Deletions of the derivative chromosome 9 have recently been reported in chronic myeloid leukemia. These deletions are large, occur at the time of the Philadelphia (Ph) translocation, span the translocation breakpoint, and represent a powerful prognostic indicator. However, the molecular mechanisms responsible for the poor prognosis associated with deletions are obscure, and several possible models are investigated here. First, we demonstrate that all derivative chromosome 9 deletions detected by fluorescence in situ hybridization were associated with an absence ofABL-BCR expression. However, loss ofABL-BCR expression also occurred without an overt deletion, suggesting the existence of other mechanisms by whichABL-BCR transcription can be abolished. Furthermore, analysis of survival in 160 patients demonstrated that loss ofABL-BCR expression, in contrast to deletion status, was not an indicator of poor prognosis. Second, we addressed the possibility that concomitant small deletions of the Ph chromosome modulateBCR-ABL transcription. Real-time reverse-transcription polymerase chain reaction was used to demonstrate that derivative chromosome 9 deletions were not accompanied by altered levels of BCR-ABL transcripts. Third, deletions may represent a consequence of genetic instability within the target cell at the time of the Ph translocation, with the poor prognosis reflecting a predisposition to subsequent additional genetic alterations. However, patients with deletions do not exhibit an increased frequency of secondary cytogenetic changes following disease progression. Taken together, these data support a model in which deletions of the derivative chromosome 9 result in rapid disease progression as a result of the loss of one or more genes within the deleted region.
Introduction
Chronic myeloid leukemia (CML) is characterized by the formation of the BCR-ABL fusion gene, usually as a consequence of the Philadelphia (Ph) translocation between chromosomes 9 and 22.1-3 Transgenic and retroviral transduction studies have shown that the expression of BCR-ABL in murine bone marrow cells results in leukemia, with some cases resembling CML,4-10 and suggest that BCR-ABL fusion is the initiating event. Furthermore, in one recent transgenic model, the leukemia could be reversed by down-regulatingBCR-ABL.11
In contrast to the apparent molecular homogeneity of chronic phase CML, the clinical disease is heterogeneous with marked differences between patients in laboratory and clinical characteristics, including the rate of progression to blast crisis. However, we and others have recently reported previously unsuspected deletions of the derivative chromosome 9.12-17 These deletions are large, have varying breakpoints, and occur at the time of the Ph translocation, thus resulting in considerable genetic heterogeneity from the beginning of the disease.17 Importantly, deletion status is a powerful and independent prognostic factor, and one that is considerably more potent than the Sokal18 or Hasford19 scoring systems.17
In many cases, the derivative chromosome 9 deletions span the translocation breakpoint and are, therefore, likely to result in loss of expression of ABL-BCR, the reciprocal fusion gene formed by the Ph translocation. This observation raises the possibility that loss of ABL-BCR might modulate disease progression, a concept reminiscent of the proposed role of the reciprocal fusion gene in acute promyelocytic leukemia.20,21 In contrast to the wealth of information available on the role of BCR-ABL,relatively little attention has been paid to ABL-BCR. ABL-BCR expression has been reported in about 60% of patients with CML22 and did not correlate with cytogenetic response to interferon-α.23 However, there are a number of ways in which ABL-BCR might contribute to the biology of CML. The carboxy terminus of BCR, which is present in the predicted ABL-BCR fusion protein, encodes a domain with the ability to activate guanosine 5′-triphosphatases involved in RAS signaling pathways.24 It has also been suggested that BCR can function as a negative regulator of BCR-ABL.25
The pathogenetic consequences of derivative chromosome 9 deletions may reflect loss of one or more other critical genes within the deleted region, particularly since the deletions are large and frequently span several megabases. Alternatively, it is also possible to envisage 2 other mechanisms by which derivative chromosome 9 deletions may affect prognosis. First, a deletion on the derivative chromosome 9 may act as a surrogate marker for smaller intronic deletions on the Ph chromosome, which could influence the level of BCR-ABL expression. Several lines of evidence suggest that the level of BCR-ABLexpression is important in determining the phenotype ofBCR-ABL–positive leukemias: an extra Ph chromosome is the most common secondary change seen with development of blast crisis in CML26; the p185 BCR-ABL tyrosine kinase associated with acute lymphocytic leukemia has increased kinase activity when compared with the standard CML p210 protein,27 and an increase in p210 BCR-ABL expression precedes progression to accelerated phase or blast crisis in patients with CML.28 Second, deletions may be associated with poor outcome because they represent a consequence of genetic instability within the target cell at the time of the Ph translocation. In this case, the poor prognosis would reflect a predisposition to subsequent additional genetic alterations within the malignant clone. Patients with chronic phase CML do not exhibit genomic instability as assessed by microsatellite analysis,29 30 but these data do not exclude other levels of genetic instability.
In this manuscript, we have examined these various mechanisms. We provide the first direct comparison of ABL-BCRexpression with deletions of the derivative chromosome 9. Our results show that not all ABL-BCR–negative patients have a detectable deletion, suggesting the existence of other mechanisms by which ABL-BCR expression may be abolished, and we demonstrate that ABL-BCR expression itself is not a marker of poor prognosis. Furthermore, we show that deletions are not associated with increased levels of BCR-ABL expression and are not accompanied by increased cytogenetic instability during progression to blast crisis.
Patients, materials, and methods
Patient samples, patient data, and cell lines
Peripheral blood, bone marrow, fixed cytogenetic preparations, and peripheral leukapheresis products were obtained from patients in accordance with local institutional ethical protocols. These patients were diagnosed with BCR-ABL–positive CML between December 1984 and March 2001 and attended the Haematology Departments of Addenbrooke's Hospital Cambridge, United Kingdom; The City Hospital Nottingham, United Kingdom; The University Hospital of Poitiers, France; and The University Hospital, Leipzig, Germany. Clinical outcome and laboratory data were available for 160 of these patients. Risk categories were determined by means of standard methods.18,19 Length of chronic phase was defined as the time from diagnosis until progression to accelerated phase or blast crisis, which were determined as previously.31 32 All patients included in the quantitative BCR-ABL analysis were 100% Ph+ on cytogenetic analysis and a 5 cell–type manual differential was performed on the leukapheresis product in 15 of 16 of these. We studied 3BCR-ABL–positive cell lines: MEG-01, BV173 (which demonstrates no deletions), and MC3 (which carries a deletion). All of these cell lines were pseudodiploid and possessed 2 copies of the Ph chromosome (data not shown).
Expression analysis of ABL-BCR
RNA extraction from peripheral blood granulocytes and bone marrow and complementary DNA (cDNA) preparation were performed as previously described33 with the following modifications: random hexamers were used (25 mM final concentration), and the final concentration of MgCl2 was 3 mM. Multiplex polymerase chain reaction (PCR) amplification of the undiluted cDNA, with dual amplification of a housekeeping gene, nonerythropoietic porphobilinogen deaminase (PBGD), and ABL-BCR was as described33 with the following modifications: final concentrations of MgCl2 and deoxynucleoside 5′-triphosphates were 2.5 mM and 500 μM, respectively, with 1.5 U Amplitaq Gold (Applied Biosystems, Warrington, United Kingdom) used. We performed 35 cycles of PCR at an annealing temperature of 62°C for 45 seconds.
In 65 patients, ABL-BCR PCR was performed with the use of forward primers from both ABL exon 1b, primer 1bAB (CACGAATTCTGGAAAGGGGTACCTA TTA), and ABL exon 1a, primer 1aAB (TACGGAATTCATGTT GGAGATCTGCCTGAA), together with a reverse primer from BCR exon b5/e16, primer e16AB (CACAGTATCCTCAGGGTCTGGGA) (Figure 1). In the remaining 128 patients, PCR was performed only for theABL1b–BCR transcript, as expression of anABL1a–BCR transcript was not seen without concomitant expression of ABL1b–BCR, as has previously been described.22,23 34 The primers used were forward primer ABL1b (TACTTGGGGACCAAAGAAGG) and reverse primerBCR b4 (CAGCTGTGTCCCTGTAGACG) from exon b4/e15 (Figure 1). Primers used for PBGD were PBGD-F (GTCTGGTAACGGCAATGCG) from exon 1 and PBGD-R (CAAACTGCAGGCCAGGGTAC) from exon 4. PCR products were visualized on 2.5% agarose gels following ethidium bromide staining.
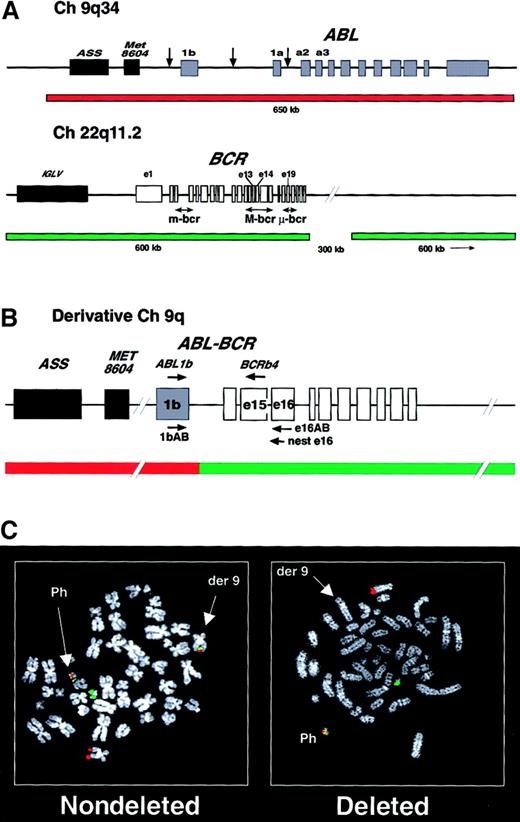
Probe sets and images for the detection of deletions of the derivative chromosome 9.
(A) Structure of the ABL and BCR loci showing the common breakpoints (arrows) and the probes used in the dual-color, dual-fusion detection system. ASS indicates arginine succinate synthetase; Met8604, Met8604 gene; IGLV, immunoglobulin lambda light chain locus. (B) A representativeABL-BCR (1b-b4) rearrangement is shown on the derivative chromosome 9 with the position of the ABL1b,BCRb4, 1bAB, e16AB, and neste16 primers and the FISH fusion signal. (C) Fluorescence in situ hybridization (FISH) analysis of nondeleted and deleted Ph+ metaphase cells. In both cells, the normal chromosome 9 and 22 demonstrate a single red and green signal, respectively, with a fusion signal present on the Ph+ cell. In the nondeleted cell, the derivative chromosome 9 also demonstrates a fusion signal, but this is missing from the derivative chromosome 9 in the deleted cell.
Probe sets and images for the detection of deletions of the derivative chromosome 9.
(A) Structure of the ABL and BCR loci showing the common breakpoints (arrows) and the probes used in the dual-color, dual-fusion detection system. ASS indicates arginine succinate synthetase; Met8604, Met8604 gene; IGLV, immunoglobulin lambda light chain locus. (B) A representativeABL-BCR (1b-b4) rearrangement is shown on the derivative chromosome 9 with the position of the ABL1b,BCRb4, 1bAB, e16AB, and neste16 primers and the FISH fusion signal. (C) Fluorescence in situ hybridization (FISH) analysis of nondeleted and deleted Ph+ metaphase cells. In both cells, the normal chromosome 9 and 22 demonstrate a single red and green signal, respectively, with a fusion signal present on the Ph+ cell. In the nondeleted cell, the derivative chromosome 9 also demonstrates a fusion signal, but this is missing from the derivative chromosome 9 in the deleted cell.
Samples showing no PCR product after one round of amplification were further analyzed by nested PCR (the initial 65 patients), with the use of the same forward primer and a nested reverse primer, also from exon b5/e16, neste16 (CACAGTATCCTC AGGGTCTGGGA) (Figure 1), or by Southern hybridization (remaining 128 patients) by means of standard methods35 and an ABL-BCR probe generated by PCR from the cell line MEG-1 cDNA with primers ABL1b andBCRb4.
Quantitative reverse-transcription PCR forBCR-ABL
Quantitative reverse-transcription PCR (RT-PCR) forBCR-ABL was performed by means of the Taqman method on a 7700 sequence detection system (Applied Biosystems). Total RNA was extracted from peripheral blood leukapheresis samples and cell lines with the use of TRI-reagent, as above. Then, 2 μg RNA was reverse transcribed with the use of 25 mM random hexamers and 200 U M-MLV RT at 42°C for 45 minutes. Real-time PCR was then performed in a final volume of 25 μL containing 2 μL cDNA, 1.3 μM each primer, 400 nM the fluorescent internal probe, and 1 × Taqman universal master-mix (Applied Biosystems). Primers and probe forBCR-ABL were BCR-F (TGACCAACTCGTGTGTGAAACTC),ABL-R (GGGTCCAGCGAGAAGGTTTT), and ABL-P (AGCGGCCAGTAGCATCTGACTTTGAGC), respectively. Primers and probe for the control gene β2-microglobulin, B2M, wereB2M-F (CCTGCCGTGTTGAACCATGT), B2M-R (CGGCATCTTCAAACCTCCAT), and B2M-P (ACATGTCTCGATCCCACTTAACTATCTTGGGCT), respectively. The threshold was set at a ΔRn value of 0.2, between 3 and 12 cycles, for each Taqman plate.
To correct for potential differences in the cDNA, BCR-ABLexpression in each sample was compared with the expression of the control gene B2M for that sample by subtracting the threshold cycle, CT, of the control gene from that ofBCR-ABL to obtain the Δ CT. This allowed the quantification of BCR-ABL transcripts, expressed as a percentage relative to the control gene, by the equation, % of control gene = (1 + E)−ΔCT, where E equals the efficiency of the PCR reaction (which was deduced from a K562 standard curve and was 0.93). Each sample value was determined in triplicate for BCR-ABL and in duplicate for B2M.
Fluorescence in situ hybridization detection of deletions and cytogenetic comparison of secondary changes occurring with blast crisis
Fluorescence in situ hybridization (FISH) detection of deletions was performed and interpreted as previously described17with the use of the BCR/ABL dual-color, dual-fusion probe (Vysis, Downers Grove, IL) (Figure 1) following the manufacturer's instructions. Cytogenetic analysis, performed on a minimum of 20 metaphases by means of conventional methods,36 was available for a total of 84 patients at the time of their progression to blast crisis. The deletion status of these patients was known, with 37 patients taken from the above cohort of 193 patients and the remaining 47 from our previously reported cohort.17
Statistical analysis
All calculations were performed with the SPSS (Chicago, IL) statistical package. Survival time was calculated, with a median follow-up of 39 months (range, 3-206 months), with patients censored as previously.17 Survival data and length of chronic phase comparisons were as previously used.17
In the patients for whom cytogenetic analysis was available at the time of progression to blast crisis, the proportion of patients with secondary changes who carried deletions and the proportion with changes lacking deletions were compared by Fisher exact test. The mean and median number of total secondary changes and double-stranded breaks, along with the interquartile range, were calculated for patients with and without deletions. Any differences were compared by the Mann-Whitney U test. For BCR-ABL expression, the proportion of CD34+ cells, blasts, promyelocytes, or myelocytes in the leukapheresis product were compared by the Mann-Whitney U test between patients who carried or lacked deletions. The median and mean levels of BCR-ABLtranscripts, along with the interquartile range, were calculated for patients with deletions and for those who lacked them. Any differences in expression levels were compared by the Mann-Whitney Utest.
Results
Comparison of ABL-BCR expression with deletion of the derivative chromosome 9
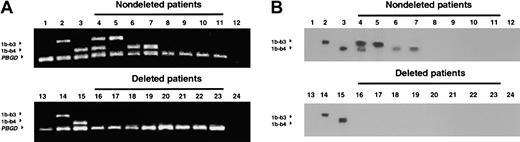
Expression of the ABL-BCR transcript was determined by RT-PCR in samples from 193 patients with CML (56% in chronic phase, 27% in accelerated phase, and 17% in blast crisis). Patients with a major or complete cytogenetic remission were excluded from analysis.BCR-ABL transcripts were detected in each sample. Expression of ABL-BCR was detected in 128 patients (66%) and was absent in 65 patients (34%) (Figure 2), a prevalence similar to previous results.22 23 AllABL-BCR samples negative on first-round PCR were confirmed either by Southern blotting of the PCR products (Figure 2) or by a second round of PCR with nested primers.
Detection of the ABL-BCR transcript by RT-PCR and Southern blotting.
Ethidium bromide–stained agarose gels (panel A) and Southern blots (panel B) from the same patients were run on different gels. Lanes 4-11 show nondeleted patients; lanes 16-23, deleted patients. Lanes 1 and 13 show HL6O (BCR-ABL–negative) cell line; lanes 2 and 14, Meg-1 cell line; lanes 3 and 15, LAMA cell line, lanes 12 and 24, water control. 1b-b3 indicatesABL1b-BCRb3 transcript; 1b-b4,ABL1b-BCRb4 transcript; PBGD, porphobilinogen deaminase transcript.
Detection of the ABL-BCR transcript by RT-PCR and Southern blotting.
Ethidium bromide–stained agarose gels (panel A) and Southern blots (panel B) from the same patients were run on different gels. Lanes 4-11 show nondeleted patients; lanes 16-23, deleted patients. Lanes 1 and 13 show HL6O (BCR-ABL–negative) cell line; lanes 2 and 14, Meg-1 cell line; lanes 3 and 15, LAMA cell line, lanes 12 and 24, water control. 1b-b3 indicatesABL1b-BCRb3 transcript; 1b-b4,ABL1b-BCRb4 transcript; PBGD, porphobilinogen deaminase transcript.
To compare ABL-BCR expression with the presence of derivative chromosome 9 deletions, FISH analysis was performed on all cytogenetic preparations available from patients lackingABL-BCR expression (n = 55) and also on samples from 50ABL-BCR–positive patients by means of a dual-color system that detects both reciprocal translocation products (Figure 1). Of the 55 ABL-BCR–negative patients, 19 (35%) exhibited deletions detectable by FISH, whereas 36 (65%) lacked such deletions. Fourteen patients demonstrated deletions of both chromosome 9 and chromosome 22 sequences; 3 patients had deletions of only 9 sequences; and 2 patients had deletions of only 22 sequences (data not shown). By contrast, of 50ABL-BCR–positive patients, none had a deletion detectable by FISH. The presence of ABL-BCR transcripts can therefore be used to identify patients lacking such deletions (negative predictive value, 100%; 95% confidence interval [CI], 93%-100%).
Lack of expression of ABL-BCR does not explain the poor prognosis associated with deletions
All patients with a derivative chromosome 9 deletion lackedABL-BCR expression, which might account for the poor prognosis associated with such deletions. To address this issue directly, we compared the survival and length of chronic phase ofABL-BCR–negative and ABL-BCR–positive patients. Clinical outcome data were available for 160 patients (103ABL-BCR positive and 57 ABL-BCR negative). The clinical and laboratory characteristics at diagnosis are shown in Table1 and are similar for the 2 groups. The only significant difference is in the proportion of patients with splenomegaly, which is greater in those who did not express the transcript (P = .006). However, this difference did not translate into a significant increase in the number of patients classified as high risk by either the Sokal or the Hasford scoring systems.
Patient characteristics at diagnosis and at end of study
| Patient characteristics (n = 160) . | Patients who express ABL-BCR (n = 103) . | Patients who do not express ABL-BCR (n = 57) . |
|---|---|---|
| Sex (M/F) | 58/45 | 30/27 |
| Median age, y (interquartile range) | 47 (38-58) | 47 (37-57) |
| Patients with splenomegaly, no. (%)* | 33/76 (43) | 23/33 (70)† |
| Median platelet count × 109/L, no. (interquartile range)* | 382 (266-603) | 335 (213-498) |
| Median peripheral blood blasts as % of WCC (interquartile range)* | 1 (0-2) | 2 (0-3) |
| Sokal score (%) | ||
| No. patients assessed | 76 | 33 |
| High score, no. | 22 (29) | 12 (36) |
| Intermediate score, no. | 28 (37) | 12 (36) |
| Low score, no. | 26 (34) | 9 (28) |
| Hasford score (%) | ||
| No. patients assessed | 75 | 31 |
| High score, no. | 15 (20) | 9 (29) |
| Intermediate score, no. | 32 (43) | 14 (45) |
| Low score, no. | 28 (37) | 8 (26) |
| Treatment type (%) | ||
| IFN-α ± oral chemotherapy score, no. | 103 (100) | 53 (93) |
| Other chemotherapy | 35 (34) | 14 (24) |
| STI571 | 85 (82) | 44 (77) |
| BMT | ||
| Allogeneic | 15 (14) | 10 (18) |
| Autologous | 3 (3) | 1 (2) |
| Karyotype (%) | ||
| Classical Ph | 99 (96) | 51 (89) |
| Complex/variant Ph | 4 (4) | 6 (11) |
| Patients at end of study, no. (%) | ||
| Alive | 76 (74) | 38 (66) |
| Censored during study | 15 (14) | 10 (18) |
| Dead | 12 (12) | 9 (16) |
| Patient characteristics (n = 160) . | Patients who express ABL-BCR (n = 103) . | Patients who do not express ABL-BCR (n = 57) . |
|---|---|---|
| Sex (M/F) | 58/45 | 30/27 |
| Median age, y (interquartile range) | 47 (38-58) | 47 (37-57) |
| Patients with splenomegaly, no. (%)* | 33/76 (43) | 23/33 (70)† |
| Median platelet count × 109/L, no. (interquartile range)* | 382 (266-603) | 335 (213-498) |
| Median peripheral blood blasts as % of WCC (interquartile range)* | 1 (0-2) | 2 (0-3) |
| Sokal score (%) | ||
| No. patients assessed | 76 | 33 |
| High score, no. | 22 (29) | 12 (36) |
| Intermediate score, no. | 28 (37) | 12 (36) |
| Low score, no. | 26 (34) | 9 (28) |
| Hasford score (%) | ||
| No. patients assessed | 75 | 31 |
| High score, no. | 15 (20) | 9 (29) |
| Intermediate score, no. | 32 (43) | 14 (45) |
| Low score, no. | 28 (37) | 8 (26) |
| Treatment type (%) | ||
| IFN-α ± oral chemotherapy score, no. | 103 (100) | 53 (93) |
| Other chemotherapy | 35 (34) | 14 (24) |
| STI571 | 85 (82) | 44 (77) |
| BMT | ||
| Allogeneic | 15 (14) | 10 (18) |
| Autologous | 3 (3) | 1 (2) |
| Karyotype (%) | ||
| Classical Ph | 99 (96) | 51 (89) |
| Complex/variant Ph | 4 (4) | 6 (11) |
| Patients at end of study, no. (%) | ||
| Alive | 76 (74) | 38 (66) |
| Censored during study | 15 (14) | 10 (18) |
| Dead | 12 (12) | 9 (16) |
The patient age, treatment and survival data were available from 160 patients, with the length of chronic phase available from 159 patients. Information sufficient to perform Sokal and Hasford scores was available in 109 and 106 patients, respectively. Many patients received more than one type of therapy, so the sum of the patients in the treatment section is greater than 100%.
WCC indicates white cell count; IFN, interferon; BMT, bone marrow transplantation; Ph, Philadelphia chromosome.
Calculated from 76 patients who expressed ABL-BCRand 33 patients who did not.
Significant (P = .006).
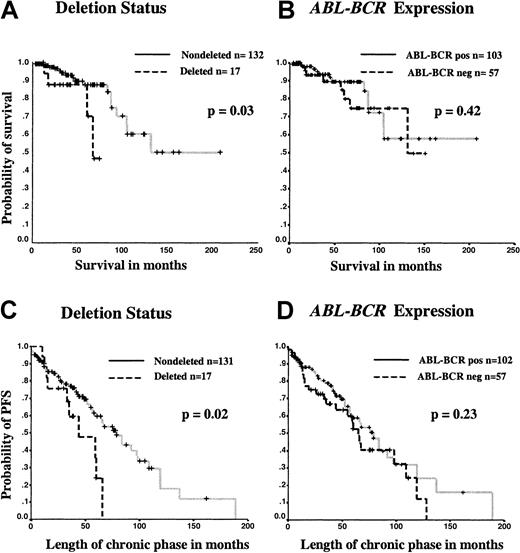
Consistent with previous reports,15-17 in the 149 patients for whom the deletion status was known (or inferred from the presence of ABL-BCR transcripts), there was a significant difference in the survival of the patients with deletions compared with those lacking a detectable deletion (Figure3A). The estimated median survival for patients with a deletion was 67 months (95% CI, 53-73 months) whereas the median survival for patients without a deletion could not be calculated, as more than 50% of patients were still alive. This difference was significant by log-rank analysis (P = .03). By contrast, the estimated median survival for patients who did not express the ABL-BCR transcript was 131 months (95% CI, 64-198 months), while the estimated median survival for patients who expressed the transcript could not be calculated, as more than 50% were still alive. This difference was not significant by log-rank analysis (P = .42) (Figure 3B).
Kaplan-Meier analysis of survival and length of chronic phase comparing patients by deletion status and ABL-BCRexpression.
For ABL-BCR expression, calculations were performed for survival and length of chronic phase with the use of data for 160 and 159 patients, respectively; for deletion status, calculations were performed for 149 and 148 patients, respectively. The significance of any survival difference was assessed by log-rank analysis. Patients who underwent allogeneic stem cell transplantation and patients who died of causes unrelated to CML were censored at the time of the procedure or death, respectively. Separate Kaplan-Meier graphs are shown for survival according to deletion status (panel A) or ABL-BCRexpression (panel B) and for length of chronic phase according to deletion status (panel C) or ABL-BCR expression (panel D).
Kaplan-Meier analysis of survival and length of chronic phase comparing patients by deletion status and ABL-BCRexpression.
For ABL-BCR expression, calculations were performed for survival and length of chronic phase with the use of data for 160 and 159 patients, respectively; for deletion status, calculations were performed for 149 and 148 patients, respectively. The significance of any survival difference was assessed by log-rank analysis. Patients who underwent allogeneic stem cell transplantation and patients who died of causes unrelated to CML were censored at the time of the procedure or death, respectively. Separate Kaplan-Meier graphs are shown for survival according to deletion status (panel A) or ABL-BCRexpression (panel B) and for length of chronic phase according to deletion status (panel C) or ABL-BCR expression (panel D).
The estimated median length of chronic phase for patients with deletions was 44 months (95% CI, 14-73 months), compared with 84 months (95% CI, 65-103 months) for patients without deletions. This difference was significant by log-rank analysis (P = .02) (Figure 3C). By contrast, the estimated median length of chronic phase was 65 months (95% CI, 55-75 months) for ABL-BCR–negative patients and 79 months (95% CI, 63-94 months) forABL-BCR–positive patients (Figure 3D). This difference was not significant by log-rank analysis (P = .23). Exclusion of patients with a deletion from the latter comparison resulted in an increase in the estimated length of chronic phase inABL-BCR–negative patients to 92 months (95% CI, 53-130 months).
These results demonstrate that lack of ABL-BCR expression itself is not an indicator of poor prognosis and therefore suggest that deletions of the derivative chromosome 9 confer a poor prognosis by an alternative mechanism.
Derivative chromosome 9 deletions are not associated with increased levels of BCR-ABL transcripts
BCR-ABL transcripts were quantitated in diagnostic samples and compared between patients according to deletion status. Real-time RT-PCR was used to measure BCR-ABL andB2M transcript levels in 3 cell lines (1 deleted and 2 nondeleted) and in samples obtained from 16 patients (4 deleted and 12 nondeleted). There was no significant difference between samples from deleted and nondeleted patients in the proportion of CD34+ cells, blasts, promyelocytes, or myelocytes (Mann-Whitney U test, P = .90,P = .84, P = .43, and P = .56, respectively). As shown in Table 2 the levels of BCR-ABL transcripts were not increased in the cell line carrying a derivative 9 deletion compared with the 2 cell lines that lacked a deletion. Moreover, there was no significant difference in the levels of BCR-ABL transcripts (expressed as a percentage of the control gene) or in the ΔCT values obtained for patients with a deletion compared with those lacking a deletion by Mann-Whitney U test (P = 1.00) (Table 2). These results demonstrate that deletion of the derivative chromosome 9 is not consistently accompanied by an increase inBCR-ABL expression.
There are no differences in expression of theBCR-ABL transcript between patients with and without deletions
| Patient/cell line . | Translocation type . | Transcript type . | BCR-ABLCT . | B2MCT . | ΔCT . | BCR-ABLtranscripts, % of control gene . |
|---|---|---|---|---|---|---|
| Deleted cell line | ||||||
| MC3 | Cl | b3a2 | 24.5 (± 0.1) | 20.5 (± 0.01) | 4 | 6.9 |
| Patients with deletions | ||||||
| DS63 | Cl | b3a2 | 34.5 (± 0.5) | 27.0 (± 0.3) | 7.5 | 0.7 |
| JD66 | Co | b2a2 | 28.3 (± 0.1) | 22.2 (± 0.04) | 6.1 | 1.9 |
| MF68 | Cl | b3a2 | 38.5 (± 0.7) | 30.6 (± 0.1) | 7.9 | 0.5 |
| SP68 | Cl | b2a2 | 29.2 (± 0.2) | 24.7 (± 0.2) | 4.5 | 5.2 |
| Mean for patients with deletions | 6.5 | 1.4 | ||||
| Median for patients with deletions | 6.8 (4.9-7.8)* | 1.1 (4.0-0.6)* | ||||
| Nondeleted cell lines | ||||||
| MEG-01 | Cl | b2a2 | 24.1 (± 0.06) | 22.7 (± 0.1) | 1.4 | 39.6 |
| BV173 | Cl | b2a2 | 23.7 (± 0.2) | 19.5 (± 0.5) | 4.2 | 6.3 |
| Patients without deletions | ||||||
| DS70 | Cl | b3a2 | 37.4 (± 0.2) | 24.9 (± 0.2) | 12.5 | 0.03 |
| JF69 | Co | b3a2 | 33.4 (± 0.03) | 27.9 (± 0.2) | 5.5 | 2.7 |
| JH55 | Cl | b2a2 | 30.6 (± 0.2) | 23.3 (± 0.02) | 7.3 | 0.9 |
| JK67 | Cl | b2a2 | 31.3 (± 0.1) | 24.7 (± 0.1) | 6.6 | 1.3 |
| KC61 | Cl | b2a2 | 30.8 (± 0.2) | 24.4 (± 0.06) | 6.4 | 1.5 |
| MA64 | Co | b3a2 | 35.9 (± 0.4) | 28.4 (± 0.4) | 7.5 | 0.7 |
| MT72 | Cl | b2a2 | 31.0 (± 0.3) | 24.3 (± 0.01) | 6.7 | 1.2 |
| RS89 | Cl | b2a2 | 34.0 (± 0.2) | 28.8 (± 0.2) | 5.2 | 3.5 |
| SA65 | Cl | b3a2 | 28.5 (± 0.2) | 21.6 (± 0.1) | 6.9 | 1.1 |
| SB60 | Cl | b3a2 | 30.4 (± 0.02) | 23.8 (± 0.04) | 6.6 | 1.3 |
| SG68 | Cl | b3a2 | 30.9 (± 0.2) | 24.3 (± 0.04) | 6.6 | 1.3 |
| TJ44 | Cl | b2a2 | 29.4 (± 0.1) | 24.1 (± 0.04) | 5.3 | 3.0 |
| Mean for patients without deletions | 6.9 | 1.1 | ||||
| Median for patients without deletions | 6.6 (5.7-7.2)* | 1.3 (2.4-0.9)* |
| Patient/cell line . | Translocation type . | Transcript type . | BCR-ABLCT . | B2MCT . | ΔCT . | BCR-ABLtranscripts, % of control gene . |
|---|---|---|---|---|---|---|
| Deleted cell line | ||||||
| MC3 | Cl | b3a2 | 24.5 (± 0.1) | 20.5 (± 0.01) | 4 | 6.9 |
| Patients with deletions | ||||||
| DS63 | Cl | b3a2 | 34.5 (± 0.5) | 27.0 (± 0.3) | 7.5 | 0.7 |
| JD66 | Co | b2a2 | 28.3 (± 0.1) | 22.2 (± 0.04) | 6.1 | 1.9 |
| MF68 | Cl | b3a2 | 38.5 (± 0.7) | 30.6 (± 0.1) | 7.9 | 0.5 |
| SP68 | Cl | b2a2 | 29.2 (± 0.2) | 24.7 (± 0.2) | 4.5 | 5.2 |
| Mean for patients with deletions | 6.5 | 1.4 | ||||
| Median for patients with deletions | 6.8 (4.9-7.8)* | 1.1 (4.0-0.6)* | ||||
| Nondeleted cell lines | ||||||
| MEG-01 | Cl | b2a2 | 24.1 (± 0.06) | 22.7 (± 0.1) | 1.4 | 39.6 |
| BV173 | Cl | b2a2 | 23.7 (± 0.2) | 19.5 (± 0.5) | 4.2 | 6.3 |
| Patients without deletions | ||||||
| DS70 | Cl | b3a2 | 37.4 (± 0.2) | 24.9 (± 0.2) | 12.5 | 0.03 |
| JF69 | Co | b3a2 | 33.4 (± 0.03) | 27.9 (± 0.2) | 5.5 | 2.7 |
| JH55 | Cl | b2a2 | 30.6 (± 0.2) | 23.3 (± 0.02) | 7.3 | 0.9 |
| JK67 | Cl | b2a2 | 31.3 (± 0.1) | 24.7 (± 0.1) | 6.6 | 1.3 |
| KC61 | Cl | b2a2 | 30.8 (± 0.2) | 24.4 (± 0.06) | 6.4 | 1.5 |
| MA64 | Co | b3a2 | 35.9 (± 0.4) | 28.4 (± 0.4) | 7.5 | 0.7 |
| MT72 | Cl | b2a2 | 31.0 (± 0.3) | 24.3 (± 0.01) | 6.7 | 1.2 |
| RS89 | Cl | b2a2 | 34.0 (± 0.2) | 28.8 (± 0.2) | 5.2 | 3.5 |
| SA65 | Cl | b3a2 | 28.5 (± 0.2) | 21.6 (± 0.1) | 6.9 | 1.1 |
| SB60 | Cl | b3a2 | 30.4 (± 0.02) | 23.8 (± 0.04) | 6.6 | 1.3 |
| SG68 | Cl | b3a2 | 30.9 (± 0.2) | 24.3 (± 0.04) | 6.6 | 1.3 |
| TJ44 | Cl | b2a2 | 29.4 (± 0.1) | 24.1 (± 0.04) | 5.3 | 3.0 |
| Mean for patients without deletions | 6.9 | 1.1 | ||||
| Median for patients without deletions | 6.6 (5.7-7.2)* | 1.3 (2.4-0.9)* |
The cycle threshold (CT) for BCR-ABL and the control gene B2M are shown. The ΔCT value for a sample corrects for any differences in complementary DNA between samples, and these values are directly comparable between samples. The mean, median, and interquartile range are shown for the ΔCT values of patients with and without deletions. The fold differences between the control gene andBCR-ABL were calculated from the equation, fold difference = (1 + E)−ΔCT, where E represents the efficiency of the PCR reaction. The fold differences are represented by BCR-ABL as a percentage of expression of the control gene. There were no differences in levels of transcripts between patients with deletions and those who lacked them by Mann-Whitney U test (P = 1.00).
CT indicates cycle threshold; Cl, classical Philadelphia translocation; Co, complex/variant translocation.
Interquartile range.
Derivative chromosome 9 deletions are not associated with increased karyotypic instability
To address the possibility of an increased karyotypic instability accompanying derivative chromosome 9 deletions, we have analyzed the karyotype of 84 patients following progression to blast crisis (16 patients with deletions and 68 patients without deletions). A summary of the results is presented in Table 3. The proportion of patients with secondary cytogenetic changes was 51 of 68 (75%) for patients without deletions and 12 of 16 (75%) for patients with deletions (Fisher exact test, P = 1.00). There was also no difference in the prevalence of individual specific chromosomal changes (Table 3), although the numbers in each case are small. The mean and median total number of changes per patient was 2.2 and 1.0, respectively (interquartile range, 0.25-3.0), for patients without deletions and 1.9 and 2.0, respectively (interquartile range, 0.25-2.75) for patients with deletions. There was no significant difference by Mann-Whitney U test (P = .86). The mean and median number of double-stranded breaks was 0.9 and 1.0 (interquartile range, 0-1) for patients without deletions and 1.1 and 1.0 (interquartile range, 0-1) for patients with deletions. Again, there was no significant difference by Mann-Whitney U test (P = .62). These data therefore suggest that patients with deletions do not have an increased tendency to develop additional cytogenetic alterations.
Incidence of chromosomal abnormalities in patients with and without deletions upon development of blast crisis
| Cytogenetic characteristics . | Deletion status . | |
|---|---|---|
| Nondeleted (%) (n = 68) . | Deleted (%) (n = 16) . | |
| Any abnormality | 51 (75) | 12 (75) |
| Extra Ph+ | 12 (18) | 2 (12) |
| Trisomy 8 | 9 (14) | 3 (18) |
| Loss of 17p | 8 (13) | 2 (12) |
| Trisomy 19 | 5 (8) | 1 (6) |
| Trisomy 21 | 5 (8) | 0 |
| del13(q) | 3 (5) | 0 |
| Marker | 5 (8) | 3 (18) |
| Total changes per patient, mean/median | 2.2/1.0 | 1.9/2.0 |
| Double-stranded breaks per patient, mean/median | 0.9/1.0 | 1.1/1.0 |
| Cytogenetic characteristics . | Deletion status . | |
|---|---|---|
| Nondeleted (%) (n = 68) . | Deleted (%) (n = 16) . | |
| Any abnormality | 51 (75) | 12 (75) |
| Extra Ph+ | 12 (18) | 2 (12) |
| Trisomy 8 | 9 (14) | 3 (18) |
| Loss of 17p | 8 (13) | 2 (12) |
| Trisomy 19 | 5 (8) | 1 (6) |
| Trisomy 21 | 5 (8) | 0 |
| del13(q) | 3 (5) | 0 |
| Marker | 5 (8) | 3 (18) |
| Total changes per patient, mean/median | 2.2/1.0 | 1.9/2.0 |
| Double-stranded breaks per patient, mean/median | 0.9/1.0 | 1.1/1.0 |
The prevalence of abnormalities was similar in patients with and without deletions, and the type of changes are consistent with those previously described following progression to blast crisis.26 There were no differences in the proportion of patients with secondary changes by Fisher exact test (P= 1.00) or in the total number of changes or double-stranded breaks by Mann-Whitney U test (P = .86 andP = .62, respectively) between patients with and without deletions.
Ph indicates Philadelphia chromosome.
Discussion
Large deletions of the derivative chromosome 9 occur at the time of the Ph translocation,17 usually span the translocation breakpoint,15 and represent a powerful indicator of poor prognosis.16 17 In this paper, we have investigated several potential mechanisms by which such deletions might result in rapid disease progression.
Our results demonstrate that all patients with a deletion detectable by dual-fusion FISH lacked expression of ABL-BCR. This result is consistent with our previous demonstration that deletions span the translocation breakpoint on the derivative chromosome 9.15However, our data also show that 65% of patients lackingABL-BCR expression do not have a deletion detectable by FISH. This observation suggests the existence of other mechanisms by which ABL-BCR transcription can be abolished. It is likely that small deletions occur that would abolish ABL-BCRtranscription but that would be below the threshold of detection for the dual-fusion FISH and other, similar FISH-based techniques that use large probes. This would be consistent with previous evidence for small deletions adjacent to the translocation breakpoints in CML and other leukemias.37-42 In addition, approximately 10% of patients have a breakpoint on the derivative chromosome 9 that is upstream of ABL exon 1b, and that therefore removes both sites at whichABL transcription is normally initiated.43,44Finally, some patients with a variant Ph translocation have 5′ABL and 3′ BCR sequences present on separate chromosomes, suggesting that an ABL-BCR transcript would not be formed.45
We also show that loss of ABL-BCR transcription is not associated with reduced survival or reduced length of chronic phase. This demonstrates that lack of ABL-BCR expression is not sufficient for the poor prognosis associated with overt derivative chromosome 9 deletions. However, our results do not exclude the possibility that lack of ABL-BCR expression may be necessary for deletions to confer a poor outcome. It is conceivable that the ABL-BCR protein could directly or indirectly modulate activity of the BCR-ABL protein.24,25 However, existence of a stable ABL-BCR protein product has not yet been demonstrated.46
One important practical aspect of our results is that it demonstrates that all patients with ABL-BCR transcripts lack a deletion detectable by dual fusion FISH. RT-PCR for ABL-BCRcan therefore be used as a rapid and inexpensive screen, with a high predictive value, to identify the majority of patients who lack an overt derivative chromosome 9 deletion and who therefore fall into a good prognostic category. FISH could be subsequently used to study the 30% to 40% of patients who lack ABL-BCR transcripts, in order to identify the subset who have an overt deletion and who therefore have a poor prognosis.
In addition to loss of ABL-BCR expression, we have also investigated 2 other potential mechanisms that could account for the association between derivative chromosome 9 deletions and rapid disease progression. First, it is possible that translocations that result in deletions on the derivative chromosome 9 will also be accompanied by deletions of the Ph chromosome. Such deletions would need to be small enough to permit formation of a BCR-ABL transcript but could nonetheless remove regulatory elements and thereby modulateBCR-ABL transcription. There are several examples of small deletions adjacent to the breakpoint of reciprocal translocations in CML and other leukemias,37-42 and small deletions on both derivative chromosomes have also been shown in constitutional reciprocal translocations.47 Moreover, increased levels ofBCR-ABL transcripts are associated with progression to blast crisis.26 28 However, our results argue against this model since patients with and without deletions do not have discernibly different levels of BCR-ABL transcripts.
Second, it is also possible that large deletions may represent the consequence of a translocation occurring in a target cell with an inherent predisposition to chromosome rearrangements. In this model, the deletion would not represent the cause of the poor prognosis but would reflect the underlying chromosome instability. The fact that deletions occur in a subset of patients could reflect heterogeneity within the stem cell compartment and/or differences among individuals. However, our results do not provide any evidence for increased chromosome instability in patients with derivative chromosome 9 deletion.
Taken together, the results presented here strongly suggest that derivative chromosome 9 deletions are associated with a poor prognosis as a consequence of the loss of one or more genes within the deleted region. The biological consequence of the deletion could be a direct effect of haploinsufficiency or a consequence of one or more “second hits” affecting the remaining normal alleles. The deletions are large, extending up to 5.5 megabase (Mb) on the chromosome 9 side of the translocation breakpoint, and up to 17 Mb on the chromosome 22 side. Existing data do not allow us to ascertain whether the critical region involves chromosome 9 sequences, chromosome 22 sequences, or both. Each of these regions is gene rich, and between them, the regions contain approximately 300 genes. Our data therefore lay the foundation for a systematic search for critical target genes in the corresponding regions of chromosome 9 and 22.
We are grateful to Emma Frith, Kevin Jestice, Ian Carter, Kate Martin, Francoise Brizard, Laurence Daheron, and Eveline Hennig for assistance with samples.
Supported by a Medical Research Council (United Kingdom) Clinical Training Fellowship (B.J.P.H.); the Association pour la Recherche sur le Cancer (E.D.), the Fondation de France (Comite Leucemie) (E.D.); and the Pre-Leukaemia Society (E.D.); work in the authors' laboratories is funded by the Leukemia Research Fund and the Kay Kendall Leukemia Fund.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Anthony R. Green, Dept of Hematology, University of Cambridge, Wellcome Trust/MRC Bldg, Hills Rd, Cambridge, CB2 2XY, United Kingdom; e-mail: arg1000@cam.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal