregation.43 Our data demonstrate that the association of TM4SF proteins with c-kit may alter the function of the c-kit receptor tyrosine kinase.
Gain-of-function mutations of c-kit in vivo render both constitutive phosphorylation and kinase activation.44,45 In contrast, the elevated basal phosphorylation of c-kit in the TM4SF complex did not lead to kinase activation as estimated by immune complex kinase assay. Moreover, the large enhancement of phosphorylation and kinase activation by SLF was severely impaired in the c-kit associated with TM4SF proteins. How these characteristics of c-kit in the complex are engineered remains to be determined. Not all mutants of c-kit generated in vitro with increased tyrosine phosphorylation in the absence of SLF exhibited elevated kinase activity.45 Some mutants in the kit juxtamembrane region showed a similar phenotype to that of the TM4SF-associated c-kit in our study in terms of tyrosine phosphorylation and kinase activity in the absence or presence of SLF.46 Thus, it is possible that association of TM4SF proteins may cause a conformational change leading to spontaneous phosphorylation but not to kinase activation. On the other hand, CD63 has been reported to be associated with tyrosine kinase activity in neutrophils.11 Therefore, another possibility is that TM4SF proteins may recruit other tyrosine kinases to the complex. Tyrosine phosphatase activity, which has been reported to be associated with TM4SF complex,47 may contribute to deficiency of induced phosphorylation in response to SLF.
Our surface cross-linking study suggested the possibility that the c-kit in the TM4SF complex might have a higher affinity for SLF. However, SLF binding to the c-kit protein that was associated with TM4SF proteins did not lead to kinase activation, suggesting signaling was dampened in the complex. We hypothesize that this phenotype may be related to a threshold for SLF stimulation and that some TM4SF proteins may be negative regulators of c-kit function. In agreement with a previous report by others, which showed that the tyrosine kinase activity of c-kit was essential for ligand-induced internalization,37 the c-kit in the TM4SF complex was neither internalized nor degraded after SLF treatment. These results suggest constitutive presence of the c-kit/TM4SF complex on the cell surface independent of SLF stimulation. This constant surface expression may be important for maintaining a buffering effect in the constitutive presence of low levels of SLF. Our results suggest that interaction of SLF and TM4SF-associated c-kit may reach equilibrium without receptor consumption and downstream signaling at physiologic conditions (ie, very low SLF concentrations). CD34+/c-kit+ hematopoietic progenitor cells are reported to transcribe low amounts of SLF messenger RNA.36,48 An autocrine mechanism has been suggested in normal hematopoiesis as well as leukemogenesis.48 49 The existence of a c-kit/TM4SF complex may lessen or modulate an autocrine loop generated by low level expression of endogenous SLF.
With regard to TM4SF association with cytokine receptors, only CD82 association with EGF receptor (EGFR) tyrosine kinase has been reported.50 In contrast to the c-kit/TM4SF interaction in this study, EGF stimulation destabilized the EGFR/CD82 complex. In addition, CD82 overexpression accelerated ligand-induced clearance of EGFR from the cell surface, resulting in attenuation of EGFR signaling. Thus, the regulatory mechanism of the EGFR tyrosine kinase appears to be quite different from that of c-kit presented here.
Little is known about the organization of the TM4SF complex. Other studies used different detergent conditions to determine specificity and proximity of TM4SF associations.51,52 The fact that stringent detergent conditions (1% NP40 + 0.1% SDS + 0.5% sodium deoxycholate) did not disrupt the CD63/c-kit and CD81/ c-kit associations completely indicate that at least some of these associations may be direct. CD9/c-kit association was abrogated under these conditions. However, prior chemical cross-linking before cell lysis yielded comparable levels of c-kit amounts in CD9 immunoprecipitates within CD63 or CD81 immunoprecipitates, indicating that CD9 is sterically close to the c-kit. We failed to detect TM4SF proteins in reciprocal experiments under these conditions suggesting that only small amounts of c-kit may be in close proximity to these TM4SF molecules. After 3 successive rounds of precleaning with a single mAb to TM4SF, none of the mAbs toTM4SF had much capacity to coimmunoprecipitate c-kit (data not shown). Thus, it is possible that any single complex may contain all the components (CD9, CD63, CD81, c-kit). Clarifying the actual amounts and structural aspects of the c-kit/TM4SF complex is a subject for further investigation. It remains to be determined if the complex exists in raftlike membrane microdomains.52
The precleaning procedure mentioned above reduced c-kit protein amount by 23.3% ± 5.0% (average ± SD), as judged by Western blotting (data not shown). Thus, we grossly estimate that the percentage of c-kit complexes with TM4SF proteins in the absence of SLF may be approximately 20% in MO7e cells. The fact that the percentage of c-kit, which remained on the cell surface after SLF stimulation was nearly 20% as judged by the level of surface biotinylation (Figure 8), may support this estimation. Although colocalization of 2 surface molecules by cocapping is clearly demonstrated on the cell line, and on primary CD34+ cells, we believe that the purely qualitative nature of the technique precludes any interpretation regarding relative amounts of the complexes present. TM4SF has been proposed to function as a molecular facilitator organizing the relative position of other cell surface molecules and modulating their function.8 We speculate that size or composition of the complex may change dynamically during development of hematopoietic progenitors in relation to other associated partner molecules. Our observations that there is no apparent colocalization of CD63 with c-kit in cord blood CD34+ cells (data not shown) may be in line with the above hypothesis. We also observed that transduced erythropoietin receptor became associated with TM4SF molecules in MO7e cells, causing a reduction in the amount of TM4SF proteins coimmunoprecipitated with anti–c-kit antibody (N.A. and H.E.B., unpublished data, April 2001). Taking into the account the importance of SLF synergy with other growth factors, especially with erythropoietin for erythroid development,53 54 this phenomenon seems intriguing, and experiments are needed to determine the contribution of TM4SF to cytokine receptor interactions.
Integrins are recognized as components of TM4SF complexes in a variety of cell systems including MO7e.9-18 SLF is a potent stimulator of cell adhesion and migration through integrin functional modification, the underlying mechanism of which is only partially understood.26-28,55 Studies with mast cells derived from W/Wv mutant mice showed minimal requirement of c-kit tyrosine kinase activity for cell adhesion and migration.56 In other respects, mobilization mediated by blocking antibodies to integrin α4β1 and its cellular ligand vascular cell adhesion molecule 1 required functional c-kit, indicating the importance of c-kit for cell migration and the existence of integrin/c-kit receptor cross-talk.29TM4SF may be an unidentified link between c-kit and integrins and the c-kit/TM4SF complex may have a role coupled to integrin functions. This type of role for TM4SF has been proposed in the case of the B-cell costimulatory molecule CD19 and integrins.57 Although our results suggest that integrin/TM4SF and c-kit/TM4SF complexes may exist as discrete entities in steady-state conditions, the possibility remains that TM4SF links c-kit with integrin on activation.
The role of TM4SF in hematopoiesis has been only marginally elucidated. CD9 ligation with a specific mAb inhibited production of myeloid cells in long-term cultures.58 The mechanism proposed for this effect was that CD9 on stromal cells might regulate physical interaction with hematopoietic progenitors, which allows these cells to proliferate and differentiate.59 It was also reported that engagement of CD9 on pre–B cells with a specific mAb induced adhesion of these cells to bone marrow fibroblasts.60 CD81 has recently been proposed to serve as a marker for defining developmental stages of lymphohematopoietic stem cells.61 No functional study of other TM4SF members in hematopoiesis has been made, although expression of some members such as CD63 and CD82 on hematopoietic progenitors have been reported.62 63 Our results suggest that TM4SF molecules may associate with cytokine receptors as well as integrins in hematopoietic progenitors. TM4SF molecules through their unique network formation may provide a novel functional key in the complexity of regulation of hematopoiesis.
Supported by Public Health Service grants RO1HL56416, RO1HL67384, and RO1DK5674 from the National Institutes of Health (to H.E.B.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hal E. Broxmeyer, Walther Oncology Center, Indiana University School of Medicine, 1044 W Walnut St, Indianapolis, IN 46202-5254; e-mail: hbroxmey@iupui.edu.

![Fig. 4. Immune complex kinase activity of TM4SF-associated and total c-kit fractions in the absence or the presence of SLF. / MO7e cells were stimulated with or without 50 ng/mL SLF at 37°C for 10 minutes. Cells were solubilized with lysis buffer containing 1% CHAPS. The cell lysates were immunoprecipitated with indicated first antibodies. Immunoprecipitated materials were eluted with 1% NP-40 lysis buffer and supernatants were immunoprecipitated with anti–c-kit mAb (cloneYB5.B8). In the case of anti–c-kit mAb as a first immunoprecipitation antibody, immunoprecipitated proteins were treated with NP-40 in the same way. The immune complexes were incubated with 10 μCi (0.37 MBq) [32P-γ] adenosine triphosphate for 10 minutes at room temperature. Immune complexes were separated by SDS-PAGE and transferred onto Immobilon-P membrane. [32P] incorporation into c-kit protein was visualized by autoradiography (upper panel). The same membrane was immunoblotted with anti–c-kit polyclonal antibody (lower panel). These are the representative results from 2 separate experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/99/12/10.1182_blood.v99.12.4413/4/m_h81222688004.jpeg?Expires=1765911470&Signature=y2dBg0phxP1kT7OIS5xwWPQCcIqo6pApsywMGIw8rczhCXKZxXmGQOyLrq7dltoQ1p6FU4gkBiMNAzjHUOsPJaMmcJe4X-EHOlRWjI6hG0c8klUKeCL8m-7FOzI~qtLlPfJIst0D3kuGqoOM6YkUAW3iNcnzF1L9LwsyOjDYev-W6gMe9m5XzpdVV5G0fx2bDsTX8~zouT4L8bTvRukk3Vfc-R-Ap2niToqBsvwWEToCAUpJmqONVVLS8zIQoiEdkZQ21NFO5FR6JXXkYss8JODEMCUkCvKd4M6ipl5uxkCympLRnhG2JuQ2NU~~4Ivbw66NivRQvK1HrR3BW-lwQQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal