Abstract
α1-Microglobulin is a 26-kd protein, widespread in plasma and tissues and well-conserved among vertebrates. α1-Microglobulin belongs to the lipocalins, a protein superfamily with highly conserved 3-dimensional structures, forming an internal ligand binding pocket. The protein, isolated from urine, has a heterogeneous yellow-brown chromophore bound covalently to amino acid side groups around the entrance of the lipocalin pocket. α1-Microglobulin is found in blood both in free form and complex-bound to immunoglobulin A (IgA) via a half-cystine residue at position 34. It is shown here that an α1-microglobulin species, which we name t–α1-microglobulin (t = truncated), with a free Cys34 thiol group, lacking its C-terminal tetrapeptide, LIPR, and with a more polar environment around the entrance of the lipocalin pocket, is released from IgA–α1-microglobulin as well as from free α1-microglobulin when exposed to the cytosolic side of erythrocyte membranes or to purified oxyhemoglobin. The processed t–α1-microglobulin binds heme and the α1-microglobulin–heme complex shows a time-dependent spectral rearrangement, suggestive of degradation of heme concomitantly with formation of a heterogeneous chromophore associated with the protein. The processed t–α1-microglobulin is found in normal and pathologic human urine, indicating that the cleavage process occurs in vivo. The results suggest that α1-microglobulin is involved in extracellular heme catabolism.
Introduction
Hemoglobin, the major oxygen carrier system in the blood, has a number of toxic, potentially dangerous side effects.1,2 Most of these have their origin in the auto-oxidation of oxyhemoglobin. Hemoglobin is a tetramer consisting of 4 globin subunits (α2β2), each carrying a heme group in its active center.3 Heme consists of protoporphyrin IX and a ferrous (Fe2+) iron atom which has high affinity for free oxygen (O2). Ferrous hemoglobin binding to O2 is called oxyhemoglobin. Auto-oxidation of oxyhemoglobin is a spontaneous intramolecular oxidation-reduction reaction eventually leading to production of ferric (Fe3+) hemoglobin (methemoglobin), ferryl (Fe4+) hemoglobin, free heme (“heme” and “hemin” are sometimes used to designate free protoporphyrin IX with a bound Fe2+ or Fe3+atom, respectively; in this article, “heme” is used regardless of the iron oxidation state), and various reactive oxygen species, including free radicals.4 These compounds present considerable oxidative stress, leading to tissue damage and cell destruction.
The overwhelming part of hemoglobin is found strictly compartmentalized within erythrocytes. The auto-oxidation of oxyhemoglobin and downstream free-radical formation is largely prevented by the intracellular inhibitors superoxide dismutase, catalase, and glutathione peroxidase.5,6 In spite of this, slow auto-oxidation occurs intracellularly. Oxidized hemoglobin forms which are unstable and easily denatured are found deposited together with free heme and iron on the cytosolic face of the erythrocyte membrane.7Hemoglobin is also found extracellularly in plasma at normal concentrations up to around 5 mg/L, mainly as a result of hemolysis.8 Plasma contains haptoglobin,9 a high-affinity binder of oxyhemoglobin and inhibitor of auto-oxidation; iron- and heme-binding proteins such as transferrin, albumin, and hemopexin; and antioxidants such as vitamin E and ascorbic acid. However, it is generally agreed that these systems are not sufficient to protect against hemoglobin-mediated oxidative cell and tissue damage during increased extravascular hemolysis and general hemolytic pathologic disorders.
α1-Microglobulin (α1m) is a plasma and tissue protein with unknown biologic functions. It is evolutionarily well-conserved and has so far been found in mammals, birds, fish, and amphibians. α1m is a glycoprotein with a relatively low molecular mass, 26 kd.10 Hence, it is filtered through the glomerular membranes of the kidney and was originally isolated from urine.11 α1m, also called protein HC,12 has many intriguing properties.13,14 It belongs to the lipocalin superfamily, a group of proteins from animals, plants, and bacteria with a well-conserved 3-dimensional structure. The lipocalins consist of a 160 to 190 amino-acid polypeptide folded into an 8-stranded β-barrel surrounding a pocket that can bind hydrophobic ligands.15 The ligand of α1m is unidentified. α1m carries an extremely heterogeneous yellow-brown chromophore, covalently bound to lysyl residues (Lys 92, 118, and 130) surrounding the entrance of the lipocalin pocket.16 α1m has one unpaired cysteine residue (Cys 34) which can interact with a second free Cys exposed at the surface of other proteins. Thus, a circulating covalent complex between α1m and immunoglobulin A (IgA), involving about 50% of plasma-α1m, was shown to be mediated by Cys 34 of α1m and the penultimate Cys residue of the IgA α-chain.17
The Cys-Cys interchain cross-link of IgA-α1m has so far been impossible to reduce in vitro. Therefore, the original observation of this work, the cleavage of IgA-α1m into free α1m and IgA by ruptured erythrocytes, was surprising. It prompted an investigation of the role of α1m in hemolysis. The results indicate that an activated form of α1m, which participates in degradation of heme, is released from free α1m and its IgA complex by exposed erythrocyte membranes and hemoglobin. The results suggest a possible biologic function of α1m in protection against unsequestered heme/hemoglobin that could explain its widespread distribution in tissues and species as well as some of its enigmatic biochemical properties.
Materials and methods
Proteins and reagents
Urine and blood from apparently healthy donors and urine from patients were collected. Information about the medical condition and serologic data for the patient and control groups was obtained from the Hematology Department and Blood Center at the University Hospital in Lund, Sweden. Approval was obtained from the institutional review board for these studies. Informed consent was provided according to the Declaration of Helsinki. Blood was drawn with addition of heparin anticoagulant. The urine and plasma, prepared from the blood, were frozen at −30°C within 1 hour. Oxyhemoglobin was prepared by diethylaminoethyl (DEAE)–Sephadex ion-exchange chromatography of erythrocyte hemolysates according to Winterbourn.4IgA-α1m, plasma-α1m, urine α1m, and pepsin-α1m were purified in our laboratory as described earlier.16,18,19 Rabbit anti-α1m was prepared by immunization with urine α1m as described.20 Rabbit anti-LIPR was prepared by AgriSera AB (Vännäs, Sweden) by immunization with the synthetic peptide CKKLIPR conjugated to keyhole limpet hemocyanin (KLH). Mouse monoclonal anti-α1m, BN11.10, was prepared and purified as described21 and immobilized to Affigel Hz (Bio-Rad Labs, Hercules, CA) at 20 mg/mL, following instructions from the manufacturer. Goat antihuman heme oxygenase I (cat. no. sc-1796) was from Santa Cruz Biotechnology (Santa Cruz, CA). Orosomucoid, ovalbumin, and human serum albumin were purchased from Sigma Chemicals (St Louis, MO).
Erythrocytes and membranes
Blood was centrifuged at 1200g for 10 minutes, plasma- and buffy coat–aspirated, and the red blood cells washed 4 times with 10 volumes of phosphate buffered saline (PBS): 10 mM phosphate buffer, pH 7.4, 120 mM NaCl, 3 mM KCl. The packed erythrocytes were lysed by resuspension in 1 volume of cold, hypotonic buffer (H2O:PBS, 20:1) on ice. Membrane pellets were separated from cytosol by centrifuging the lysed suspension at 14 000g for 20 minutes with subsequent washing. The pellets were then resuspended in PBS to one-tenth of the original blood volume. Hemoglobin-free membranes were prepared by repeated washing as described by Dodge et al.22 Membrane proteins were quantitated by the BCA Protein Assay Kit (Pierce, Rockford, IL). For further fractionation of membrane constituents, the lysed erythrocytes were centrifuged at 32 000g for 30 minutes, the membrane pellet solubilized in 50 mM Tris-HCl, pH 8.0 + 1% Nonidet P-40, and centrifuged at 8000g for 10 minutes. The supernatant was then applied to Sephacryl S-300 gel chromatography, eluting with 20 mM Tris-HCl, pH 8.0, 0.15 M NaCl, and 0.02% NaN3, and analyzing the eluted fractions for ultraviolet (UV) absorbance and α1m cleavage activity as described below.
Cleavage of α1-microglobulin
Freeze-dried α1m or IgA-α1m was incubated for indicated time periods at 37°C with the lysed, nonfractionated erythrocytes, cytosol, or membranes at a final α1m concentration of 40 μM. For cleavage with purified oxyhemoglobin, 0.04 μmol freeze-dried plasma-α1m was incubated for 3 hours at 37°C with 0.2 μmol oxyhemoglobin tetramer in 0.2 mL PBS. After incubation, α1m was purified from the incubation mixtures by affinity chromatography on monoclonal anti-α1m Affigel columns. After washing the columns with PBS, adsorbed α1m was eluted by the addition of 0.1 M glycine-HCl, pH 2.3. Eluted fractions were immediately neutralized with one-tenth volume of 1 M Tris-HCl, pH 8.5, and analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) under reducing and nonreducing conditions. The α1m proteins were sometimes alkylated with iodoacetamide (IAA) before the cleavage, incubating 1 mg/mL protein and 0.17 M IAA in 20 mM Tris-HCl, pH 8.5, for 30 minutes at room temperature and dialysing against PBS. For cleavage of 125I-α1m or IgA-α1m, 10 μL plasma, lysed erythrocytes, membranes, cytosol, or gel chromatography fractions were incubated with .5 μCi (20 kBq) 125I-proteins 14 μCi (.5 MBq)/μg in PBS. The reaction proceeded for 3 hours at 37°C. One microliter of the incubations was diluted 10 times with PBS and applied to SDS-PAGE.
Time-dependence of α1m cleavage
A quantity of 0.5 μg of plasma-α1m was added to 4 μL lysed erythrocytes or 15 μL erythrocyte membranes. In both cases the total sample volume was adjusted to 20 μL with PBS. The samples were incubated at 37°C for 1, 5, 30, 60, and 240 minutes. Cleavage was determined by Western blotting using antibodies against α1m and LIPR.
Heme binding experiments
The binding of heme by α1m was studied by incubation with (1) methemoglobin-Sepharose, (2) purified oxyhemoglobin, and (3) [14C]heme. Oxyhemoglobin was immobilized to CNBr-activated Sepharose 4B at 10 mg/mL (Amersham Pharmacia Biotech, Uppsala, Sweden) according to the description from the manufacturer and with modifications made by Gattoni et al.23 The immobilized oxyhemoglobin was oxidized to the ferric form (methemoglobin) by incubating with 1.2 equivalents of potassium ferricyanide (K3Fe[CN]6) at room temperature for 30 minutes and then washed extensively with 0.1 M phosphate buffer, pH 7.4. Methemoglobin-Sepharose was incubated with α1m or control proteins in PBS. The reaction proceeded for 60 minutes at room temperature with careful agitation and was terminated by pelleting the hemoglobin-Sepharose and removing the supernatants for absorbance spectrum analysis. Oxyhemoglobin (7 μM) and α1m or control proteins (5 μM) were incubated in PBS at 37°C for 1 hour. α1m was purified by affinity chromatography on a column of monoclonal anti-α1m Affigel, concentrated and analyzed by spectrophotometry. α1m or control proteins (0.1 nmol-0.2 nmol) in 10 μL PBS were incubated with 25 pmol [14C]heme diluted in PBS plus 1% Tween-20. The incubation was performed at room temperature in dim light. The reaction was stopped at different time intervals by adding SDS-PAGE sample buffer, separating by SDS-PAGE, staining, and phosphoimaging analysis (see below).
Heme degradation
Oxyhemoglobin (3 μM) was incubated alone or with t-α1m or control proteins (24 μM) in 20 mM phosphate buffer, pH 7.4, at 37°C. The absorbance at 410 nm was read at 5-minute intervals for up to 24 hours, blanking with the reaction solutions themselves at time zero.
Preparation of [14C]heme
Radiolabeled, 14C-substituted heme was produced using the E coli strain AN344 containing the plasmid pTYR13. This strain/plasmid combination (kindly donated by Dr Lars Hederstedt, Department of Microbiology, Lund University) requires the heme-precursor δ-aminolevulinic acid (ALA) for growth and was cultured with LB-medium (10 g/L Tryptone-Peptone, Difco, Becton Dickinson, Sparks, MD, 5 g/L yeast extract, Merck, Darmstadt, Germany, 5 g/L sodium chloride) containing [4-14C]ALA, 50 Ci/mol (New England Nuclear, Boston, MA) as described by Schiött et al.24 The yield of [14C]heme was about 20 nmol per 60-mL culture with a specific activity of 2.5 Ci/mmol (90GBq/mmol).
Gel electrophoresis and immunoblotting
SDS-PAGE was performed using either 10% or 12% slab gels in the buffer system described by Laemmli,25 sometimes including 2% vol/vol β-mercaptoethanol in the sample buffers. High-molecular-mass standards (rainbow markers; Amersham Pharmacia Biotech, Uppsala, Sweden) were used. The polyacrylamide gels were stained with Coomassie Brilliant Blue R-250 and in some cases dried. For immunoblotting, the gels were transferred to polyvinylidenefluoride (PVDF) membranes (Immobilon, Millipore, Bedford, MA) as described.26 The membranes were then incubated with antiserum as previously described.27 Radioactive samples (14C or125I) in dried gels and membranes were analyzed by phosphoimaging in a Fujix BAS 2000 Bioimaging analyzer (Fujifilm Sverige AB, Stockholm, Sweden).
Protein radiolabeling
Proteins were labeled with 125I (Svensk Radiofarmaka AB, Stockholm, Sweden) using the chloramine-T method.28 The labeled proteins were separated from free iodide by desalting the reaction mixture on prepacked 9-mL Sephadex G-25 columns (PD10, Amersham Pharmacia Biotech). The specific activity of the labeled proteins was approximately 14 μCi(0.5 MBq)/μg protein.
Spectrophotometric methods
Absorbance spectra were measured on a Beckman (Beckman Instruments, Fullerton, CA) DU 640i spectrophotometer using a scan rate of 240 nm/minute and protein concentrations between 1 μM and 50 μM. Fluorescence spectra were recorded with a Perkin Elmer (Sollentuna, Sweden) LB50 fluorescence spectrometer. The excitation was made at 280 nm with an excitation slit-width of 5 nm and an emission slit-width of 3 nm. The scan speed was 150 nm/minute. The proteins used in this experiment were diluted with PBS to a concentration of 4 μM.
Carbohydrate analysis of α1-microglobulin
Detection of O-linked and N-linked oligosaccharides was done by glycosidase cleavage and lectin blotting as described.18
Amino acid sequence analysis
Amino acid sequencing was done by Dr Bo Ek at Statens Lantbruksuniversitet, Uppsala, Sweden. Briefly, proteins were separated by SDS-PAGE, specific bands cut from the gel and digested by trypsin incubation. Trypsin digests were characterized by electrospray tandem mass spectrometry. This made it possible to make identifications based both on peptide mass data as well as on sequence information (peptide fragmentation data). All measurements were made on a Q-tof instrument (Micromass, Manchester, United Kingdom) essentially according to the manufacturer's instructions.
Alkylation with iodo[14C] acetamide
Proteins were incubated with iodo[14C] acetamide ([14C]IAA) (Amersham Life Science, specific activity 59.0 mCi(2.2 GBq)/mmol). The reaction mixtures contained 4 μM protein in 0.2 M Tris-HCl, pH 8.5, and 1 mM [14C]IAA. The reaction proceeded for 75 minutes at 25°C in the dark. To determine the amounts of bound [14C]IAA, the alkylated proteins were subjected to SDS-PAGE and phosphoimaging.
Gel chromatography
Proteins were separated by gel chromatography on a 50-mL Sephacryl S-300 column (Pharmacia, Uppsala, Sweden), equilibrated with 20 mM Tris-HCl, 150 mM NaCl, 0.02% NaN3, pH 8.0, at 4°C. The column was eluted at a flow rate 12 mL/hour and the eluted fractions were analyzed by absorbance at 280 nm and 410 nm.
Results
Cleavage of α1-microglobulin by erythrocyte membranes and hemoglobin
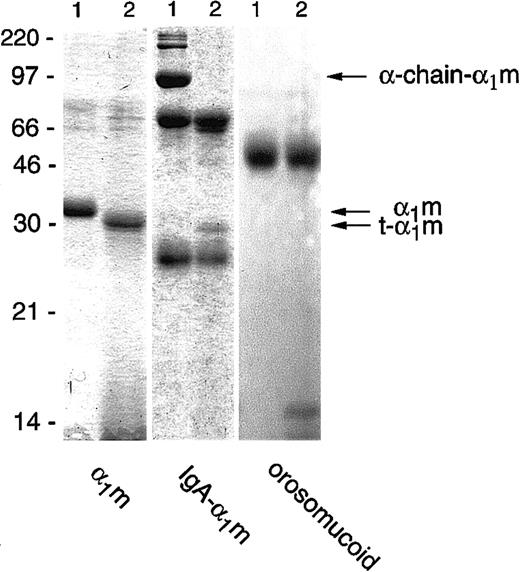
Incubation of free plasma–α1m with lysed erythrocytes, or the membrane or cytosolic fractions separately, resulted in a reduction in size of the protein when analyzed by SDS-PAGE. The truncated form of α1m, which we henceforth call t-α1m, had an apparent molecular mass of 30 kd, approximately 3 kd less than uncleaved α1m (Figure1). The IgA-α1m complex was also cleaved by the erythrocyte fractions. In this complex α1m is covalently cross-linked by a nonreducible disulfide bond to one of the heavy (α)-chains of IgA. The IgA-α1m complex appears as 3 major bands on SDS-PAGE: the 90-kd α1m–α-chain, the 60-kd α-chain, and the 25-kd light chain. In addition, various less-abundant high-molecular-weight bands can be seen, representing polymeric forms of IgA-α1m containing the 90-kd IgA-α1m–α-chain component. After incubation with the erythrocyte fractions, the 90-kd IgA–α-chain band disappeared, and was replaced by free α1m with a molecular mass of 30 kd, that is, t-α1m (Figure 1). The same incubation conditions did not affect the migration of control proteins orosomucoid (Figure1), human serum albumin, IgG, IgA, and ovalbumin (not shown) upon SDS-PAGE. Cleavage of α1m and IgA-α1m was achieved using up to 80 nmol protein/mL erythrocytes. The cleavage could be detected after purification by anti-α1m affinity chromatography followed by SDS-PAGE (see below), by Western blotting of the erythrocyte/α1m mixture, or by using125I-labeled protein (see “Materials and methods”).
Cleavage of α1m and control proteins by erythrocyte membranes.
Plasma-α1m, IgA-α1m, and orosomucoid were incubated with purified erythrocyte membranes for 2 hours at 37°C. A quantity of 1 μg to 3 μg of each protein was separated by SDS-PAGE (T = 12%, C = 3.3%) without prior affinity chromatography purification of the α1m-components and stained with Coomassie Brilliant Blue. Lane 1 shows the proteins alone and lane 2 shows the proteins with added erythrocyte membranes. The electrophoresis was performed in the presence of mercaptoethanol. Molecular masses of standards are given in kilodaltons.
Cleavage of α1m and control proteins by erythrocyte membranes.
Plasma-α1m, IgA-α1m, and orosomucoid were incubated with purified erythrocyte membranes for 2 hours at 37°C. A quantity of 1 μg to 3 μg of each protein was separated by SDS-PAGE (T = 12%, C = 3.3%) without prior affinity chromatography purification of the α1m-components and stained with Coomassie Brilliant Blue. Lane 1 shows the proteins alone and lane 2 shows the proteins with added erythrocyte membranes. The electrophoresis was performed in the presence of mercaptoethanol. Molecular masses of standards are given in kilodaltons.
Erythrocytes were lysed and separated by centrifugation into membrane and cytosolic fractions that were suspended in the original blood cell volume. As stated above, both fractions cleaved plasma-α1m into t-α1m. Whole, nonruptured erythrocytes had no cleaving activity at all. Gel filtration on Sephacryl S-300 of solubilized membranes showed that the cleaving activity was associated with medium-sized molecules (not shown). Hemoglobin is a major macromolecular constituent of both the cytosolic and membrane portions of the lysed erythrocytes. Therefore, we also tested cleaving of α1m by purified hemoglobin. Mixing oxyhemoglobin with plasma-α1m resulted in the same decrease in size as the erythrocyte fractions (Figure2A). The specific cleaving activity was estimated by dilution of the membrane and cytosolic fractions and purified hemoglobin (hemoglobin tetramer concentration 0.02 mM, 4 mM, and 1 mM, respectively). The membrane fraction had much higher specific cleaving activity than the other 2; that is, t-α1m was formed from plasma-α1m by the membranes at more than 10-fold higher dilution than the cytosol or purified hemoglobin. Hemoglobin-free membranes had no cleaving activity (not shown). These results suggest that hemoglobin in synergy with unknown factors in erythrocyte membranes, alternatively a minor form of hemoglobin found enriched in erythrocyte membranes, mediate the cleavage of α1m.
Release of the C-terminal tetrapeptide from α1m after cleavage with erythrocyte membranes and oxyhemoglobin.
Plasma-α1m (lanes 1 and 3) was incubated with erythrocyte membranes (lane 2) or oxyhemoglobin (lane 4) as described in “Materials and methods.” After incubation for 3 hours at 37°C, α1m was purified from the reaction mixtures by affinity chromatography and gel chromatography on Sephacryl S-300. Purified α1m, 1 μg to 3 μg, was separated by SDS-PAGE in the presence of mercaptoethanol, staining, and Western blotting with antibodies against α1m or LIPR.
Release of the C-terminal tetrapeptide from α1m after cleavage with erythrocyte membranes and oxyhemoglobin.
Plasma-α1m (lanes 1 and 3) was incubated with erythrocyte membranes (lane 2) or oxyhemoglobin (lane 4) as described in “Materials and methods.” After incubation for 3 hours at 37°C, α1m was purified from the reaction mixtures by affinity chromatography and gel chromatography on Sephacryl S-300. Purified α1m, 1 μg to 3 μg, was separated by SDS-PAGE in the presence of mercaptoethanol, staining, and Western blotting with antibodies against α1m or LIPR.
C-terminal proteolysis of α1-microglobulin
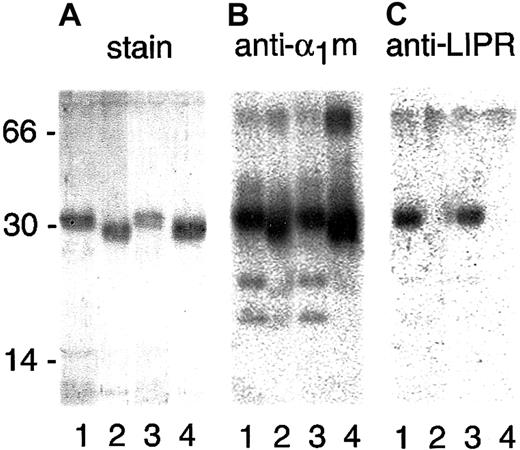
The t-α1m formed by cleavage of plasma, recombinant, or IgA-α1m was purified by applying the mixture of α1m and lysed erythrocytes or hemoglobin to a column of immobilized monoclonal antibodies against α1m followed by gel chromatography on Sephacryl S-300 (Figure 2, lanes 2 and 4, respectively). Hemoglobin was coeluted with α1m from the anti-α1m column but was removed by the gel chromatography step. The cleavage of IgA-α1m yielded an intact IgA molecule, besides t-α1m, as judged by SDS-PAGE (eg, Figure 1) or Sephacryl S-300, underscoring the specificity of the cleavage mechanism. The amino acid sequence of purified t-α1m was analyzed by trypsin cleavage followed by mass spectrometry, and it was shown that α1m was cleaved C-terminally; that is, the C-terminal tetrapeptide LIPR (amino acid pos. 180-183) was missing in t-α1m whether it was produced from free plasma–α1m or IgA-α1m, whereas the N-terminus was intact (Table1). Polyclonal rabbit antibodies were raised against the synthetic peptide LIPR and used to confirm the absence of the tetrapeptide in t-α1m. Thus, after SDS-PAGE and blotting, anti-LIPR recognized intact full-length α1m but not t-α1m (Figure 2C), whereas a conventional anti-α1m antiserum recognized both intact, full-length α1m and t-α1m (Figure2B).
Molecular composition of α1m before and after cleavage by erythrocyte membranes and purified oxyhemoglobin
| Source of α1m . | N-terminus . | C-terminus . | −SH group* . | Carbohydrates . | Chromophore . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Before . | After . | Before . | After . | Before . | After . | Before . | After . | Before . | After . | |
| Plasma | G1PVP … | G1PVP … | … PEPILIPR183 | … PEPI179 | + | + | 3 | 3 | + | +++† |
| IgA-α1m | G1PVP … | G1PVP … | … PEPILIPR183 | … PEPI179 | 0 | + | nd | 3 | − | +++ |
| Source of α1m . | N-terminus . | C-terminus . | −SH group* . | Carbohydrates . | Chromophore . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Before . | After . | Before . | After . | Before . | After . | Before . | After . | Before . | After . | |
| Plasma | G1PVP … | G1PVP … | … PEPILIPR183 | … PEPI179 | + | + | 3 | 3 | + | +++† |
| IgA-α1m | G1PVP … | G1PVP … | … PEPILIPR183 | … PEPI179 | 0 | + | nd | 3 | − | +++ |
nd indicates not determined.
Free thiol groups were determined as absent (0) or present (+) before and after cleavage by erythrocyte membranes by alkylation with [14C]iodoacetamide.
The amount of chromophore was estimated by absorbance spectrum analysis and is graded from − to +++.
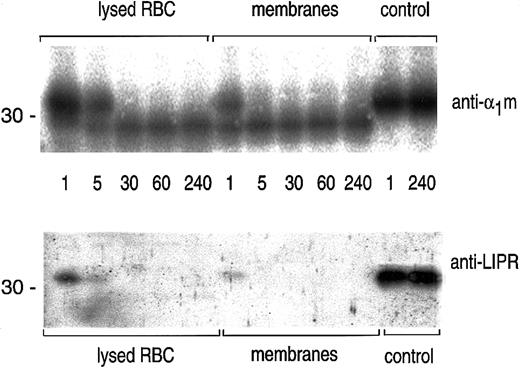
The time course of the cleavage of plasma-α1m by lysed erythrocytes and erythrocyte membranes was studied by SDS-PAGE followed by blotting with anti-α1m and anti-LIPR (Figure3). The results shows that the cleavage by erythrocyte membranes was completed after 5 minutes.
Time studies on the cleavage of α1m.
A quantity of 0.5 μg plasma-α1m was incubated with lysed erythrocytes, erythrocyte membranes, or PBS (control) at 37°C for indicated time points: 1, 5, 30, 60, and 240 minutes. The cleavage reaction was stopped by adding SDS-PAGE sample buffer and was analyzed by SDS-PAGE in the presence of mercaptoethanol and Western blotting with anti-α1m and anti-LIPR.
Time studies on the cleavage of α1m.
A quantity of 0.5 μg plasma-α1m was incubated with lysed erythrocytes, erythrocyte membranes, or PBS (control) at 37°C for indicated time points: 1, 5, 30, 60, and 240 minutes. The cleavage reaction was stopped by adding SDS-PAGE sample buffer and was analyzed by SDS-PAGE in the presence of mercaptoethanol and Western blotting with anti-α1m and anti-LIPR.
The results thus suggest that the C-terminal tetrapeptide of α1m is released from plasma-α1m and the IgA-α1m complex when exposed to hemoglobin and other unknown factors of the cytosol and cytosolic surface of erythrocyte membranes.
Biochemical properties of t–α1-microglobulin
Characterization of purified t-α1m from plasma-α1m and IgA-α1m showed that the cleavage involved the C-terminus (see above), the unpaired Cys34 residue (in the case of IgA-α1m), and the chromophores of α1m, whereas the N-terminus as well as the carbohydrate moieties were intact (Table 1).
The reactivity of the unpaired sulfhydryl group (-SH) of Cys34 was investigated by alkylation with iodo[14C]acetamide (Figure 4; Table 1). Plasma-α1m could be alkylated with iodo[14C]acetamide (Figure 4A, lane 1), indicating that the sulfhydryl group of Cys34 is free. The protein was still alkylable after cleavage (Figure 4A, lane 2). IgA-α1m was first alkylated with an excess of nonlabeled iodoacetamide to block all remaining free sulfhydryl groups on the IgA part of the molecule (Figure 4B, lane 2) and then subjected to cleavage by erythrocyte membranes. Free sulfhydryl groups then appeared on both the α1m- and α-chain parts (Figure 4B, lane 3). The α1m part of IgA-α1m, called pepsin-α1m, can be prepared by pepsin cleavage.17 It is composed of the C-terminal nonapeptide of the IgA–α-chain linked to the intact α1m polypeptide by an unusual, nonreducable bond involving Cys34 of α1m and the penultimate Cys of the α-chain.16 17 As expected, pepsin-α1m could not be alkylated with iodo[14C]acetamide (Figure4C, lane 1). However, incubation of pepsin-α1m with erythrocyte membranes yielded a free, alkylable sulfhydryl group (Figure 4C, lane 2). The results thus suggest that the cleavage of IgA-α1m involves both a reduction of the bond between IgA and α1m and a C-terminal truncation of the α1m part.
Appearance of free thiol groups on α1m after cleavage with erythrocyte membranes.
(A) A quantity of 3 μg free plasma–α1m (lane 1) was incubated with erythrocyte membranes and purified by affinity chromatography on a column with anti-α1m (lane 2). The samples were treated with [14C]IAA, separated by SDS-PAGE, stained, and analyzed by phosphoimaging. (B) A quantity of 10 μg IgA-α1m was either left untreated (lane 1), alkylated with cold IAA (lane 2), or alkylated with cold IAA and incubated with lysed erythrocytes (lane 3). The proteins were then incubated with [14C]IAA, separated by SDS-PAGE and stained, or analyzed by phosphoimaging. (C) The α1m-fragment of IgA-α1m was prepared by pepsin digestion. A quantity of 3 μg of the α1m-fragment was either left untreated (lane 1) or incubated with lysed erythrocytes (lane 2). The proteins were incubated with [14C]IAA, separated by SDS-PAGE and stained, or analyzed by autoradiography. All electrophoreses were performed in the presence of mercaptoethanol.
Appearance of free thiol groups on α1m after cleavage with erythrocyte membranes.
(A) A quantity of 3 μg free plasma–α1m (lane 1) was incubated with erythrocyte membranes and purified by affinity chromatography on a column with anti-α1m (lane 2). The samples were treated with [14C]IAA, separated by SDS-PAGE, stained, and analyzed by phosphoimaging. (B) A quantity of 10 μg IgA-α1m was either left untreated (lane 1), alkylated with cold IAA (lane 2), or alkylated with cold IAA and incubated with lysed erythrocytes (lane 3). The proteins were then incubated with [14C]IAA, separated by SDS-PAGE and stained, or analyzed by phosphoimaging. (C) The α1m-fragment of IgA-α1m was prepared by pepsin digestion. A quantity of 3 μg of the α1m-fragment was either left untreated (lane 1) or incubated with lysed erythrocytes (lane 2). The proteins were incubated with [14C]IAA, separated by SDS-PAGE and stained, or analyzed by autoradiography. All electrophoreses were performed in the presence of mercaptoethanol.
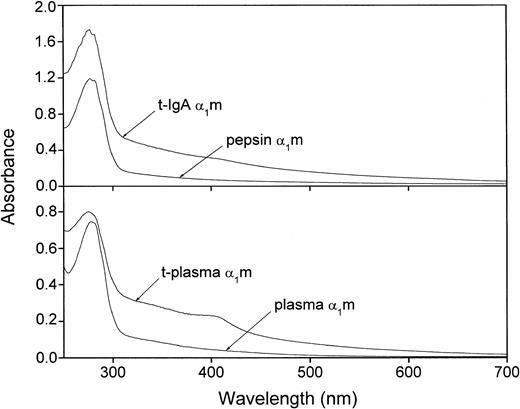
The extremely heterogeneous chromophore of α1m gives the protein a characteristic absorbance spectrum with a slowly declining absorbance throughout the upper UV and visible regions. It has been shown previously that urinary α1m carries more chromophoric material than plasma-α1m, whereas IgA-α1m has almost no chromophore.16 27Figure 5 shows that t-α1m from both plasma- and IgA-α1m had a higher absorbance in the upper UV and visible regions as compared with the uncleaved proteins. A shoulder in the absorbance was sometimes seen around 410 nm. t-α1m was also brown-colored in the test tube, whereas plasma-α1m is light yellow and IgA-α1m and pepsin-α1m are uncolored at the same concentrations.
Absorbance spectrum of IgA-α1m and free plasma–α1m before and after cleavage with lysed erythrocytes.
Uncleaved IgA-α1m is represented by pepsin-α1m (eg, the α1m part of IgA-α1m). Approximately 20 μM solutions in PBS were used.
Absorbance spectrum of IgA-α1m and free plasma–α1m before and after cleavage with lysed erythrocytes.
Uncleaved IgA-α1m is represented by pepsin-α1m (eg, the α1m part of IgA-α1m). Approximately 20 μM solutions in PBS were used.
α1m contains 4 tryptophan residues, 3 of which are located near the entrance of the lipocalin pocket.29 The tryptophan fluorescence spectra of free plasma-α1m, pepsin-α1m, and t-α1m displayed emission maxima at 337, 338, and 345 nm, respectively (not shown). This shows that the local environment around the 4 tryptophan residues is more polar in t-α1m as compared with free plasma-α1m and pepsin-α1m (ie, the α1m part of IgA-α1m).
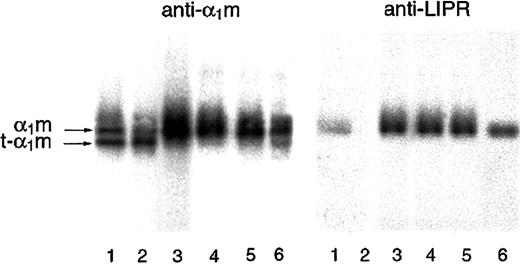
Cleaved α1-microglobulin in urine
Due to its small size, α1m is passed from the blood to the primary urine via glomerular filtration. A small part escapes tubular reabsorption and is excreted in the urine. It was shown that normal urine contains t-α1m, lacking C-terminal LIPR, and full-length α1m (Figure 6, lanes 4-6). Furthermore, an increased t-α1m fraction was seen in urine from patients with hemolytic disorders (Figure 6, lanes 1-3), suggesting a relationship to pathologic erythrocyte destruction. This indicates that the cleavage of α1m is a normally occurring biologic process and that t-α1m may be produced during hemolysis in vivo.
SDS-PAGE of urinary α1m.
Urinary α1m was purified by affinity chromatography on a column with monoclonal antibodies against α1m. The eluate was analyzed by SDS-PAGE in the presence of mercaptoethanol (T = 12%, C = 3,3), staining, and Western blotting with anti-α1m and anti LIPR. Urine from apparently healthy donors (lanes 4-6) as well as patients having different disorders associated with the following changes in erythrocyte formation and destruction were used: lane 1, suspected mechanical hemolytic anemia associated with mitral valve reconstruction following endocarditis; lane 2, autoimmune hemolytic anemia (warm type, IgG-mediated); lane 3, paroxysmal nocturnal hemoglobinuria and myelodysplastic syndrome.
SDS-PAGE of urinary α1m.
Urinary α1m was purified by affinity chromatography on a column with monoclonal antibodies against α1m. The eluate was analyzed by SDS-PAGE in the presence of mercaptoethanol (T = 12%, C = 3,3), staining, and Western blotting with anti-α1m and anti LIPR. Urine from apparently healthy donors (lanes 4-6) as well as patients having different disorders associated with the following changes in erythrocyte formation and destruction were used: lane 1, suspected mechanical hemolytic anemia associated with mitral valve reconstruction following endocarditis; lane 2, autoimmune hemolytic anemia (warm type, IgG-mediated); lane 3, paroxysmal nocturnal hemoglobinuria and myelodysplastic syndrome.
Binding and degradation of heme
The shoulder in the absorbance spectrum of freshly prepared t-α1m (Figure 5) even after gel chromatography suggests the presence of heme, which has a characteristic absorbance peak at 405 nm to 415 nm called the Soret band. This prompted an investigation of a possible interaction among α1m, t-α1m, and heme. Equimolar amounts of plasma-α1m, t-α1m, human serum albumin, and orosomucoid were incubated with [14C]heme for various time periods, separated by SDS-PAGE, and analyzed by autoradiography (Figure7). Both plasma-α1m and t-α1m displayed a binding of radiolabeled heme. Interestingly, more heme was bound to α1m, especially to t-α1m, than to human serum albumin, a physiologic heme-binding protein.31 Orosomucoid, a negative control from the Lipocalin superfamily, did not show any binding.
Time-dependence of [14C]heme-binding to α1m.
Approximately 0.2 nmol plasma-α1m, t-α1m, human serum albumin (HSA), and orosomucoid was incubated with 25 pmol [14C]heme for 1, 3, 15, 60, and 180 minutes. The binding of [14C]heme to the proteins was then determined by SDS-PAGE, staining (A), and phosphoimaging (B).
Time-dependence of [14C]heme-binding to α1m.
Approximately 0.2 nmol plasma-α1m, t-α1m, human serum albumin (HSA), and orosomucoid was incubated with 25 pmol [14C]heme for 1, 3, 15, 60, and 180 minutes. The binding of [14C]heme to the proteins was then determined by SDS-PAGE, staining (A), and phosphoimaging (B).
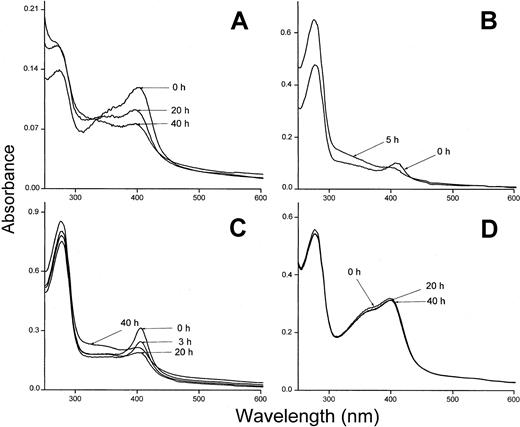
The heme binding was also investigated using hemoglobin insolubilized to Sepharose and oxidized to methemoglobin. Methemoglobin-Sepharose has been shown to bind heme less tightly than oxyhemoglobin and the metheme-group is transferred to albumin and other heme-binding proteins.23 Plasma-α1m and t-α1m were incubated with methemoglobin-Sepharose and the absorbance spectrum of the supernatants showed a binding of metheme to both proteins (Figure 8A,D). The binding of heme was seen as a pronounced Soret band for both plasma-α1m and t-α1m. However, the time-dependence of the binding was radically different between the 2 α1m-forms using this method. The proteins were incubated with methemoglobin-Sepharose for one hour, removed by centrifugation and their absorbance spectra then analyzed at different times. After 40 hours the Soret band of heme had disappeared almost completely from t-α1m (Figure 8A). Instead, the absorbance at lower wavelengths increased (300 nm-400 nm), yielding a heterogeneous spectrum that resembles the highly brown-colored urinary α1m and recombinant α1m from baculovirus-infected insect cells.27 On the other hand, the absorbance spectrum of free plasma–α1m was stable during this time period (Figure 8D). The rearrangement of the spectrum was also observed when t-α1m was incubated with soluble oxyhemoglobin (Figure 8B). Spectral analysis of t-α1m freshly isolated after incubation of plasma-α1m with lysed erythrocytes also revealed a decrease of the Soret band concomitant with an increase at 300 nm to 400 nm (Figure 8C). These results suggest a simultaneous heme degradation and chromophore formation in t-α1m.
Spectral analysis of t-α1m after incubation with hemoglobin.
(A) Methemoglobin-Sepharose (75 μM) and t-α1m (3 μM) were incubated in PBS at 37°C for 1 hour. The sample was centrifuged and the absorbance spectrum of the supernatant measured at the indicated time intervals. The spectrum of a control sample (methemoglobin-Sepharose only) was subtracted. (B) Oxyhemoglobin (7 μM) and t-α1m (5 μM) were incubated in PBS at 37°C for 1 hour. The t-α1m was purified by affinity chromatography on a column of monoclonal anti-α1m Affigel, concentrated, and the absorbance spectrum read after 0 and 5 hours. (C) Lysed erythrocytes (original cell volume diluted 1:1) and plasma-α1m (40 μM) were mixed and incubated at 37°C for 3 hours. The t-α1m formed was purified by affinity chromatography on a column of monoclonal anti-α1m Affigel and gel chromatography on Sephacryl S-300, concentrated, and the absorbance spectrum read after the indicated time intervals. (D) Methemoglobin-Sepharose (75 μM) and plasma-α1m (10 μM) were incubated for 1 hour at 37°C. The sample was centrifuged and the absorbance spectrum of the supernatant measured at the indicated time intervals.
Spectral analysis of t-α1m after incubation with hemoglobin.
(A) Methemoglobin-Sepharose (75 μM) and t-α1m (3 μM) were incubated in PBS at 37°C for 1 hour. The sample was centrifuged and the absorbance spectrum of the supernatant measured at the indicated time intervals. The spectrum of a control sample (methemoglobin-Sepharose only) was subtracted. (B) Oxyhemoglobin (7 μM) and t-α1m (5 μM) were incubated in PBS at 37°C for 1 hour. The t-α1m was purified by affinity chromatography on a column of monoclonal anti-α1m Affigel, concentrated, and the absorbance spectrum read after 0 and 5 hours. (C) Lysed erythrocytes (original cell volume diluted 1:1) and plasma-α1m (40 μM) were mixed and incubated at 37°C for 3 hours. The t-α1m formed was purified by affinity chromatography on a column of monoclonal anti-α1m Affigel and gel chromatography on Sephacryl S-300, concentrated, and the absorbance spectrum read after the indicated time intervals. (D) Methemoglobin-Sepharose (75 μM) and plasma-α1m (10 μM) were incubated for 1 hour at 37°C. The sample was centrifuged and the absorbance spectrum of the supernatant measured at the indicated time intervals.
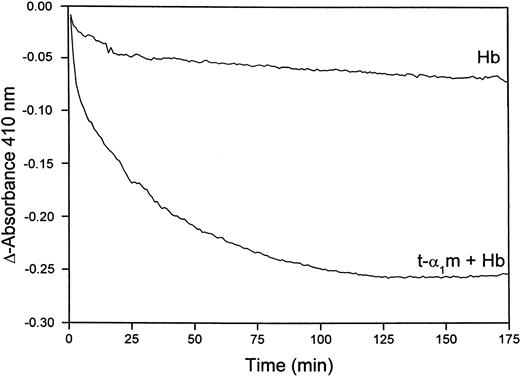
Figure 8B shows that heme is transferred from oxyhemoglobin to t-α1m and degraded. The time-dependence of the degradation was investigated more closely by continuously reading the absorbance at 410 nm in a t-α1m and oxyhemoglobin mixture using an excess of α1m (molar ratio 8:1). Figure9 shows that most of the absorbance at 410 nm disappeared within one hour, suggesting that the degradation is a rapid process.
Rate of heme-degradation in a t-α1m and oxyhemoglobin mixture.
Oxyhemoglobin (3 μM) was incubated alone or mixed with t-α1m (24 μM) at 37°C in 0.5 mL 20 mM phosphate buffer, pH 7.4. The absorbance at 410 nm was read at 5-minute intervals for 10 hours, blanking with the incubation solution itself at time zero.
Rate of heme-degradation in a t-α1m and oxyhemoglobin mixture.
Oxyhemoglobin (3 μM) was incubated alone or mixed with t-α1m (24 μM) at 37°C in 0.5 mL 20 mM phosphate buffer, pH 7.4. The absorbance at 410 nm was read at 5-minute intervals for 10 hours, blanking with the incubation solution itself at time zero.
Discussion
The results in this work show that plasma-α1m is proteolytically processed by ruptured erythrocytes and the C-terminal tetrapeptide LIPR is released. The cleavage is induced by hemoglobin and factors in erythrocyte membranes. The released, processed α1m (t-α1m) binds and degrades heme with a concomitant formation of a yellow-brown chromophore strongly linked to the protein. The results thus suggest that α1m has a role in heme catabolism after exposure of hemoglobin and erythrocyte membranes.
The processed t-α1m was found in urine together with the unprocessed form. In blood, α1m is rapidly lost from the circulation by glomerular filtration.32Therefore, the t-α1m found in urine most likely has been filtered from plasma. This suggests that the cleavage described in this paper actually occurs in vivo. The finding is supported by a previous report that urinary α1m is a mixture of full-length α1m with 183 amino acids and a C-terminally truncated 179-amino acid form.33 The proportion of t-α1m in urine should reflect to what extent α1m is cleaved C-terminally in blood and/or tissues. It is therefore of interest to measure the ratio of full-length α1m/t-α1m in urine of patients with for example hemolytic disorders as compared with healthy individuals. An initial approach to such an investigation is shown in Figure 6. As expected, the ratio varies individually but a relatively higher amount of t-α1m is indicated in 2 of the 3 patients. Indeed, α1m from the patient with mainly extravascular hemolysis is almost completely cleaved. Although it must be emphasized that this is not a clinical study, the preliminary findings are encouraging. Larger patient groups as well as a methodological fine-tuning are needed to better evaluate the potential clinical applications of the t-α1m/α1m ratio. Interestingly, the chromophore of urinary α1m has a much more pronounced yellow-brown color and absorbance spectrum than plasma-α1m. The explanation for this could be that urinary α1m partly consists of t-α1m, since it was shown here that the absorbance spectrum and color of the latter is similar to urinary α1m.
The C-terminal processing of α1m is apparently performed by factors found inside the erythrocytes. No sign of transport of α1m across the erythrocyte membranes has been detected (not shown). It must therefore be concluded that in vivo processing only takes place after rupture of the red blood cells and exposure of the interior of the cell to α1m. Purified hemoglobin displayed a processing activity at high concentrations and may be responsible for the cleavage activity found in the cytosol, where it is found in very high amounts. The membrane fraction showed a stronger specific processing activity than the cytosolic fraction. Hemoglobin is found deposited in various aggregated and oxidized forms on the cytosolic face of the erythrocyte membranes.7 Therefore, it is possible that a variant of hemoglobin and other unknown membrane factors cause the C-terminal processing of α1m.
The results suggest that the processed t-α1m induces a degradation of heme which is accompanied by a chromophore formation in the protein. It has been shown that the chromophore consists of covalent modifications of Lys 92, 118, and 130. According to molecular modeling of α1m these residues are located semiburied at the entrance of the lipocalin pocket.16 29 It is possible that heme binds to the lipocalin pocket and induces the chromophore formation either as a direct precursor or by a reaction mechanism in which the chromophore is a by-product. Fluorescence analysis suggests that the tryptophan residues are more exposed to the hydrophilic environment in t-α1m. Interestingly, 3 of the 4 tryptophan residues are located around the entrance of the lipocalin pocket. It may be speculated that the C-terminal tetrapeptide is located close to the entrance of the pocket and that its proteolytic removal initiates heme degradation by exposing reactive side groups.
IgA-α1m may be regarded as a depot of α1m from which α1m is released and processed locally as soon as the erythrocyte ruptures. The size of IgA-α1m prevents the molecule from glomerular filtration and loss from the circulation. The release of α1m from IgA and the C-terminal processing are probably 2 separate reactions requiring separate cofactors. The former reaction involves a reduction of the disulfide bond between Cys34 on α1m and the Cys residue on the α-chain. The bond is unusually reduction-resistant and it has previously not been possible to break the bind in vitro. It was shown here that Cys34 on t-α1m is reduced and presents a free thiol group. According to the structural α1m-model this residue is exposed and located on a flexible omega-loop near the entrance to the pocket. Cys34 has previously been shown to be attached to chromophore substances,34 and it is possible that it is involved in the heme-binding and chromophore formation. Other heme-binding proteins have been shown to bind the heme group via an unpaired Cys residue; for instance, proteins carrying a so-called heme regulatory motif (HRM), such as the bacterial iron response regulator protein.35
This work indicates that α1m has a role in heme catabolism. Heme oxygenase, first described by Tenhunen et al,36 is an intracellular enzyme that catalyzes the degradation of heme to biliverdin, CO, and free iron.37 Heme oxygenase is membrane-bound and found in microsomes and it may be speculated that its diverse functions—heme degradation, iron utilization, and production of the powerful antioxidant biliverdin—are to be executed within the cells. α1m, on the other hand, is found extracellularly in most organs38-40 and is therefore expected to execute its tentative heme-degradation functions outside the cells. The presence of contaminations of heme oxygenase in the t-α1m preparations would have explained their heme-degradative properties. However, no trace of heme oxygenase was found using commercial antibodies to the protein in Western blotting, suggesting that the effects indeed were caused by t-α1m.
Several heme-binding and antioxidative substances are present in plasma, for example, albumin, hemopexin, vitamin E, and ascorbic acid. It is therefore not likely that the heme-binding and degradation properties of α1m are primarily intended for free hemoglobin in plasma. More likely targets for these properties of α1m are extravascular ruptured erythrocyte membranes. The oxidized forms of precipitated hemoglobin, free heme, and iron found on the cytosolic face of erythrocyte membranes are highly toxic to neighboring tissue. A role of α1m could therefore be to protect against the exposed heme on the erythrocyte membranes, especially during extravascular hemolysis or massive intravascular hemolysis, where these plasma antiheme factors are not available in sufficient amounts.
Both free monomeric α1m and high-molecular-weight α1m have been found widely distributed in extravascular compartments.38-40 IgA-α1m has been isolated from the placenta.40 Interestingly, a truncated form of α1m with an apparent molecular mass around 30 kd was observed associated with a placenta membrane fraction. This α1m variant may be identical to t-α1m, the activation product of erythrocyte membranes, described in this paper. Indeed, it can be speculated that an activation of free α1m and IgA-α1m takes place in ruptured tissue cells, for instance during inflammation and necrosis. Instead of hemoglobin, however, which is confined to erythrocytes, α1m may interact with other heme-binding proteins such as cytochrome c in tissue cells. The t-α1m found in urine may very well originate from extravascular tissue rather than blood, since the protein was shown to be rapidly transported between the 2 compartments.41 It remains to be shown, however, whether α1m can be processed and activated by all cells.
Victoria Rydengård and Kerstin Nilsson are acknowledged for excellent technical performance. The authors are indebted to Dr Lars Hederstedt for generous help with the preparation of radiolabeled heme.
Supported by grants from the Swedish Medical Research Council (project no. 7144), EU-Biotech (project no. BIO4-CT98-0420), King Gustav V's 80-year foundation, the Swedish Society for Medical Research, the Royal Physiographic Society in Lund, Swedish Society of Medicine, and the Foundations of Greta and Johan Kock, Magnus Bergvall, and Crafoord.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Bo Åkerström, Department of Cell and Molecular Biology, B14, BMC, S-221 84 Lund, Sweden; e-mail:bo.akerstrom@medkem.lu.se.

![Fig. 4. Appearance of free thiol groups on α1m after cleavage with erythrocyte membranes. / (A) A quantity of 3 μg free plasma–α1m (lane 1) was incubated with erythrocyte membranes and purified by affinity chromatography on a column with anti-α1m (lane 2). The samples were treated with [14C]IAA, separated by SDS-PAGE, stained, and analyzed by phosphoimaging. (B) A quantity of 10 μg IgA-α1m was either left untreated (lane 1), alkylated with cold IAA (lane 2), or alkylated with cold IAA and incubated with lysed erythrocytes (lane 3). The proteins were then incubated with [14C]IAA, separated by SDS-PAGE and stained, or analyzed by phosphoimaging. (C) The α1m-fragment of IgA-α1m was prepared by pepsin digestion. A quantity of 3 μg of the α1m-fragment was either left untreated (lane 1) or incubated with lysed erythrocytes (lane 2). The proteins were incubated with [14C]IAA, separated by SDS-PAGE and stained, or analyzed by autoradiography. All electrophoreses were performed in the presence of mercaptoethanol.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/99/6/10.1182_blood.v99.6.1894/6/m_h80622232004.jpeg?Expires=1763610996&Signature=iEX9HcO9VZerAKdZAvzG16oSBhuU73zPWPkBPfFhJHfA-Id~EEbX4bF~XuT2nXBobWY760uarz3ViQrxoJSYRNolYBOA3DrQmnFrohei8pTmlo78QHsYJi6KaMb7RAYNvSASlQ5n50oIYBMb9KJA4pWF~ZVlYLlgqEp8yzbLkLCH82KJeRzznjwDW5SGs0D4NxewF6QPxb5WjoFJgDpvxvcp8h8Igg840W51k7SvrLtNH4lIqsK0nQkuKtbjidz83kg6VYiddKQMhb2ZSkvwnjUbZEINA76EKYN~ozpRIeeBEyPG7Xi990r4thTCizwgFxs8DrQTHjaFeOZZDXV95g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 7. Time-dependence of [14C]heme-binding to α1m. / Approximately 0.2 nmol plasma-α1m, t-α1m, human serum albumin (HSA), and orosomucoid was incubated with 25 pmol [14C]heme for 1, 3, 15, 60, and 180 minutes. The binding of [14C]heme to the proteins was then determined by SDS-PAGE, staining (A), and phosphoimaging (B).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/99/6/10.1182_blood.v99.6.1894/6/m_h80622232007.jpeg?Expires=1763610996&Signature=XP65X2RAmiJyXO6UPNDtPt0k9~R86wTwZsjTdtUbV83Z6c4IAqVpp9ROaPLM5ggwqGQo44ru5XzyPCULhyR1Z33FPFaOfWeaOSisca34uEP8e9pI-2wKJxxaez3kWyogUdamloVGSFNscm4Iat~WcWbNre3ZgXM6ytoQwd4e6IgkDD6Z54dRIsSJWO6ufSPkg8K2DYZjFBGVkzLZkOt5Uia1N9pEm9~isTjA~OVWZQ~u-3227g5erwzDPbV13RDJc~3ZEZLwEvLIm4bfpecSWdPEgP~WuQIvekMST19AjqKyWOOTn3fw~GqoFKARvDybxifTqNrqiviU0DcfNUSKFA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal