Abstract
Relative proportions of peripheral blood (PB) B lymphocytes (B220%) as well as CD4 (CD4%) and CD8 (CD8%) T lymphocytes differ significantly among inbred mouse strains: B220% is high in C57BL/6J (B6) and C57BR/cdJ, intermediate in BALB/cByJ (BALB) and DBA/2J (D2), and low in NOD/LtJ (NOD) and SJL/J (SJL) mice, whereas CD4% and CD8% are high in NOD and SJL mice and low in the other 4 strains. By following segregating genetic markers linked to these traits in (B6 × D2) recombinant inbred (BXD RI) mice, the study defined 2 quantitative trait loci (QTLs) for the B220% phenotype:Pbbcp1 (peripheral blood B cell percentage 1, logarithm of odds [LOD] 4.1, P < .000 01) and Pbbcp2(LOD 3.7, P < .000 04) on chromosome 1 (Chr 1) at about 63 cM and 48 cM; one suggestive locus for the CD4% phenotype (LOD 2.6,P < .000 57) on Chr 8 at about 73 cM; and one QTL for the CD8% phenotype: Pbctlp1 (peripheral blood cytotoxic T lymphocyte percentage 1, LOD 3.8, P < .000 02) on Chr 19 at about 12 cM. The study further segregated PB lymphocyte proportions in B6SJLF2 mice by using DNA markers adjacent to these mapped QTLs and found that the Pbbcp1 locus (LOD 5.6,P < .000 01) was also important in this mouse population. In both BXD RI and B6SJLF2 mice, QTLs regulating B-cell proportions showed no significant effect on T-cell proportions and vice versa. Thus, PB B- and T-lymphocyte proportions are regulated separately by different genetic elements.
Introduction
Mature leukocytes in mouse and human peripheral blood (PB) express specific cell surface molecules (markers) that can be detected by fluorescence-activated cell staining (FACS) using specific antibodies.1,2 The major categories of PB leukocytes include B lymphocytes that express B220, T-helper lymphocytes that express CD4, cytotoxic T lymphocytes that express CD8, and granulocytes that express Gr1. Under physiologic conditions, proportions of PB leukocyte subsets are relatively constant in mice from a particular genotype, showing a precise regulation of hematopoietic lineage commitment, differentiation, maturation, recruitment, and elimination.3 Proportions of PB leukocyte subsets can be dramatically altered by spontaneous and induced mutations; for example, a mutation in the DNA-dependent protein kinase gene in severe combined immune-deficient mice results in the absence of mature B and T lymphocytes in blood circulation.4-6 In addition, stromal cells, cytokines, receptors, ligands, signaling molecules, and transcription factors have been shown to affect levels of B and T lymphocytes.7-10
Previous studies identified molecular factors that play important roles in the regulation of B-and T-cell commitment and balance such as cytokines interleukin-3 (IL-3) and IL-7, signaling receptor and ligandNotch and Jagged, paired-box genePax5, and the transcriptional factors E2A andEBF.3,11-13 Mutations that change the relative levels of other leukocyte subsets may also indirectly affect B- and T-cell proportions.14 Earlier studies found that the CD4/CD8 ratio is under genetic control in the mouse as well as in humans.15-17 A study using inbred mouse strains identified a quantitative trait locus (QTL) that regulates peripheral B220% on mouse chromosome 15 (Chr 15).18 Differences in the percentage of pre-B cells (BP-1+B220+) in the bone marrow of SL/Kh and NFS/N mice helped to map another regulatory QTL on mouse Chr 3.19 These studies illustrated that there are specific genetic loci that regulate CD4/CD8 ratio, B-cell apoptosis, and pre–B-cell expansion.
We and others have found significant strain differences in primitive immunohematopoietic progenitor cell functions between B6, BALB, and D2 mice and have defined QTLs that regulate hematopoietic stem cell development, proliferation, and senescence.20-23 It is important to also define strain differences in mature blood cell proportions. The current study is focused on the genetic elements that regulate strain differences in blood cell proportions and tests whether the same QTL affects both B- and T-cell proportions.
We examined PB B220%, CD4%, and CD8% in healthy young mice of 6 inbred strains from 3 separate original stocks and found significant strain differences. We then segregated these strain differences in 35 strains of (C57BL/6 × DBA/2) recombinant inbred (BXD RI) mice and mapped 2 QTLs for the B220% phenotype, one QTL for the CD8% phenotype, and one suggestive locus for the CD4% phenotype. We further analyzed 98 intercross F2 mice derived from C57BL/6J (B6) and SJL/J (SJL) inbred strains (B6SJLF2) and found that one of the QTLs for the B220% phenotype was also mapped to the same location in the B6SJLF2 mice. In both studies, PB B- and T-lymphocyte proportions are regulated separately by different QTLs.
Materials and methods
Mice
Mice of the B6, C57BR/cdJ, BALB/cByJ (BALB), DBA/2J (D2), NOD/LtJ (NOD), and SJL inbred strains, of 35 BXD RI strains, and of the B6SJLF2 intercross were all produced and raised at The Jackson Laboratory (Bar Harbor, ME) using standard animal care and nutrition.24 Mice of both genders were used as specified in each experiment.
Sample collection and FACS analysis
Procedures for FACS analysis were adapted from previous studies.25 In brief, 2 micro-hematocrit tubes (75 μL) of blood were taken from the orbital sinus of each mouse and mixed with 1350 μL Hanks balanced salt solution in the presence of 5 mM ethylenediaminetetraacetic acid. Diluted blood samples were incubated in Gey solution twice for 10 minutes each time to lyse erythrocytes. Leukocytes were stained with specific antibody cocktails in FACS buffer for 30 minutes. When a biotinylated antibody was used, samples were stained with streptavidin red 670 for an additional 30 minutes. Cells were kept on ice or at 4°C during incubation, staining, and centrifugation procedures. Monoclonal antibodies specific for mouse B220 (clone RA3-6B2), CD4 (clone GK1.5), CD8 (clone 53-6.72), Gr1 (clone RB6-8C5), and the streptavidin-conjugated dye red 670 were purchased from Pharmingen (San Diego, CA). Stained cells were analyzed by either FACScan II or FACScalibur flow cytometry (Becton Dickinson Immunocytometry Systems, Mansfield, MA). We collected 10 000 to 20 000 leukocytes for each sample.
Polymerase chain reaction
Genomic DNA samples were prepared from mouse tail tips, and DNA markers were analyzed by polymerase chain reaction (PCR) for each of the 98 B6SJLF2 mice. Primers for the Gli2 locus,Gli2-pA: TTCAGGCAGACCAAAGATAGAACATT and Gli2-pB: CACTGACATATGTACCATTTTCAT and primers for the Bcl2 locus, 24.MMBCL2A: CATTATCAATGATGTAC CATG and 24.MMBCL2B: GCAGTAAATAGCTGATTCGAC were purchased from One Trick Pony Oligos (Ramona, CA). Primers for regular Mit DNA microsatellite markers were purchased from Research Genetics (Huntsville, AL). PCRs were carried out in a GeneAmp PCR system 9600 (Perkin Elmer) using Taq DNA polymerase. We used a program with a 97°C touchdown for 30 seconds followed by 40 cycles of 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds + 1 second. A 10-minute enlongation at 72°C was added at the end of the reaction. Each sample was amplified in 10 μL volume, electrophoresed in a 3% agarose gel, and stained with ethidium bromide.
QTL mapping and statistical analysis
Genotype data for the 35 BXD RI mouse strains were retrieved online from the Mouse Genome Database, which is maintained at The Jackson Laboratory (http://www.bioinformatics.jax.org). We used Map Manager QTXb11 software, also available online at the same Web site, for QTL mapping analyses.26 Significance of linkage was judged by using the LOD (logarithm of odds) score in both the RI lines and the B6SJLF2 intercross mice.26-28 The B220%, CD4%, and CD8% of B6SJLF2 mice were also tested by variance analysis, using the JMP statistical discovery software (SAS Institute, Cary, NC) on the Fit Model platform to define allelic difference.29
Results
Strain differences in PB lymphocyte proportions
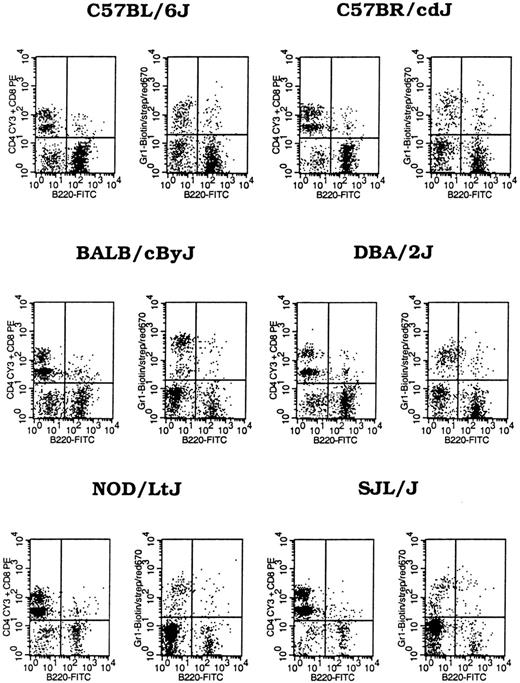
We analyzed PB leukocyte proportions in healthy young (2-3 months) B6, C57BR/cdJ, BALB, D2, NOD, and SJL mice. Data presented in Table1 show that concentrations of circulating white blood cells (WBCs) were high in B6 and C57BR/cdJ mice, slightly lower in BALB and D2 mice, and significantly lower (P < .01) in NOD and SJL mice. Percentages of B cells (B220%) were significantly different among the 6 inbred strains (P < .01): high in B6 (67%) and C57BR/cdJ (60%) mice, intermediate in BALB (46%) and D2 (46%) mice, and low in NOD (18%) and SJL (16%) mice. Percentages of CD4 (CD4%) and CD8 (CD8%) T lymphocytes were significantly higher (P < .01,P < .01) in NOD (50%, 17%) and SJL (58%, 20%) mice than in the other 4 strains (CD4, 13%-30%; CD8, 6%-11%), whereas percentages of granulocytes (Gr1%) were similar in the 6 strains (Table 1, Figure 1). Thus, there are significant strain differences in mouse PB B220%, CD4%, and CD8%, suggesting that PB lymphocyte proportions are genetically regulated.
Strain effects on peripheral blood lymphocyte composition
| Strain . | WBCs (106/mL) . | B220% . | CD4% . | CD8% . | Gr1% . |
|---|---|---|---|---|---|
| C57BL/6J | 11.2 ± 0.5 | 67.0 ± 1.1 | 13.3 ± 0.6 | 7.7 ± 0.3 | 11.1 ± 0.3 |
| C57BR/cdJ | 14.4 ± 0.3 | 59.6 ± 1.3 | 23.2 ± 1.2 | 10.4 ± 1.5 | 13.2 ± 0.6 |
| BALB/cByJ | 10.3 ± 0.6 | 45.8 ± 2.7 | 29.3 ± 1.7 | 10.8 ± 0.6 | 13.3 ± 1.2 |
| DBA/2J | 8.6 ± 1.1 | 46.1 ± 4.6 | 18.3 ± 0.1 | 5.9 ± 0.2 | 15.5 ± 2.9 |
| NOD/LtJ | 3.2 ± 0.6 | 17.6 ± 0.9 | 49.5 ± 1.5 | 17.1 ± 0.8 | 12.9 ± 2.8 |
| SJL/J | 5.7 ± 1.0 | 16.1 ± 1.3 | 57.5 ± 1.1 | 20.1 ± 1.6 | 8.3 ± 1.2 |
| B6D2F1 | 9.6 ± 0.2 | 63.7 ± 1.0 | 14.9 ± 0.3 | 8.9 ± 0.4 | 12.1 ± 0.6 |
| CByB6F1 | 8.6 ± 1.3 | 59.3 ± 2.0 | 12.8 ± 0.7 | 6.2 ± 1.4 | 11.9 ± 1.6 |
| Strain effect | P < .01 | P < .01 | P < .01 | P < .01 | NS |
| Strain . | WBCs (106/mL) . | B220% . | CD4% . | CD8% . | Gr1% . |
|---|---|---|---|---|---|
| C57BL/6J | 11.2 ± 0.5 | 67.0 ± 1.1 | 13.3 ± 0.6 | 7.7 ± 0.3 | 11.1 ± 0.3 |
| C57BR/cdJ | 14.4 ± 0.3 | 59.6 ± 1.3 | 23.2 ± 1.2 | 10.4 ± 1.5 | 13.2 ± 0.6 |
| BALB/cByJ | 10.3 ± 0.6 | 45.8 ± 2.7 | 29.3 ± 1.7 | 10.8 ± 0.6 | 13.3 ± 1.2 |
| DBA/2J | 8.6 ± 1.1 | 46.1 ± 4.6 | 18.3 ± 0.1 | 5.9 ± 0.2 | 15.5 ± 2.9 |
| NOD/LtJ | 3.2 ± 0.6 | 17.6 ± 0.9 | 49.5 ± 1.5 | 17.1 ± 0.8 | 12.9 ± 2.8 |
| SJL/J | 5.7 ± 1.0 | 16.1 ± 1.3 | 57.5 ± 1.1 | 20.1 ± 1.6 | 8.3 ± 1.2 |
| B6D2F1 | 9.6 ± 0.2 | 63.7 ± 1.0 | 14.9 ± 0.3 | 8.9 ± 0.4 | 12.1 ± 0.6 |
| CByB6F1 | 8.6 ± 1.3 | 59.3 ± 2.0 | 12.8 ± 0.7 | 6.2 ± 1.4 | 11.9 ± 1.6 |
| Strain effect | P < .01 | P < .01 | P < .01 | P < .01 | NS |
Data shown as mean ± SEM of 3 to 4 males measured for each strain at 2 to 3 months of age.
WBC indicates white blood cell.
Peripheral blood leukocyte composition.
Peripheral blood from 2- to 3-month-old mice were stained with B220-FITC + CD4-Cy3 + CD8-PE +Gr1-biotin and streptavidin-red 670 after erythrocytes were lysed with Gey solution. Data shown are dot plots of representatives from 3 to 4 male mice measured for each strain.
Peripheral blood leukocyte composition.
Peripheral blood from 2- to 3-month-old mice were stained with B220-FITC + CD4-Cy3 + CD8-PE +Gr1-biotin and streptavidin-red 670 after erythrocytes were lysed with Gey solution. Data shown are dot plots of representatives from 3 to 4 male mice measured for each strain.
To define the pattern of inheritance, we measured B220%, CD4%, CD8%, Gr1%, and total WBCs in B6D2F1 and CByB6F1 hybrid mice. Interestingly, both hybrid F1 stocks had leukocyte proportions similar to those of B6 mice (Table 1), indicating that B6 genetic elements are dominant to those of the BALB or D2 genetic elements in the regulation of PB lymphocyte proportions.
Mapping QTLs for PB lymphocyte proportions in BXD RI mice
To segregate the genetic factors that regulate lymphocyte proportions, we measured B220%, CD4%, and CD8% in PB of healthy young mice from 35 BXD RI strains. Individuals from the same RI strain share the same genotype and are homozygous at almost all (> 99.99%) loci, with about half of the loci carrying alleles from each parental strain.30 31 Many polymorphic loci and DNA markers have been analyzed in the BXD RI strains, and the allele information is available online through the Mouse Genome Database. The mean B220%, CD4%, and CD8% values for the 35 BXD RI strains shown in Table2 were used in mapping analyses, which relied heavily on data from BXD 1-32 (Table 2) because many marker loci have not been defined in BXD 33-42.
Concurrence of B220%, CD4%, and CD8% with marker loci in BXD RI mice
| BXD . | Pbbcp2 . | B% . | Pbbcp1 . | CD4% . | D8Mit156 . | CD8% . | Pbctlp1 . |
|---|---|---|---|---|---|---|---|
| 1 | B | 60 | B | 20 | D | 7.7 | B |
| 2 | B | 64 | B | 9 | B | 6.1 | B |
| 5 | D | 49 | D | 25 | D | 11.5 | B |
| 6 | D | 38 | D | 18 | D | 9.0 | D |
| 8 | B | 58 | B | 14 | D | 6.4 | D |
| 9 | B | 61 | B | 14 | D | 11.7 | B |
| 11 | D | 44 | D | 18 | B | 7.1 | D |
| 12 | B | 61 | B | 16 | D | 7.1 | B |
| 13 | D | 55 | D | 12 | D | 5.6 | D |
| 14 | B | 67 | B | 6 | B | 2.5 | B |
| 15 | D | 53 | D | 16 | D | 7.5 | B |
| 16 | D | 34 | D | 19 | D | 14.6 | D |
| 18 | D | 44 | D | 11 | B | 7.1 | B |
| 19 | B | 60 | B | 10 | B | 6.4 | B |
| 20 | D | 9 | D | 25 | D | 25.4 | D |
| 21 | B | 48 | B | 13 | D | 6.9 | B |
| 22 | B | 58 | B | 19 | D | 10.0 | B |
| 23 | D | 39 | D | 15 | B | 9.0 | D |
| 24 | D | 44 | D | 20 | D | 8.0 | B |
| 25 | B | 69 | B | 11 | D | 4.5 | B |
| 27 | — | 53 | B | 11 | D | 13.5 | D |
| 28 | — | 56 | — | 10 | B | 6.9 | B |
| 29 | D | 52 | D | 15 | D | 11.7 | B |
| 30 | D | 49 | D | 16 | B | 11.7 | D |
| 31 | B | 61 | B | 7 | B | 6.7 | B |
| 32 | D | 46 | D | 12 | D | 5.4 | B |
| 33 | — | 58 | — | 12 | B | 8.4 | D |
| 34 | — | 46 | — | 20 | D | 7.3 | B |
| 35 | — | 42 | — | 18 | D | 15.7 | D |
| 36 | — | 54 | — | 16 | B | 9.6 | D |
| 37 | — | 44 | — | 21 | D | 14.9 | D |
| 38 | — | 43 | — | 18 | D | 15.7 | D |
| 39 | — | 49 | — | 19 | D | 9.0 | D |
| 40 | — | 63 | — | 11 | B | 7.3 | B |
| 42 | — | 50 | — | 12 | D | 7.6 | B |
| BXD . | Pbbcp2 . | B% . | Pbbcp1 . | CD4% . | D8Mit156 . | CD8% . | Pbctlp1 . |
|---|---|---|---|---|---|---|---|
| 1 | B | 60 | B | 20 | D | 7.7 | B |
| 2 | B | 64 | B | 9 | B | 6.1 | B |
| 5 | D | 49 | D | 25 | D | 11.5 | B |
| 6 | D | 38 | D | 18 | D | 9.0 | D |
| 8 | B | 58 | B | 14 | D | 6.4 | D |
| 9 | B | 61 | B | 14 | D | 11.7 | B |
| 11 | D | 44 | D | 18 | B | 7.1 | D |
| 12 | B | 61 | B | 16 | D | 7.1 | B |
| 13 | D | 55 | D | 12 | D | 5.6 | D |
| 14 | B | 67 | B | 6 | B | 2.5 | B |
| 15 | D | 53 | D | 16 | D | 7.5 | B |
| 16 | D | 34 | D | 19 | D | 14.6 | D |
| 18 | D | 44 | D | 11 | B | 7.1 | B |
| 19 | B | 60 | B | 10 | B | 6.4 | B |
| 20 | D | 9 | D | 25 | D | 25.4 | D |
| 21 | B | 48 | B | 13 | D | 6.9 | B |
| 22 | B | 58 | B | 19 | D | 10.0 | B |
| 23 | D | 39 | D | 15 | B | 9.0 | D |
| 24 | D | 44 | D | 20 | D | 8.0 | B |
| 25 | B | 69 | B | 11 | D | 4.5 | B |
| 27 | — | 53 | B | 11 | D | 13.5 | D |
| 28 | — | 56 | — | 10 | B | 6.9 | B |
| 29 | D | 52 | D | 15 | D | 11.7 | B |
| 30 | D | 49 | D | 16 | B | 11.7 | D |
| 31 | B | 61 | B | 7 | B | 6.7 | B |
| 32 | D | 46 | D | 12 | D | 5.4 | B |
| 33 | — | 58 | — | 12 | B | 8.4 | D |
| 34 | — | 46 | — | 20 | D | 7.3 | B |
| 35 | — | 42 | — | 18 | D | 15.7 | D |
| 36 | — | 54 | — | 16 | B | 9.6 | D |
| 37 | — | 44 | — | 21 | D | 14.9 | D |
| 38 | — | 43 | — | 18 | D | 15.7 | D |
| 39 | — | 49 | — | 19 | D | 9.0 | D |
| 40 | — | 63 | — | 11 | B | 7.3 | B |
| 42 | — | 50 | — | 12 | D | 7.6 | B |
Two males 3 to 5 months of age were measured for each BXD RI line.
Pbbcp2 indicates peripheral blood B cell percentage 2;Pbbcp1, peripheral blood B cell percentage 1;Pbctlp1, peripheral blood cytotoxic T lymphocyte percentage 1; B, B6 allele; D, D2 allele; and —, genotype data not available.
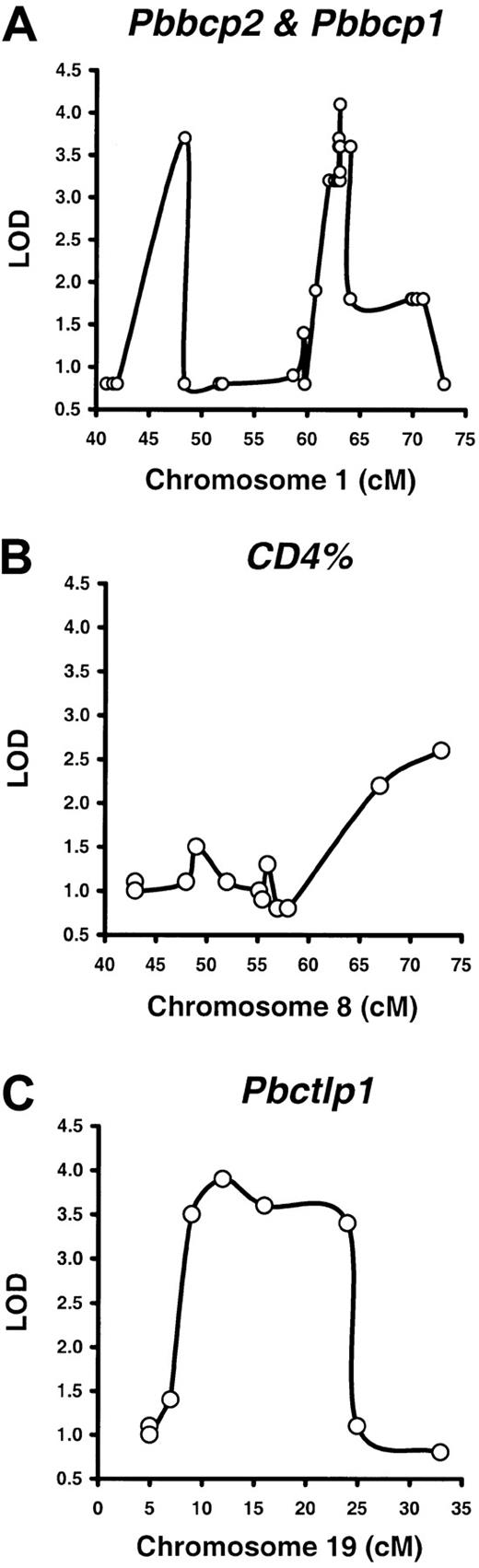
For the B220% phenotype, we mapped 2 QTLs. One of these QTLs showed linkage to the 62- to 64-cM segment of Chr 1 with the strongest linkage to the D1Mit188 marker at 63.1 cM (LOD 4.1,P < .000 01). We have provisionally designated this locus peripheral blood B cell percentage 1, or Pbbcp1(Figure 2A). The other locus also mapped to Chr 1 with the strongest linkage to the D1Ncvs45 marker at 48.4 cM (LOD 3.7, P < .000 04). We designated this locus Pbbcp2 (Figure 2A). Both P values passed the P < .0001 threshold and are considered significant in genome-wide mapping analyses.27 32 The Bcl2locus is located in between the 2 QTLs at 59.8 cM.
QTLs for the B220%, CD4%, and CD8% phenotypes in BXD RI mice.
Peripheral blood B220%, CD4%, and CD8% were measured in 35 BXD RI strains for QTL mapping by using predetermined genotype data and the Map Manager QTXb11 software, both available from The Jackson Laboratory (http://www.bioinformatics.jax.org) Web site.56Two QTLs were defined for the B220% phenotype (A), designated peripheral blood B cell percentage 1 (Pbbcp1, LOD 4.1,P < .00001) and Pbbcp2 (LOD 3.7,P < .00004), on chromosome 1 (Chr 1) at 63.1 cM and 48.4 cM, respectively. A suggestive locus was found for the CD4% phenotype (LOD 2.6, P < .000 57) at 73 cM on Chr 8 (B). A QTL for the CD8% phenotype, peripheral blood cytotoxic T lymphocyte percentage 1 (Pbctlp1, LOD 3.8, P < .000 02), was mapped to the 9- to 24-cM region of Chr 19 (C).
QTLs for the B220%, CD4%, and CD8% phenotypes in BXD RI mice.
Peripheral blood B220%, CD4%, and CD8% were measured in 35 BXD RI strains for QTL mapping by using predetermined genotype data and the Map Manager QTXb11 software, both available from The Jackson Laboratory (http://www.bioinformatics.jax.org) Web site.56Two QTLs were defined for the B220% phenotype (A), designated peripheral blood B cell percentage 1 (Pbbcp1, LOD 4.1,P < .00001) and Pbbcp2 (LOD 3.7,P < .00004), on chromosome 1 (Chr 1) at 63.1 cM and 48.4 cM, respectively. A suggestive locus was found for the CD4% phenotype (LOD 2.6, P < .000 57) at 73 cM on Chr 8 (B). A QTL for the CD8% phenotype, peripheral blood cytotoxic T lymphocyte percentage 1 (Pbctlp1, LOD 3.8, P < .000 02), was mapped to the 9- to 24-cM region of Chr 19 (C).
For the CD4% phenotype, we found a putative linkage to a DNA markerD8Mit156 (LOD 2.6, P < .000 57) at 73 cM on Chr 8 (Figure 2B). This P value is considered suggestive and reportable but not fully significant.27 32 For the CD8% phenotype, we mapped a QTL to the 9- to 24-cM region of Chr 19 with the strongest linkage to the D19Mit28 marker at 12 cM (LOD 3.8,P < .000 02). We have designated this locus peripheral blood cytotoxic T lymphocyte percentage 1, or Pbctlp1(Figure 2C).
From the mapping analyses, we noted that the QTL for the B220% phenotype showed no linkage to either the CD4% or the CD8% phenotype. However, the Pbbctlp1 locus had no linkage to the B220% phenotype either. Thus, B- and T-cell proportions are regulated by different QTLs in the BXD RI mice.
Mapping of the Pbbcp1 locus to the same location in B6SJLF2 mice
The existence of RI lines with preanalyzed genotype data provides a good resource for QTL mapping. However, other genetic methods should be used to test QTLs defined by RI lines. Toward this end, we produced a panel of B6SJLF2 mice and measured PB B220%, CD4%, and CD8% at 2 to 3 months of age. We then tested DNA markers on each mouse near each putative locus defined earlier by using the BXD RI lines. When a marker of interest was not polymorphic between B6 and SJL mice, a nearby polymorphic marker was analyzed.
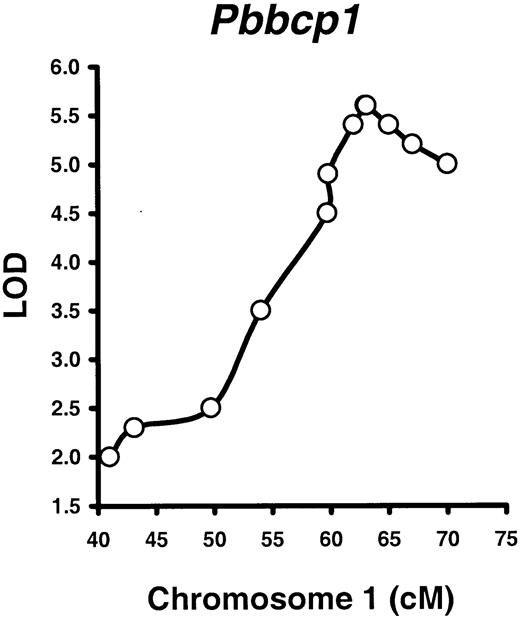
We first tested the hypothesis that Pbbcp1 andPbbcp2 are important loci in the regulation of B220% difference between B6 and SJL mice. This test was done by analyzing 12 polymorphic markers in the 41- to 70-cM region of Chr 1 on 98 B6SJLF2 mice, followed by a mapping analysis. Three markers adjacent to thePbbcp1 locus, D1Mit387 (62 cM), Gli2(63 cM), and D1Mit419 (63.1 cM), all showed strong linkage to the B220% phenotype (LOD 5.4, 5.6, and 5.6, all atP < .000 01), whereas the linkages to the B220% were lower but still significant at Bcl2 (59.8 cM, LOD 4.9).D1Mit139 (65 cM, LOD 5.4), D1Mit286 (67 cM, LOD 5.2), and D1Mit446 (70 cM, LOD 4.9) loci (Figure3). Note that the Gli2 andD1Mit419 loci overlap on Figure 3. At the Gli2locus where Pbbcp1 showed the strongest linkage (LOD 5.6), B220% was significantly higher (P < .01) in the carriers of the B6 (36.6% ± 1.8%) and F1 (32.6% ± 1.0%) alleles than in the carriers of the SJL (6.3% ± 1.3%) allele. Thus, we mapped thePbbcp1 locus to the same 60- to 70-cM Chr 1 location in the B6SJLF2 intercross mouse population. It is of particular interest to note that the Pbbcp1 locus had no effect on either the CD4% or the CD8% phenotype in B6SJLF2 mice, further indicating that PB B- and T-lymphocyte proportions are regulated by different genetic elements.
Mapping the
Pbbcp1 locus in B6SJLF2 mice. A total of 98 B6SJLF2 mice (51 males, 47 females) were produced; measured for PB B220%, CD4%, and CD8% phenotypes at 2 to 3 months of age; tested for selected polymorphic markers adjacent to the Pbbcp1,Pbbcp2, and Pbctlp1 QTL regions; and proceeded for linkage analyses with the use of the Map Manager QTXb11 software. Only the Pbbcp1 locus was mapped to the same Chr 1 region with statistic significance (LOD 5.6,P < .000 01).
Mapping the
Pbbcp1 locus in B6SJLF2 mice. A total of 98 B6SJLF2 mice (51 males, 47 females) were produced; measured for PB B220%, CD4%, and CD8% phenotypes at 2 to 3 months of age; tested for selected polymorphic markers adjacent to the Pbbcp1,Pbbcp2, and Pbctlp1 QTL regions; and proceeded for linkage analyses with the use of the Map Manager QTXb11 software. Only the Pbbcp1 locus was mapped to the same Chr 1 region with statistic significance (LOD 5.6,P < .000 01).
The other 5 Chr 1 markers we have tested on B6SJLF2 mice that cover thePbbcp2 locus region showed decreasing significance levels in the linkage analysis: D1Mit135 (59.7 cM, LOD 4.5,P < .000 01), D1Mit48 (54.0 cM, LOD 3.5,P < .000 05), D1Mit216 (49.7 cM, LOD 2.5,P < .000 87), D1Mit46 (43.1 cM, LOD 2.3,P < .001 18), and D1Mit24 (41.0 cM, LOD 2.0,P < .002 30). Thus, on the basis of the markers we have tested, Pbbcp2 was not an independent locus for the B220% phenotype in the B6SJLF2 mouse population.
For the suggestive CD4% locus, we tested 12 standard Mit markers in the 69- to 73-cM region of Chr 8: D8Mit140 andD8Mit324 at 69 cM; D8Mit122 at 70 cM;D8Mit42, D8Mit52, D8Mit92, D8Mit279, and D8Mit325 at 71 cM; D8Mit93,D8Mit245, and D8Mit326 at 72 cM; andD8Mit156 at 73 cM. None of these markers showed detectable polymorphism between B6 and SJL mice by using our assay.
For the Pbctlp1 locus, we tested 5 polymorphic markers in the 8- to 41-cM region of mouse Chr 19 on B6SJLF2 mice:D19Mit69 (8 cM), D19Mit61 (9 cM),D19Mit106 (22 cM), D19Mit40 (25 cM),D19Mit11 (41 cM), and we performed a mapping analysis. None of the 5 tested markers was linked to the CD8% phenotype in the B6SJLF2 intercross mouse population. It is possible that the SJL strain may not differ from the B6 strain at the Pbctlp1 locus.
Overall, we mapped 2 QTLs for the B220% phenotype, one QTL for the CD8% phenotype and one suggestive locus for the CD4% phenotype by using the BXD RI mice (Table 3). ThePbbcp1 locus was also mapped to the same Chr 1 region in the B6SJLF2 intercross mouse population.
Quantitative trait loci for peripheral blood lymphocyte proportions
| Name . | Phenotype . | Chromosome . | Location . | LOD . | P . | In B6SJLF2 . |
|---|---|---|---|---|---|---|
| Pbbcp1 | B220% | 1 | 63 cM | 5.6 | < .00001 | Yes |
| Pbbcp2 | B220% | 1 | 48 cM | 3.7 | < .00004 | No |
| Pbctlp1 | CD8% | 19 | 12 cM | 3.8 | < .00002 | No |
| — | CD4% | 8 | 73 cM | 2.6 | < .00057 | No |
| Name . | Phenotype . | Chromosome . | Location . | LOD . | P . | In B6SJLF2 . |
|---|---|---|---|---|---|---|
| Pbbcp1 | B220% | 1 | 63 cM | 5.6 | < .00001 | Yes |
| Pbbcp2 | B220% | 1 | 48 cM | 3.7 | < .00004 | No |
| Pbctlp1 | CD8% | 19 | 12 cM | 3.8 | < .00002 | No |
| — | CD4% | 8 | 73 cM | 2.6 | < .00057 | No |
LOD indicates logarithm of odds.
Discussion
Genetic regulation of PB lymphocyte proportions
Mouse PB B220%, CD4%, and CD8% are genetically regulated, as shown by the consistent differences among inbred mouse strains derived from different original stocks (Table 1). The B6 and C57BR/cdJ inbred strains both originated from the mating of female 57 with male 52 from Miss Abbie Lathrop's stock,24 and both have high levels of B cells and low levels of CD4 and CD8 T cells. The BALB and D2 inbred strains both originated from Castle mice received from Lathrop,24 and both have intermediate levels of B cells and low levels of CD4 and CD8 cells. The NOD and SJL inbred strains both originated from Swiss mice at the Pasteur Institute. NOD was derived from outbred ICR/Jcl mice in the 1970s, and SJL was derived from noninbred Swiss Webster mice in the 1960s.33Both of these Swiss-derived strains have low levels of B cells and high levels of CD4 and CD8 cells (Table 1). With low concentrations of circulating WBCs (Table 1), NOD and SJL mice have significantly lower levels of circulating B cells than the other 4 strains.
Importance in the regulation of PB lymphocyte proportions
Regulation of PB lymphocyte proportions may have biomedical significance. B6 and C57BR/cdJ mice have high B220%, low CD4% and CD8%, and low tumor incidences.34-39 BALB and D2 mice have intermediate B220%, CD4%, and CD8% and intermediate tumor incidences.34,35,39,40 SJL mice have low B220%, high CD4% and CD8%, and high tumor incidences, especially of the type B–reticulum cell sarcoma.41-43 NOD mice also have low B220% and high CD4% and CD8%. They develop many types of tumors when fed a specific diet to prevent diabetes.44 Thus, mouse PB B220%, CD4%, and CD8% measured early in life may be associated with tumor incidences later in life.
Regulation of PB lymphocyte proportion may also be related to average life span. In 22 strains of inbred mice and 5 F1 hybrids, we found that mean life span is positively correlated with B220% (r = 0.67, P < .0001), negatively correlated with CD4% (r = −0.54, P < .0040) and CD8% (r = −0.23, P = .26).24 These correlations may be related to tumor incidences because strains with high tumor incidences tend to die relatively early. In the BXD RI mice, however, average life spans showed no significant correlation with B220% (r = −0.16, P = .47), or CD4% (r = 0.13, P = .55), or CD8% (r = 0.29, P = .20).45 Thus, future studies are needed to clarify the relationship between B220%, CD4%, CD8%, and life expectancy.
QTL mapping
Our mapping analyses on the BXD RI mice used 3 phenotypes; thus, the threshold P value for significant linkage on genome-wide scan should be adjusted to .000 03 for each phenotype (.0001 ÷ 3). Judging by this adjusted P value, the Pbbcp1and Pbctlp1 loci are still significant, whereas thePbbcp2 locus is marginally significant. Because thePbbcp1 locus was also mapped to the same chromosomal region by using the B6SJLF2 mice, this locus regulates B220% in both the B6-D2 and B6-SJL genetic combinations.
The Pbbcp2 locus had the strongest linkage to theD1Ncvs44 marker at 48.4 cM on Chr 1 in the BXD RI mice.46 Interestingly, another locus, Ncl, also located at 48.4 cM on Chr 1,47 48 showed no linkage to the B220% phenotype in the same BXD RI strains (data not shown) because 8 of the 26 BXD RI strains showed crossover between theD1Ncvs44 locus and the Ncl locus. Whether or not the D1Ncvs44 locus is polymorphic between B6 and SJL mice is yet to be defined. Thus, further analyses are needed to clarify whetherPbbcp2 is a real locus, and whether the Pbbcp2locus is really located at 48.4 cM on mouse Chr 1.
The fact that neither the Pbctlp1 nor the suggestive CD4% locus were found in the B6SJLF2 intercross mice indicates that the B6 and SJL mice are probably not polymorphic at these loci. It is possible that the suggestive CD4% locus may not be a real locus. It is also possible, although less likely, that the Pbctlp1 locus is a false-positive QTL for regulating the CD8% phenotype.
Importantly, in both the BXD RI and the B6SJLF2 mice, B220%, CD4%, and CD8% are each regulated by different genetic elements. ThePbbcp1 and Pbbcp2 loci had no significant effect on the CD4% and CD8% phenotypes, whereas the Pbctlp1 locus had no significant effect on the B220% and CD4% phenotypes.
Potential gene candidates for the Pbbcp1 locus
There are many genes in the 60- to 70-cM Chr 1 region that are potential candidates for the Pbbcp1 locus (Table4); however, we emphasize 2 possibilities: Gli2 at 63 cM and En1 at 64.1 cM, both of which are important genes in the regulation of embryonic development. The Gli2 locus is polymorphic between B6 and D2 and between B6 and SJL mice. It encodes the vertebrate GLI2 zinc finger protein that is a putative transcription factor responding to sonic hedgehog signaling.49,50 Mutant mice lackingGli2 function exhibit defects in embryonic development.49,51,52 To date, there has been no report showing that Gli2 is involved in lymphohematopoiesis. However, the Gli2 polymorphism marked the B220% phenotype in both BXD RI (LOD 3.7, P < .000 04, Figure 2A) and B6SJLF2 mice (LOD 5.6, P < .000 01, Figure 3), suggesting that Gli2 may be a candidate gene for thePbbcp1 locus. The En1 locus encodes a homeobox-containing transcriptional factor that controls pattern formation during development of the central nervous system.53-55En1 showed significant linkage to the Pbbcp1 locus in the BXD RI lines (LOD 3.6,P < .000 05). Other potential candidate genes for thePbbcp1 locus and their locations and human homologies are listed in Table 4. However, all candidates are very tentative at the current level of genetic resolution.
Potential candidate genes and human homologies for thePbbcp1 locus
| Gene name . | Mouse gene . | Mouse position . | Human gene . | Human position . |
|---|---|---|---|---|
| B-cell leukemia/lymphoma 2 | Bcl2 | 59.8 cM | BCL2 | 18q21.33 |
| Troponin I, skeletal, slow 1 | Tnni1 | 60.0 cM | TNNI1 | 1q32-q32 |
| Troponin T2, cardiac | Tnnt2 | 60.0 cM | TNNT2 | 1q32-q32 |
| Serine (or cysteine) proteinase inhibitor, clade B (ovalbumin), member 2 | Serpinb2 | 61.1 cM | SERPINB2 | 18q22.1 |
| GLI-Kruppel family memberGli2 | Gli2 | 63.0 cM | GLI2 | 2pter-qter |
| 5-Hydroxytryptamine (serotonin) receptor 5B | Htr5b | 63.0 cM | HTR5B | 2q11-q13 |
| Engrailed 1 | Enzyme | 64.1 cM | Enzyme | 2q13-q21 |
| Inhibin beta-B | Inhbb | 64.1 cM | INHBB | 2cen-q13 |
| Gastrulation brain homeobox 2 | Gbx2 | 65.0 cM | GBX2 | 2q37-q37 |
| Chemokine (C-X-C) receptor 4 | Cmkar4 | 67.4 cM | CXCR4 | 2pter-qter |
| Gene name . | Mouse gene . | Mouse position . | Human gene . | Human position . |
|---|---|---|---|---|
| B-cell leukemia/lymphoma 2 | Bcl2 | 59.8 cM | BCL2 | 18q21.33 |
| Troponin I, skeletal, slow 1 | Tnni1 | 60.0 cM | TNNI1 | 1q32-q32 |
| Troponin T2, cardiac | Tnnt2 | 60.0 cM | TNNT2 | 1q32-q32 |
| Serine (or cysteine) proteinase inhibitor, clade B (ovalbumin), member 2 | Serpinb2 | 61.1 cM | SERPINB2 | 18q22.1 |
| GLI-Kruppel family memberGli2 | Gli2 | 63.0 cM | GLI2 | 2pter-qter |
| 5-Hydroxytryptamine (serotonin) receptor 5B | Htr5b | 63.0 cM | HTR5B | 2q11-q13 |
| Engrailed 1 | Enzyme | 64.1 cM | Enzyme | 2q13-q21 |
| Inhibin beta-B | Inhbb | 64.1 cM | INHBB | 2cen-q13 |
| Gastrulation brain homeobox 2 | Gbx2 | 65.0 cM | GBX2 | 2q37-q37 |
| Chemokine (C-X-C) receptor 4 | Cmkar4 | 67.4 cM | CXCR4 | 2pter-qter |
Pbbcp1 indicates peripheral blood B cell percentage 1.
Overall, the mapping of the Pbbcp1 locus in 2 independent studies suggests that this locus is important in the regulation of PB B220% in both the B6-D2 and B6-SJL genetic combinations. In both studies, B- and T-cell proportions are regulated independently by different QTLs. Such genetically based regulation of PB lymphocyte proportion may be associated with health and longevity. Because thePbbcp1 locus maps to a well-conserved chromosomal region that harbors genes regulating embryonic development, it is possible that the gene or genes encoded by this locus may regulate cell fate both during embryonic development and during adult lymphohematopoiesis.
We thank Drs Edward H. Leiter and Brian Soper for critical review comments, Dr Stephen Sampson for technical editing, Mr Ted Duffy for technical assistance in FACS analyses, and Ms Elaine Shown for technical assistance in PCR analyses.
Supported by R01 grant HL58820 and a core grant CA34196 from the National Institutes of Health.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jichun Chen, The Jackson Laboratory, Bar Harbor, ME 04609-1500; e-mail: jchen@jax.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal