Abstract
Paroxysmal nocturnal hemoglobinuria (PNH) is an acquired hematopoietic stem cell disorder characterized by clonal blood cells that are deficient in glycosylphosphatidylinositol-anchored proteins because of somatic mutations of the PIG-A gene. Many patients with PNH have more than one PNH clone, but it is unclear whether a single PNH clone remains dominant or minor clones eventually become dominant. Furthermore, it is unknown how many hematopoietic stem cells (HSCs) sustain hematopoiesis and how long a single HSC can support hematopoiesis in humans. To understand dynamics of HSCs, we reanalyzed the PIG-A gene mutations in 9 patients 6 to 10 years after the previous analyses. The proportion of affected peripheral blood polymorphonuclear cells (PMNs) in each patient was highly variable; it increased in 2 (from 50% and 65% to 98% and 97%, respectively), was stable in 4 (changed less than 20%), and diminished in 3 (94%, 99%, and 98% to 33%, 57%, and 43%, respectively) patients. The complexity of these results reflects the high variability of the clinical course of PNH. In all patients, the previously predominant clone was still present and dominant. Therefore, one stem cell clone can sustain hematopoiesis for 6 to 10 years in patients with PNH. Two patients whose affected PMNs decreased because of a decline of the predominant PNH clone and who have been followed up for 24 and 31 years now have an aplastic condition, suggesting that aplasia is a terminal feature of PNH.
Introduction
Paroxysmal nocturnal hemoglobinuria (PNH) is an acquired clonal hematopoietic stem cell disorder characterized by intravascular hemolytic anemia.1-3 Abnormal blood cells are deficient in glycosylphosphatidylinositol-anchored proteins (GPI-APs).4,5 In the affected hematopoietic cells from patients with PNH, the first step in biosynthesis of the GPI anchor is defective.4 At least 5 genes are involved in this reaction step,6 and one of them, an X-linked gene termedPIG-A, is mutated in affected cells.7-9 ThePIG-A gene is mutated in every patient with PNH reported to date, and deficiency of GPI in PNH has thus been considered to result solely from the PIG-A mutation(s).4 9
Many patients with PNH have more than one PNH clone.10-15How the affected stem cell clone comes to dominate hematopoiesis is a current issue. It is also unclear whether the predominant clone is maintained or a minor clone eventually overcomes. It is unknown how many hematopoietic stem cells (HSCs) sustain hematopoiesis and how long one HSC could support hematopoiesis in humans. To understand the dynamics of HSCs, we re-analyzed the PIG-Agene mutations in 9 patients 6 to 10 years after the previous analyses.9,11,15 16
Patients, materials, and methods
Patients and blood samples
We examined blood samples from 9 patients with PNH. All the patients were described elsewhere and had somatic PIG-A mutations.9,15 16 Diagnoses of PNH and aplastic anemia (AA) were made by the diagnostic criteria of the Ministry of Health and Welfare of Japan. Patients who had AA before clinically apparent PNH developed received a diagnosis of AA/PNH syndrome. The diagnosis of PNH aplasia (PNH progressed to aplasia in 2 patients) was made by our own criteria in the patients whose tri-lineage blood cell counts were reduced more than 20% from the original counts. With the informed consent of the patients, peripheral blood (PB) was obtained from each patient, and a bone marrow (BM) sample was also obtained from patient J19 6 to 10 years after the previous analysis. Polymorphonuclear cells (PMNs) and mononuclear cells (MNCs) were separated by sedimentation in 6% dextran and Ficoll-Hypaque (Pharmacia, Uppsala, Sweden) centrifugation. Approval was obtained from the Institutional Review Board at Osaka University for these studies. Informed consent was provided according to the Declaration of Helsinki.
Fluorescence-activated cell sorter analysis
The cells in the fractions of PBPMNs and BMMNCs were stained with fluorescein isothiocyanate– or phycoerythrin-conjugated anti-CD59 monoclonal antibodies (PharMingen, San Diego, CA) for the detection of GPI-APs. After staining, cells were analyzed with a FacsCalibur (Becton Dickinson, Bedford, MA). Cell types were determined by their forward and right scatter, similarly to the previous analysis.9 In 3 patients, the change of percentage GPI-AP− PMNs from previous analysis was within 20% (categorized as stable). Two patients, in whom percentage GPI-AP− PMNs increased more than 40%, were categorized as increased. Three patients, in whom the percentage GPI-AP− PMNs decreased more than 40%, were categorized as decreased.
PIG-A gene analysis
DNA was isolated from the fractions of PBPMNs and BMMNCs. The coding regions of PIG-A were amplified by polymerase chain reaction (PCR) in 5 fragments using the primer sets described previously15 and were cloned into pBluescript II. Subcloned products containing PIG-A fragments were then amplified again by PCR using the same primer sets for heteroduplex analysis with mutation detection enhancement gel (Hydrolink; AT Biochem, Malvern, PA).15 If a region containing a mutation was suspected, clones were sequenced using dideoxy chain termination and a model 377 DNA sequencer (Applied Biosystems, Foster City, CA). In each patient, the mutation ratio was counted as mutant amplified subclones (AS) out of analyzed AS.
Results
Changes in proportion of GPI-AP− cells
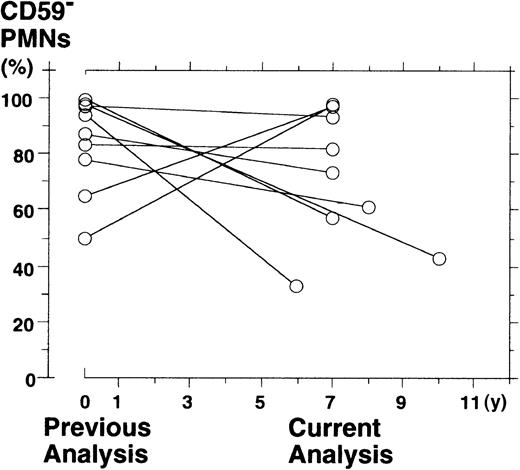
To investigate the pattern of clonal dominance, we re-analyzed the surface expression of GPI-APs on PBPMNs from 9 patients with PNH 6 to 10 years after the previous analysis. We described all the patients elsewhere9,15 16 and summarized the previous data (Tables1 and2). The percentage of PBPMNs deficient in CD59 (a measure of clonal hematopoiesis) was 74.5% ± 24.5% (medium ± range) at the previous analysis and 65.5% ± 32.5% at the current analysis (difference not significant;P = .2, Wilcoxon-signed rank test) (Figure1). However, a change in the proportion of CD59-deficient PMNs was highly variable from patient to patient; it increased in 2 (50% to 98% in J4 and 65% to 97% in J5), was stable in 4 (97% to 93% in J3, 83% to 82% in J11, 87% to 73% in J13, and 78% to 61% in J19), and diminished in 3 patients (94% to 33% in J12, 99% to 57% in J15, and 98% to 43% in J16) (Table 2).
Summary of laboratory data and clinical course in 9 patients with PNH at the previous analysis and the current analysis
| Patient . | Sex/age (y) . | Laboratory data (Previous/current) . | Clinical course . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Hb (g/dL) . | WBC (/μL) . | PMN (/μL) . | (CD59−PMN) (/μL) . | Plt (× 109/L) . | LDH (U/L) . | Duration (y) . | . | ||
| J4 | F | 7.8 | 6300 | 5090 | 2545 | 313 | 1840 | 11 | P |
| 56 | 9.4 | 5610 | 3574 | 3503 | 214 | 4342 | P | ||
| J5 | M | 12.3 | 6500 | 3835 | 2453 | 198 | 3260 | 8 | P |
| 39 | 8.5 | 5100 | 3162 | 3067 | 194 | 1830 | P | ||
| J3 | F | 6.9 | 3100 | 1783 | 1730 | 270 | 4359 | 11 | P |
| 53 | 8.0 | 3400 | 2108 | 1960 | 271 | 2281 | P | ||
| J11 | M | 4.6 | 1870 | 822 | 652 | 128 | 1084 | 12 | A-P |
| 46 | 3.8 | 1820 | 791 | 649 | 108 | 1785 | A-P | ||
| J13 | F | 7.8 | 2900 | 1102 | 758 | 289 | 4872 | 8 | P |
| 69 | 4.9 | 2600 | 468 | 342 | 333 | 2816 | P | ||
| J19 | M | 11.5 | 5780 | 3109 | 2425 | 248 | 2423 | 14 | P |
| 70 | 9.0 | 5130 | 1615 | 985 | 288 | 1689 | P | ||
| J12 | M | 10.5 | 4930 | 3879 | 3646 | 175 | 862 | 17 | A-P → P |
| 50 | 13.5 | 5010 | 3181 | 1050 | 118 | 383 | P | ||
| J15 | M | 7.1 | 3460 | 1647 | 1631 | 109 | 2772 | 31 | P |
| 58 | 4.2 | 1830 | 805 | 459 | 59 | ND | PA | ||
| J16 | F | 7.5 | 4400 | 2948 | 2889 | 90 | 5364 | 24 | A → P |
| 62 | 5.8 | 1900 | 1178 | 507 | 7 | 287 | PA | ||
| Patient . | Sex/age (y) . | Laboratory data (Previous/current) . | Clinical course . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Hb (g/dL) . | WBC (/μL) . | PMN (/μL) . | (CD59−PMN) (/μL) . | Plt (× 109/L) . | LDH (U/L) . | Duration (y) . | . | ||
| J4 | F | 7.8 | 6300 | 5090 | 2545 | 313 | 1840 | 11 | P |
| 56 | 9.4 | 5610 | 3574 | 3503 | 214 | 4342 | P | ||
| J5 | M | 12.3 | 6500 | 3835 | 2453 | 198 | 3260 | 8 | P |
| 39 | 8.5 | 5100 | 3162 | 3067 | 194 | 1830 | P | ||
| J3 | F | 6.9 | 3100 | 1783 | 1730 | 270 | 4359 | 11 | P |
| 53 | 8.0 | 3400 | 2108 | 1960 | 271 | 2281 | P | ||
| J11 | M | 4.6 | 1870 | 822 | 652 | 128 | 1084 | 12 | A-P |
| 46 | 3.8 | 1820 | 791 | 649 | 108 | 1785 | A-P | ||
| J13 | F | 7.8 | 2900 | 1102 | 758 | 289 | 4872 | 8 | P |
| 69 | 4.9 | 2600 | 468 | 342 | 333 | 2816 | P | ||
| J19 | M | 11.5 | 5780 | 3109 | 2425 | 248 | 2423 | 14 | P |
| 70 | 9.0 | 5130 | 1615 | 985 | 288 | 1689 | P | ||
| J12 | M | 10.5 | 4930 | 3879 | 3646 | 175 | 862 | 17 | A-P → P |
| 50 | 13.5 | 5010 | 3181 | 1050 | 118 | 383 | P | ||
| J15 | M | 7.1 | 3460 | 1647 | 1631 | 109 | 2772 | 31 | P |
| 58 | 4.2 | 1830 | 805 | 459 | 59 | ND | PA | ||
| J16 | F | 7.5 | 4400 | 2948 | 2889 | 90 | 5364 | 24 | A → P |
| 62 | 5.8 | 1900 | 1178 | 507 | 7 | 287 | PA | ||
Nine patients were subdivided into 3 groups: increased (J4, J5); stable (J3, J11, J13, J19); and decreased (J12, J15, J16).
In each patient, the previous laboratory data and the clinical diagnosis were described in the upper row, and the current laboratory data and the clinical diagnosis were described in the lower row.
M indicates Male; F, Female; Hb, hemoglobin; WBC, white blood cell; Plt, platelet; LDH, lactate dehydrogenase; Duration, the period between the diagnosis and the current analysis; P, PNH; A-P, AA-PNH syndrome; A, aplastic anemia; ND, not done; PA, PNH aplasia.
Summary of CD59 expressions and somatic mutations of PIG-A in 9 patients with PNH at the previous analysis and the current analysis
| Patient . | Mutation . | Previous analysis . | Current analysis . | Duration (y) . | ||||
|---|---|---|---|---|---|---|---|---|
| PB PMN . | BM Colonies/bursts DNA (AS) . | PB PMN . | BM MNC DNA (AS) . | |||||
| CD59−(%) . | mRNA (AS) . | CD59− (%) . | DNA (AS) . | |||||
| J4 | 298C to T | 50 | 11/22 | ND | 98 | 3/10 | ND | 7 |
| 273C to A | 0/10 | ND | 2/10 | ND | ||||
| J5 | 1309C del | 65 | 10/20 | ND | 97 | 9/9 | ND | 7 |
| J3 | 383A to G | 97 | 18/20 | ND | 93 | 4/10 | ND | 7 |
| J11 | 408T del | 83 | 15/20 | ND | 82 | 7/18 | ND | 7 |
| J13 | 116C to A | 87 | 18/20 | ND | 73 | 7/10 | ND | 7 |
| J19 | 987 T ins | 78 | 14/27 | 13/25 | 61 | 5/10 | 10/10 | 8 |
| 338 T to C | ND | 2/25 | 0/15 | 1/16 | ||||
| 1003 G to T | 2/27 | 4/25 | 0/10 | 0/10 | ||||
| 1028 AA del | 0/27 | 2/25 | 0/10 | 0/10 | ||||
| J12 | 936A del | 94 | 14/20 | ND | 33 | 4/16 | ND | 6 |
| 322A del | 1/12 | ND | 2/11 | ND | ||||
| J15 | Int 5 3′splice site G to A | 99 | (Major) | ND | 57 | 4/10 | ND | 7 |
| 368A ins | (Minor) | ND | 1/9 | ND | ||||
| J16 | Int 5 5′splice site T del | 98 | 5/5 | ND | 43 | 4/18 | ND | 10 |
| Patient . | Mutation . | Previous analysis . | Current analysis . | Duration (y) . | ||||
|---|---|---|---|---|---|---|---|---|
| PB PMN . | BM Colonies/bursts DNA (AS) . | PB PMN . | BM MNC DNA (AS) . | |||||
| CD59−(%) . | mRNA (AS) . | CD59− (%) . | DNA (AS) . | |||||
| J4 | 298C to T | 50 | 11/22 | ND | 98 | 3/10 | ND | 7 |
| 273C to A | 0/10 | ND | 2/10 | ND | ||||
| J5 | 1309C del | 65 | 10/20 | ND | 97 | 9/9 | ND | 7 |
| J3 | 383A to G | 97 | 18/20 | ND | 93 | 4/10 | ND | 7 |
| J11 | 408T del | 83 | 15/20 | ND | 82 | 7/18 | ND | 7 |
| J13 | 116C to A | 87 | 18/20 | ND | 73 | 7/10 | ND | 7 |
| J19 | 987 T ins | 78 | 14/27 | 13/25 | 61 | 5/10 | 10/10 | 8 |
| 338 T to C | ND | 2/25 | 0/15 | 1/16 | ||||
| 1003 G to T | 2/27 | 4/25 | 0/10 | 0/10 | ||||
| 1028 AA del | 0/27 | 2/25 | 0/10 | 0/10 | ||||
| J12 | 936A del | 94 | 14/20 | ND | 33 | 4/16 | ND | 6 |
| 322A del | 1/12 | ND | 2/11 | ND | ||||
| J15 | Int 5 3′splice site G to A | 99 | (Major) | ND | 57 | 4/10 | ND | 7 |
| 368A ins | (Minor) | ND | 1/9 | ND | ||||
| J16 | Int 5 5′splice site T del | 98 | 5/5 | ND | 43 | 4/18 | ND | 10 |
AS indicates amplified subclones; Duration, the period between the previous analysis and the current analysis; ND, not done.
Scattergraph for the fraction of CD59−PBPMNs from 9 patients with PNH during the previous analysis and the current analysis.
Scattergraph for the fraction of CD59−PBPMNs from 9 patients with PNH during the previous analysis and the current analysis.
We also investigated the absolute numbers of CD59− PMNs in each patient (Table 1). In the groups with an increased (J4 and J5) and a diminished percentage GPI-AP− (J12, J15, and J16), the absolute numbers of affected PMNs were increased and decreased, respectively. In the group with a stable percentage GPI-AP−, the numbers of affected PMNs were stable in J3 and J11, whereas they were decreased in J13 and J19. In the latter 2 patients, proportions of affected PMNs were stable because of a decline of PNH and normal clones.
Changes in proportion of PIG-A mutant clones
We then analyzed the PIG-A gene in PBPMNs after the amplification and subcloning of appropriate regions. In all patients, the previously predominant mutant clone was still dominant (Table 2). A proportion of affected PMNs in patient J5 increased because of the expansion of the predominant clone (1309delC). In patient J4, whose proportion of affected PMNs increased from 50% to 98%, the predominant clone (298 C-to-T) was detected in 3 of 10 AS of DNA, and the second clone (273 C-to-A) was newly detected in 2 of 10 AS (Table2). Because this patient is female, the result indicates that 2 mutant clones almost completely occupied the hematopoiesis and that the first clone supported approximately half the hematopoiesis for 7 years.
In 4 patients with stable proportions of affected PMNs (J3, J11, J13, and J19), the dominant clones were still dominant 7 to 8 years later. Blood samples from J19, bearing 4 independent PIG-A mutant clones, were analyzed in detail. Sixty percent of PBPMNs and 90% of BMMNCs were defective in CD59 expression (Table 2). The predominant clone (987insT) was detected in 5 of 10 AS from PBPMNs and in 10 of 10 AS from BMMNCs, indicating that proportions of abnormal phenotype and genotype are well correlated. A minor type mutation (388 T-to-C) was only detected in 1 of 16 AS from BMMNCs, and 2 other minor mutations and any additional mutations were not detected (Table 2). These 2 minor clones might have declined spontaneously, or the predominant clone might have superseded them. In every other year for 8 years, we analyzed the PBPMNs of J19 using FACS and found that 60% to 80% had complete deficiency and that none or few had partial deficiency. Because the minor clone (338 T-to-C) causes partial deficiency,15 16 the results suggest that the major clone, which has complete deficiency, maintained dominance for the 8 years.
Proportions of affected PMNs in 3 patients (J12, J15, and J16) decreased because of a decline of the major PNH clone. In patient J12 the predominant clone (936delA), which was previously detected in 14 of 20 AS, was detected in only 4 of 16 AS. A minor clone (322delA), previously detected in 1 of 12 AS, was found in 2 of 11 AS (Table 2). In patient J15, the predominant clone (3′ splice site of intron 5, G-to-A), previously detected as dominant, was detected in only 4 of 10 AS. A minor clone (368insA) was found in 1 of 9 AS (Table 2). Thus, the minor clones in 2 patients (J12 and J15) remained minor, and the decreases in affected PMNs were attributed to declines in the predominant clones.
Discussion
In all patients, the previously predominant clone was still dominant, indicating that the predominant PNH clone mainly supported hematopoiesis and manifested disease for years. However, the minor clone in 2 patients (J4 and J12) is now competing with the previously predominant clone and may overcome it in the future.
Indeed, Nafa et al17 reported a patient with PNH who underwent syngeneic bone marrow transplantation without conditioning. The patient's condition improved, but PNH relapsed 10 years later. They found that current and original PNH clones have different PIG-A mutations. Thus, the proportion of CD59-deficient PMNs was highly variable with time, indicating that the pattern of abnormal clonal expression in PNH may change quantitatively (clones increase or decrease in proportion) and qualitatively (generation of new clones, disappearance of clones). The complexity of these results reflects the marked variability of the clinical course of PNH. A demographic study of the correlation between clonal expansion or diminution and the development of or recovery from aplasia will help our understanding of the pathogenesis and natural history of PNH.
We especially investigated the clinical course in 3 patients (J12, J15, and J16), whose affected PMN levels decreased in proportion. Interestingly, de novo PNH progressed to aplasia in 2 patients (J15 and J16), and their conditions are clinically poor despite the decrease in affected PMNs. In addition, these 2 patients have been followed up significantly longer (31 and 24 years) than other patients with increased or stable proportions of GPI-AP− PMNs (8-14 years), though there is no significant disparity in age between these groups (Table 1). Thus, aplasia may be a terminal feature of PNH given that the abnormal clone declines in hematopoietic capacity, probably because of its lifespan, and other residual clones could not take its place, probably because of sustained suppression by an autoimmune mechanism thought to operate in AA. Indeed, PNH hematopoietic cells emerge in patients with severe AA at high frequency (29%-52%) during long-term survival,18-21 and the evolution to pancytopenia is associated with poor survival in patients with PNH.3 In other words, active hematopoiesis of the PNH stem cell clone, though it would result in desperate hemolysis, is better than aplasia because of a decline in the PNH stem cell clone.5 Therefore, our observations suggest that immunosuppressive therapy, such as antithymocyte or antilymphocyte globulin, may be effective in these patients. In patient J12, the normal hematopoietic stem cell clone could have enough activity to support hematopoiesis instead of the PNH stem cell clone, possibly because suppression by an autoimmune mechanism has disappeared.
Taken together, our observations provide convincing evidence that the predominant PNH stem cell clone can support hematopoiesis and manifest disease for 6 to 10 years. Furthermore, the active life of a PNH stem cell clone can be estimated to be approximately 15 years. HSCs supply all blood cells throughout life by their self-renewal and multilineage differentiation capabilities. CD34+ is a marker of human HSCs, and clinical transplantation studies that used enriched CD34+ BM cells indicated the presence of HSCs with long-term BM reconstitution ability within this fraction.22 Unlike in human, mouse primitive BM HSCs, individually having self-renewal and multilineage differentiation, were detected in the mCD34low to mCD34−fraction.23-25 Thus, the nature of HSCs is still not well characterized, especially in humans, and it is a current issue. It is also unclear how many HSCs sustain hematopoiesis and how long one HSC could support hematopoiesis in humans. Whether the long-lasting hematopoiesis of one stem cell clone found in patients with PNH is also true for normal HSCs is yet to be determined.
We thank Drs Russell E. Ware and Wendell F. Rosse for their critical reading and discussion. We also thank Yuki Murakami, Keiko Kinoshita, Fumiko Ishii, and Keiko Yamamoto for the excellent technical assistance.
Supported in part by a grant from the Japan Intractable Diseases Research Foundation and the Osaka Medical Research Foundation for Incurable Diseases.
J.N. and T.H. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jun-ichi Nishimura, Department of Immunoregulation, Research Institute for Microbial Diseases, Osaka University, 3-1 Yamada-oka, Suita, Osaka 565-0871, Japan; e-mail:junnishi@acpub.duke.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal