Key Points
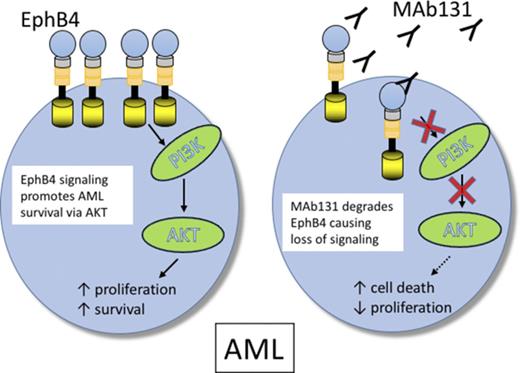
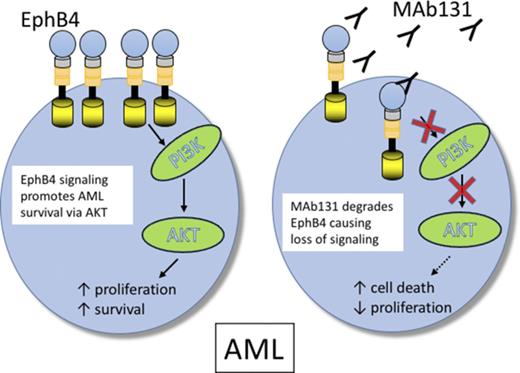
EPHB4 promotes leukemia survival via AKT activation.
EPHB4 can be therapeutically targeted in AML with monoclonal antibodies.
Abstract
EPHB4, an ephrin type B receptor, is implicated in the growth of several epithelial tumors and is a promising target in cancer therapy; however, little is known about its role in hematologic malignancies. In this article, we show that EPHB4 is highly expressed in ∼30% of acute myeloid leukemia (AML) samples. In an unbiased RNA interference screen of primary leukemia samples, we found that EPHB4 drives survival in a subset of AML cases. Knockdown of EPHB4 inhibits phosphatidylinositol 3-kinase/AKT signaling, and this is accompanied by a reduction in cell viability, which can be rescued by a constitutively active form of AKT. Finally, targeting EPHB4 with a highly specific monoclonal antibody (MAb131) is effective against AML in vitro and in vivo. EPHB4 is therefore a potential target in AML with high EPHB4 expression.
Introduction
It is estimated that ∼20 000 new cases of acute myeloid leukemia (AML) were diagnosed in the United States in 2016, and half of these patients will die of leukemia or complications of treatment.1 Therapeutic strategies for AML have not changed significantly in the past 3 decades, and thus there is a need to develop new treatments.2 Major efforts to catalog the genetic and epigenetic alterations in AML have identified targetable mutations in a subset of patients.3 We have previously reported on an RNA interference (RNAi)–based, functional kinase screen to identify genes that drive AML.4,5 One such target is the ephrin receptor, EPHB4.
Ephrins and ephrin receptors (EPH) are the largest receptor tyrosine kinase family with 15 EPH receptors and 9 membrane-bound ephrin ligands. EPH receptors interact with ephrin ligands, which are divided into 2 families: glycosylphosphatidylinositol-anchored ligands define the ephrinA family, and single-pass transmembrane ligands define the ephrinB family.6 EPH receptors can bind to >1 ephrin ligand, except for EPHB4, which only binds ephrinB2. EPH receptor and ephrin ligand binding leads to bidirectional signaling, defined as forward signaling in receptor-expressing cells and reverse signaling in ligand-expressing cells. EPH/ephrin family members are ubiquitously expressed in most adult tissues, except for EPHB4 and ephrinB2, which are expressed primarily during embryonic development in neural tissue and immature vasculature. EPH receptors typically interact with the cell surface–associated ephrin ligand at sites of cell-cell contact. EPH/ephrin signaling controls cell morphology, adhesion, migration, and invasion by modifying the organization of the actin cytoskeleton and influencing the activities of integrins and intercellular adhesion molecules. Both ligand and receptor are critical during embryonic development, and deletion of either gene causes embryonic lethality due to the failure of vasculature maturation7 ; however, conditional loss of Ephb4 in adult mice causes no abnormalities (P.S.G., unpublished data).
In the context of cancer, EPHB4/ephrinB2 signaling promotes tumor growth, invasiveness, chemoresistance and tumor angiogenesis.6,7 In bladder cancer, EPHB4 overexpression is partially regulated by epidermal growth factor receptor (EGFR) signaling and promotes cancer survival through antiapoptosis signaling.8 In esophageal tumors, the EPHB4 gene appears amplified and contributes to tumor cell survival and migration.9 Expression of EPHB4 was associated with clinically aggressive disease in gastric and gastroesophageal junction tumors.10 In colorectal cancer, EPHB4 was shown to not only promote tumor growth, but also tumor-associated angiogenesis.11,12 In an EPHB4 knockdown screen of prostate cancer, EPHB4 was shown to regulate integrin β8, a key determinant of prostate cancer invasiveness.13 Studies in ovarian cancer have shown that EPHB4 expression is associated with poorer survival, and targeting EPHB4 had promising preclinical activity.14 Overexpression of the EPHB4 ligand ephrinB2 has also been correlated with poor outcome in several tumors. Targeting EPHB4/ephrinB2 with specific antibodies was found to be effective in animal models of solid tumors.15,16 EPHB4 mutations have been reported in some epithelial cancers, such as lung cancer, whereas EPHB4 gene amplification occurs in nearly one-third of head and neck cancers.17,18 Furthermore, we have shown that EGFR activation, loss of PTEN, or Kras G12D activating mutations induce EPHB4 (Xi et al,8 Masood et al,17 Huang and Li,19 and P.S.G., unpublished data). Despite this wealth of data describing EPHB4/ephrinB2 in cancer, very little has been published about this pathway in leukemia. The few published studies describing the role of EPHB4 or ephrinB2 in leukemia are limited and mostly rely on gene expression data.20-22
To understand the function of EPHB4 in leukemia cells, we tested the activity of an anti-EPHB4–specific therapeutic antibody, MAb131. MAb131 is an agonist antibody that binds EPHB4, causing transient receptor activation followed by receptor internalization and degradation.23 Loss of EPHB4 on the surface leads to loss of phosphatidylinositol 3-kinase (PI3K)/AKT signaling and induction of apoptosis in cancer cells.24 In this study, we show that EPHB4 is expressed in AML cells, and targeting EPHB4 leads to cell death via downregulation of AKT.
Methods
RNAi screen
An RNAi screen on primary patient samples was performed as previously described.5 Briefly, patient samples were obtained with informed consent under the approval of the Institutional Review Board of Oregon Health and Science University. Cells were electroporated with small interfering RNA (siRNA; 1 μM final concentration) from a tyrosine kinase siRNA library or control siRNA in 96-well format and then replated in triplicate. Cell viability was assessed by CellTiter 96 MTS assay (Promega, Madison, WI) and normalized to a median value in the plate. Student t test was used to compare test wells to control wells, and values >2 standard deviations below the mean value on the plate and with a P value <.05 were considered significant.
Flow cytometry
Antibodies for flow cytometry for EPHB4 (MAb129, MAb131, and MAb19F4) were obtained from Vasgene. Generation and validation of MAb131 was previously described.20 MAb129 and MAb19F4 were similarly generated in mice and validated against recombinant EPHB proteins to ensure specificity for EPHB4 by a lack of binding to other EPHB family members. Antibodies were titered for flow cytometry on the EPHB4-expressing REH cell line. Antibody labeling was performed on ice for 20 minutes at the following concentrations: MAb131 (10 ng/μL), MAb129 (10 ng/μL), and MAb19F4 (1.2 ng/μL). Analysis of EPHB4 expression on primary patient samples was done with MAb131 and MAb19F4. MAb129, which recognizes a different epitope than MAb131, was used to analyze EPHB4 expression on cells after treatment with MAb131. For measurement of phospho-AKT (S472) cells were fixed with paraformaldehyde, permeabilized with methanol, and labeled with mouse anti-AKT (pS473) Clone M89-61 (BD Biosciences, San Jose, CA) according to the method of Krutzik et al.25 Cytometry data were analyzed with FlowJo software (Tree Star, Ashland, OR)
Analysis of normal bone marrow was performed on discarded excess product after bone marrow harvest. Cells were labeled with antibodies against CD34, CD38, CD19, CD33 (eBiosciences), and EPHB4 (Vasgene). Blocking was performed with Fc Block (eBiosciences).
Analysis of EPHB4 expression on putative AML stem cells was performed by labeling AML samples with antibodies against CD45, CD34, CD38 (eBiosciences), and EPHB4 (Vasgene), after Fc Block. Leukemia blasts were defined as low side scatter, low CD45. Fluorescence minus one controls were used to establish the EPHB4-negative gate.
Leukemia cell lines and primary samples
The human leukemic myeloid cell lines K562 (chronic myeloid leukemia blast crisis), KG1, MOLM14, MV-4-11, and U937 were obtained from American Type Culture Collection (Manassas, VA). HEL and ML-2 were a gift of Alan Epstein (Los Angeles, CA). All cell lines were cultured in RPMI 1640 medium (Gibco-BRL, Life Technologies, Inc, Gaithersburg, MD), supplemented with 10% fetal calf serum (Gibco-BRL) and 2 mM l-glutamine (Gibco-BRL). Primary clinical specimens were obtained from patients with newly diagnosed AML. Normal CD34+ bone marrow cells were obtained from discarded cell filters used to process bone marrow donations used for stem cell transplants. All patients granted informed consent where required and as approved by the University of Southern California Institutional Review Board. Mononuclear cells were isolated by density centrifugation by using Ficoll Paque Plus, density 1.077 (GE Healthcare Bio-Sciences, Pittsburgh, PA) followed by 2 washes with RPMI 1640 and were then frozen for analysis at later time points.
When required, primary AML/normal bone marrow samples were thawed and briefly cultured in RPMI 1640 supplemented with 10% fetal calf serum for 2 to 4 hours to allow for recovery. Cells were then analyzed for viability by trypan-blue, and if viability was <80%, dead cells were removed by Ficoll Paque Plus density centrifugation prior to further study, such as, analysis by flow cytometry, treatment with MAb131, or engraftment in mice.
Primary human AML xenograft studies
NOD-scid IL-2-γnull immunodeficient (NSG) mice (The Jackson Laboratory, Bar Harbor, ME) were irradiated with 100 rads and injected with 1 × 106 primary leukemia cells via the tail vein. Treatment with vehicle, AraC (50 mg/kg intraperitoneally [IP] weekly, Sigma-Aldrich, Saint Louis, MO), MAb131 (10 mg/kg IP, 3 times per week), or the combination, were performed. Mice were monitored daily for evidence of distress and killed when they showed evidence of disease. Engraftment was confirmed by flow cytometric analysis of bone marrow and/or spleen for evidence of human leukemic engraftment. Survival (Kaplan-Meier) curves were generated by using GraphPad Prism software with significance determined by the log-rank test. Bonferroni correction was used to adjust P values for multiple comparisons.
In vitro treatments
For RNAi studies, cells were transiently transfected with validated siRNA targeting EPHB4 (Silencer Select, Life Technologies, Carlsbad, CA). Exponentially growing cells were transfected with 10 nM anti-EPHB4 or control siRNA by using Lipofectamine RNAiMAX (Life Technologies) in accordance with the manufacturer's instructions. Cells were analyzed for viability or protein expression at designated time points.
For RNAi/AKT rescue experiments, cells were transfected with constitutively active AKT by using pcDNA3 Myr-HA-AKT1 from Addgene (Cambridge, MA), siRNA against EPHB4, or control siRNA. Viable cells were counted by hemocytometer and trypan blue exclusion.
For treatment with MAb131, leukemia cell lines or primary cells were cultured in standard media and treated with MAb131 at 10 ng/μL or isotype immunoglobulin G (IgG) as a control. Cells were assessed for EPHB4 expression by western blotting and/or flow cytometry and for viability by trypan blue exclusion or Annexin V/propidium iodide (PI) labeling.
To study the role of oncogenic drivers in the regulation of EPHB4 in myeloid leukemia, cells were treated with various concentrations of kinase inhibitors, including ruxolitinib, imatinib, and quizartinib (Selleckchem) in dimethyl sulfoxide. Control samples were treated with comparable doses of dimethyl sulfoxide containing phosphate-buffered saline (PBS). Expression of EPHB4 was measured on treated cells by using flow cytometry.
Cell viability and apoptosis assay
Cell viability was measured by using trypan blue (Sigma-Aldrich) dye exclusion assay. Treated or transfected cells were incubated with equal amounts of trypan blue for 5 minutes, and viable and dead cells were counted on a hemocytometer.
Apoptosis was measured by staining cells with Annexin V-fluorescein isothiocyanate in Annexin V staining buffer (BD Biosciences) per the manufacturer’s protocol. PI was added 15 minutes prior to flow cytometry to exclude nonviable cells.
EPHB4 internalization
K562 cells were labeled with mouse MAb131 followed by a goat antimouse secondary labeled with Alexa Flour 488 on ice. Cells were then incubated on ice or at 37°C for 1 hour. Cells were then placed on gelatin-coated slides and imaged. DAPI was used to stain nuclei.
Immunoblotting
Following treatments or transfections, cells were harvested and washed with PBS. Cells were lysed in cold radioimmunoprecipitation assay lysis buffer containing protease inhibitors (Complete; Roche Applied Science, Indianapolis, IN), followed by 3 freeze-thaw cycles. Protein content was determined by using the Bio-Rad DC protein assay (Hercules, CA). Equal amounts of protein were separated by using 10% to 15% gradient polyacrylamide gels and transferred to nitrocellulose membranes (Bio-Rad). The membranes were blocked with LICOR blocking buffer in PBS (1:1) with 0.05% Tween 20 for 1 hour at room temperature and probed with primary antibody (overnight at 4°C). Following incubation with primary antibody, membranes were washed in PBS with 0.05% Tween 20 and incubated with near infrared fluorescent–conjugated secondary antibody. Detection was performed by using the Odyssey Infrared Imaging System (LICOR Biosciences, Inc Lincoln, NB). All the antibodies were obtained from Cell Signaling Technology (Danvers, MA) except for EPHB4 antibody (Clone 2B5, Vasgene), actin (SCBT, Santa Cruz, CA), and glyceraldehyde-3-phosphate dehydrogenase (ThermoFisher Scientific, Rockford, IL).
Results
EPHB4 is expressed in AML but not in normal bone marrow progenitors
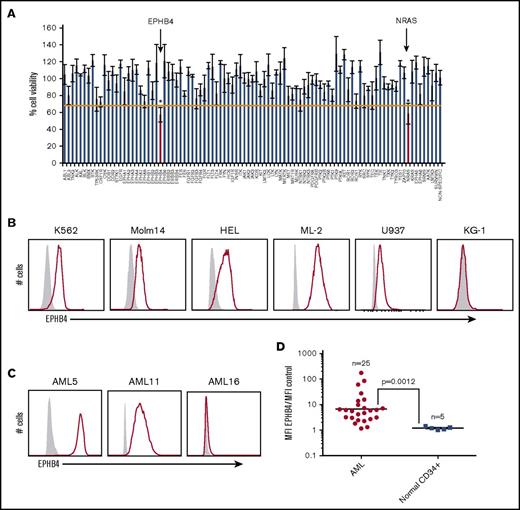
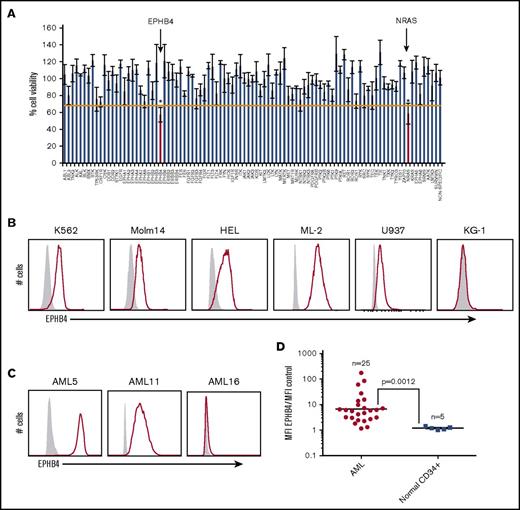
As part of an ongoing effort to identify new therapeutic targets in leukemia, we performed an RNAi screen of primary samples from patients with AML, acute lymphoblastic leukemia, myeloproliferative neoplasm, and myelodysplastic syndrome.4,5 Using this method, we identified that 17 of 281 (6%) AML samples and 33 of 539 (6%) total patient samples were sensitive to EPHB4 knockdown, suggesting that EPHB4 is a driver in a subset of leukemias and could be a potential therapeutic target in AML (Figure 1A; Table 1).
EPHB4 is expressed in AML. (A) Representative plot of 1 primary AML sample that screened positive for sensitivity to RNAi against EPHB4. This particular sample had a validated mutation in NRAS, which serves as a positive control, because AML cells demonstrate similar sensitivity to RNAi against NRAS as they do against EPHB4. *P < .05. (B) Cell surface EPHB4 expression in myeloid leukemia cell lines as measured by flow cytometry. The IgG isotype used to determine negative population is in gray. (C) Cell surface EPHB4 expression on primary AML blasts representing high (AML5), moderate (AML11), and no (AML16) expression as measured by flow cytometry. (D) Summary of EPHB4 expression on primary AML (n = 25) and normal, human CD34+ bone marrow cells obtained from healthy donors. Expression was measured by flow cytometry, and MFI was calculated and divided by the MFI of negative control. Results are plotted on log scale, therefore the expression ratio of 1 corresponds to no expression. The bar represents average MFI. P = .0012
EPHB4 is expressed in AML. (A) Representative plot of 1 primary AML sample that screened positive for sensitivity to RNAi against EPHB4. This particular sample had a validated mutation in NRAS, which serves as a positive control, because AML cells demonstrate similar sensitivity to RNAi against NRAS as they do against EPHB4. *P < .05. (B) Cell surface EPHB4 expression in myeloid leukemia cell lines as measured by flow cytometry. The IgG isotype used to determine negative population is in gray. (C) Cell surface EPHB4 expression on primary AML blasts representing high (AML5), moderate (AML11), and no (AML16) expression as measured by flow cytometry. (D) Summary of EPHB4 expression on primary AML (n = 25) and normal, human CD34+ bone marrow cells obtained from healthy donors. Expression was measured by flow cytometry, and MFI was calculated and divided by the MFI of negative control. Results are plotted on log scale, therefore the expression ratio of 1 corresponds to no expression. The bar represents average MFI. P = .0012
Encouraged by these results, we examined levels of EPHB4 protein in leukemia cell lines and primary leukemia cells. We measured the cell surface expression of EPHB4 in leukemia cell lines and primary leukemia cells by flow cytometry. Using a cutoff of threefold greater median fluorescence intensity (MFI) compared with isotype controls, we found that 5 of 6 (K562, Molm14, HEL, ML-2, U937, and KG1) leukemia cell lines and 19 of 25 primary AML samples had cell surface expression of EPHB4. (Figure 1B-D). Using a higher cutoff of 10-fold greater MFI compared with controls, 7 of 25 (28%) primary samples had high levels of EPHB4 cell surface expression (clinical details on primary patient samples are presented in supplemental Table 1). In contrast, EPHB4 was undetectable on bone marrow CD34+ progenitor cells and hematopoietic stem cells (supplemental Figure 1).
It is believed that clinical relapse of leukemia is mediated by leukemic stem cells, which are resistant to standard chemotherapy, and share an immunophenotype similar to normal bone marrow stem cells.26,27 To determine if EPHB4 was expressed in the CD34+/CD38– fraction of AML, we performed EPHB4 expression analysis in the CD34+/CD38– putative stem cell fraction of 10 primary AML samples. As has been previously described, we observed a large variation range in the percentage of the CD34+/CD38– AML blasts ranging from 0.01% to 68% (supplemental Figure 2A). For most cases that had detectable EPHB4 expression on bulk blasts, EPHB4 could be detected in the CD34+/CD38– population. Therefore, in patients with EPHB4+ AML, MAb131 could be used to target EPHB4 in bulk blasts and in the CD34+/CD38– leukemia population (supplemental Figure 2A).
Targeting EPHB4 inhibits leukemia cells
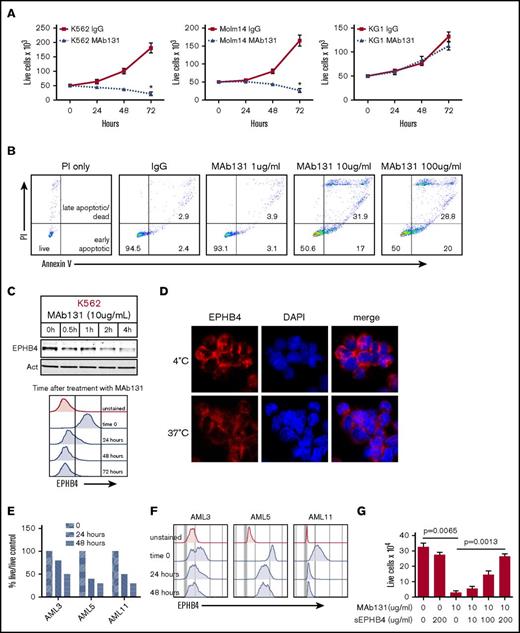
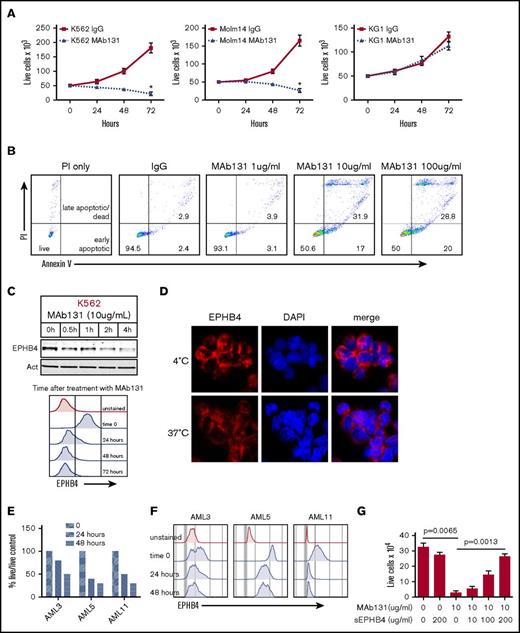
To understand the function of EPHB4 in leukemia cells, we tested MAb131, an agonist antibody that binds EPHB4, in leukemia cell lines and primary AML cells. Treatment of the EPHB4-expressing cells lines, K562 and Molm14, with MAb131 reduced cell growth and viability, but had no effect on the EPHB4-negative cell line, KG1 (Figure 2A). Previous work with MAb131 has shown that loss of EPHB4 signaling by MAb131 triggers apoptosis in cancer cells. To confirm that MAb131 induced apoptosis in leukemia cells, we treated leukemia cells with increasing doses of MAb131 and measured apoptosis by Annexin V and PI staining. Treatment with MAb131 induced apoptosis without any evidence of a dose relationship (Figure 2B). Also, consistent with prior studies, treatment with MAb131 resulted in rapid loss of EPHB4 within a few hours of treatment that was sustained for up to 72 hours, as demonstrated by immunoblotting and flow cytometry (Figure 2C). To demonstrate that treatment with MAb131 leads to internalization of EPHB4, cells were incubated with fluorescently labeled MAb131 and incubated at 4°C and 37°C. Incubation at 4°C prevented internalization of EPHB4, resulting in staining (red) that was restricted to the cell membrane (Figure 2D). Incubation at 37°C resulted in the loss of membrane staining and decreased overall signal as the antibody-labeled EPHB4 was internalized and degraded.
EPHB4 in AML can be targeted with a monoclonal antibody (MAb131). (A) EPHB4+ leukemia cells (K562 and Molm14) and EPHB4– (KG1) cells treated with 1 dose of MAb131 at time 0 and cultured under normal conditions. Live cells were counted by trypan blue exclusion at specified time points. MAb131 treatment reduced cell growth in EPHB4+ cells only. The average counts from 3 separate experiments are displayed, and error bars represent standard error of the mean. P values were calculated at 72 hours by pairwise comparison between control IgG-treated and MAb131-treated cells. *P < .01. (B) MAb131 induces apoptosis in EPHB4+ leukemia cells. K562 cells were treated with MAb131 (1, 10, and 100 μg/mL) and assayed for apoptosis by Annexin V/PI staining. The percentage of cells live (double negative), early apoptotic (Annexin V+/PI+), and late apoptotic/dead (Annexin V+/PI+) are noted. (C) K562 cells were treated with MAb131 and harvested for protein analysis by immunoblot (top) or by flow cytometry (bottom). MAb131 treatment causes loss of cell surface EPHB4 and degradation of total EPHB4 in cells. (D) K562 cells were treated with Alexa 488–labeled MAb131 and incubated at 37°C and 4°C for 1 hour, followed by imaging (40× objective). Incubation at 4°C (top) prevents the internalization and degradation of EPHB4, as evidenced by the bright cell surface staining, whereas incubation at 37°C (bottom) leads to internalization of labeled antibody and decreased overall staining. (E) EPHB4+ primary AML blasts (n = 3) were treated with MAb131. Live cells were counted on a hemacytometer by trypan blue exclusion at 24 and 48 hours and normalized to untreated controls. Treatment with MAb131 lead to increased cell death in primary AML samples. (F) Primary AML samples were treated with MAb131 and show downregulation of cell surface EPHB4 as measured by flow cytometry. (G) K562 cells were treated with increasing concentrations of soluble EPHB4, which blocks the cytotoxic effect of MAb131, confirming that MAb131 effects are due to EPHB4 binding and not off-target effects. Cells were treated and live cells counted by trypan blue exclusion at 72 hours. The average counts from 3 separate experiments are displayed, and error bars represent the standard error of the mean. P values for selected pairwise comparison are noted on the chart.
EPHB4 in AML can be targeted with a monoclonal antibody (MAb131). (A) EPHB4+ leukemia cells (K562 and Molm14) and EPHB4– (KG1) cells treated with 1 dose of MAb131 at time 0 and cultured under normal conditions. Live cells were counted by trypan blue exclusion at specified time points. MAb131 treatment reduced cell growth in EPHB4+ cells only. The average counts from 3 separate experiments are displayed, and error bars represent standard error of the mean. P values were calculated at 72 hours by pairwise comparison between control IgG-treated and MAb131-treated cells. *P < .01. (B) MAb131 induces apoptosis in EPHB4+ leukemia cells. K562 cells were treated with MAb131 (1, 10, and 100 μg/mL) and assayed for apoptosis by Annexin V/PI staining. The percentage of cells live (double negative), early apoptotic (Annexin V+/PI+), and late apoptotic/dead (Annexin V+/PI+) are noted. (C) K562 cells were treated with MAb131 and harvested for protein analysis by immunoblot (top) or by flow cytometry (bottom). MAb131 treatment causes loss of cell surface EPHB4 and degradation of total EPHB4 in cells. (D) K562 cells were treated with Alexa 488–labeled MAb131 and incubated at 37°C and 4°C for 1 hour, followed by imaging (40× objective). Incubation at 4°C (top) prevents the internalization and degradation of EPHB4, as evidenced by the bright cell surface staining, whereas incubation at 37°C (bottom) leads to internalization of labeled antibody and decreased overall staining. (E) EPHB4+ primary AML blasts (n = 3) were treated with MAb131. Live cells were counted on a hemacytometer by trypan blue exclusion at 24 and 48 hours and normalized to untreated controls. Treatment with MAb131 lead to increased cell death in primary AML samples. (F) Primary AML samples were treated with MAb131 and show downregulation of cell surface EPHB4 as measured by flow cytometry. (G) K562 cells were treated with increasing concentrations of soluble EPHB4, which blocks the cytotoxic effect of MAb131, confirming that MAb131 effects are due to EPHB4 binding and not off-target effects. Cells were treated and live cells counted by trypan blue exclusion at 72 hours. The average counts from 3 separate experiments are displayed, and error bars represent the standard error of the mean. P values for selected pairwise comparison are noted on the chart.
We next tested the effects of MAb131 on 3 primary human AML samples with varying amounts of surface EPHB4. AML 5 had high levels of EPHB4, AML 11 had moderate levels, and AML 3 had low levels. All 3 samples demonstrated decreased cell survival compared with controls, with loss of surface EPHB4 expression sustained at 48 hours (Figure 2E-F).
To determine the specificity of MAb131 activity, leukemia cells were treated with MAb131, and increasing amount of a soluble form of the EPHB4 receptor. Soluble EPHB4 was able to abrogate MAb131 activity in a dose-dependent manner (Figure 2G), showing a target-specific antibody response.
EPHB4 promotes leukemia survival via AKT signaling
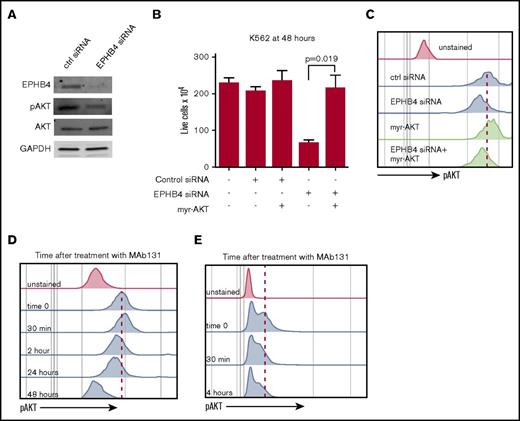
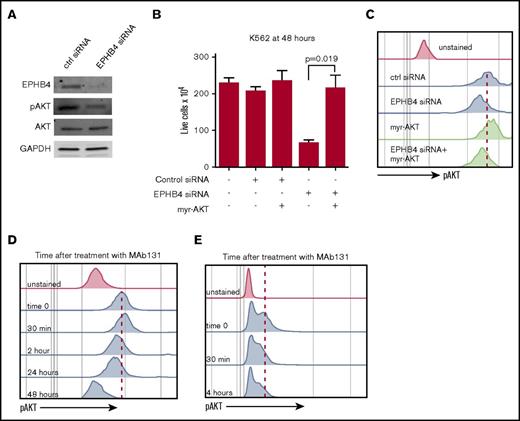
To understand the downstream effects of the loss of EPHB4 signaling in leukemia, we focused on the PI3K/AKT pathway. Knockdown of EPHB4 with siRNA decreased pAKT as measured by immunoblotting and phospho-flow MFI (Figure 3A,C). To confirm that EPHB4 regulates cell viability through the PI3K/AKT pathway, we performed rescue experiments with a constitutively active form of AKT (myr-AKT). Myr-AKT was expressed in K562 cells after EPHB4 knockdown and restored cell growth and basal AKT activity. (Figure 3B-C).
EPHB4 promotes AML survival through AKT signaling. (A) K562 cells were transduced with siRNA against EPHB4 and analyzed for EPHB4, pAKT, and total AKT at 48 hours. pAKT levels were reduced in EPHB4 knockdown cells as measured by immunoblotting, whereas total AKT levels were unchanged. (B) K562 cells were co-transduced with siRNA against EPHB4 and/or the constitutively active form of AKT (myr-AKT). Expression of myr-AKT restores cell growth and survival after EPHB4 knockdown. The average counts from 3 separate experiments are displayed, and error bars represent the standard error of the mean. (C) Levels of AKT activation in cells treated in Figure 3B as measured by phosphor-flow for pAKT. (D) K562 cells were treated with MAb131, and pAKT was measured by flow cytometry at various time points. At 30 minutes after treatment, a transient increase in pAKT was observed due to EPHB4 activation by MAb131, followed by a decrease in pAKT as EPHB4 activity was lost. (E) Primary AML blasts (unstimulated) treated with MAb131 show decreased pAKT after treatment with MAb131.
EPHB4 promotes AML survival through AKT signaling. (A) K562 cells were transduced with siRNA against EPHB4 and analyzed for EPHB4, pAKT, and total AKT at 48 hours. pAKT levels were reduced in EPHB4 knockdown cells as measured by immunoblotting, whereas total AKT levels were unchanged. (B) K562 cells were co-transduced with siRNA against EPHB4 and/or the constitutively active form of AKT (myr-AKT). Expression of myr-AKT restores cell growth and survival after EPHB4 knockdown. The average counts from 3 separate experiments are displayed, and error bars represent the standard error of the mean. (C) Levels of AKT activation in cells treated in Figure 3B as measured by phosphor-flow for pAKT. (D) K562 cells were treated with MAb131, and pAKT was measured by flow cytometry at various time points. At 30 minutes after treatment, a transient increase in pAKT was observed due to EPHB4 activation by MAb131, followed by a decrease in pAKT as EPHB4 activity was lost. (E) Primary AML blasts (unstimulated) treated with MAb131 show decreased pAKT after treatment with MAb131.
We next studied the effect of MAb131 treatment on pAKT levels in K562 using phospho-flow cytometry. Treatment with MAb131 caused an initial increase in pAKT MFI, followed by a persistent decline in pAKT from 3 to 24 hours. (Figure 3D). Treatment of primary AML cells with MAb131 similarly modulated pAKT levels (Figure 3E).
MAb131 demonstrates in vivo activity against primary AML
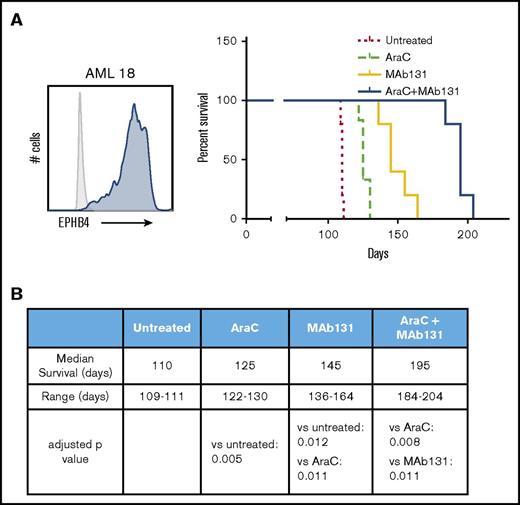
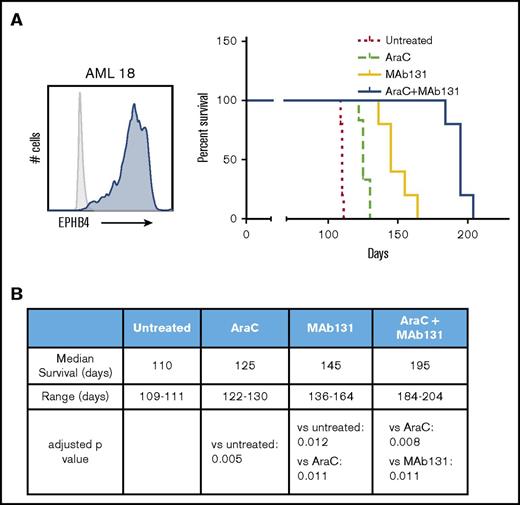
To test the activity of MAb131 in vivo, we employed orthotopic xenografts of EPHB4-positive primary human AML cells in immunodeficient mice. Mice were treated with cytarabine (a chemotherapy used in AML treatment), MAb131 alone, or a combination of cytarabine and MAb131. Vehicle controls were used for comparison. Treatment with MAb131 alone or in combination with cytarabine markedly improved survival to a median of 145 and 195 days, respectively, and compared favorably to the survival of only 110 days in vehicle controls (Figure 4A-B).
MAb131 demonstrates in vivo activity against primary AML. (A) Immunodeficient (NSG) mice were injected with 1 × 106 EPHB4+ primary AML cells (AML18) to create orthotopic xenografts. AML18 was characterized by normal cytogenetics, CEBPA mutant, and NPM1 mutation; 5 to 6 NSG mice per treatment group. Two weeks after injection, treatment mice were treated with PBS (untreated), AraC (50 mg/kg IP weekly), MAb131 10 mg/kg 3 times per week, or the combination. Treatments continued until all control mice were killed for evidence of disease. Remaining mice were observed for survival. Engraftment was confirmed at necropsy. (B) The table predicts median survival, range, and adjusted P values, after Bonferroni correction, for each survival curve.
MAb131 demonstrates in vivo activity against primary AML. (A) Immunodeficient (NSG) mice were injected with 1 × 106 EPHB4+ primary AML cells (AML18) to create orthotopic xenografts. AML18 was characterized by normal cytogenetics, CEBPA mutant, and NPM1 mutation; 5 to 6 NSG mice per treatment group. Two weeks after injection, treatment mice were treated with PBS (untreated), AraC (50 mg/kg IP weekly), MAb131 10 mg/kg 3 times per week, or the combination. Treatments continued until all control mice were killed for evidence of disease. Remaining mice were observed for survival. Engraftment was confirmed at necropsy. (B) The table predicts median survival, range, and adjusted P values, after Bonferroni correction, for each survival curve.
Mutations that drive EPHB4 expression in leukemia
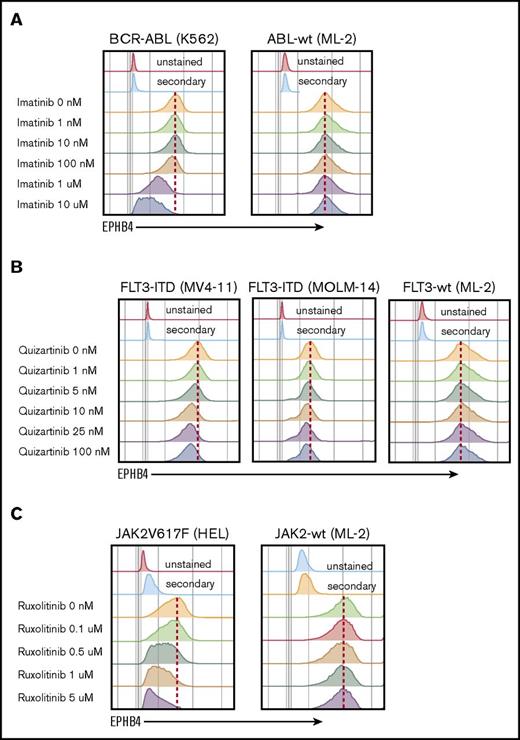
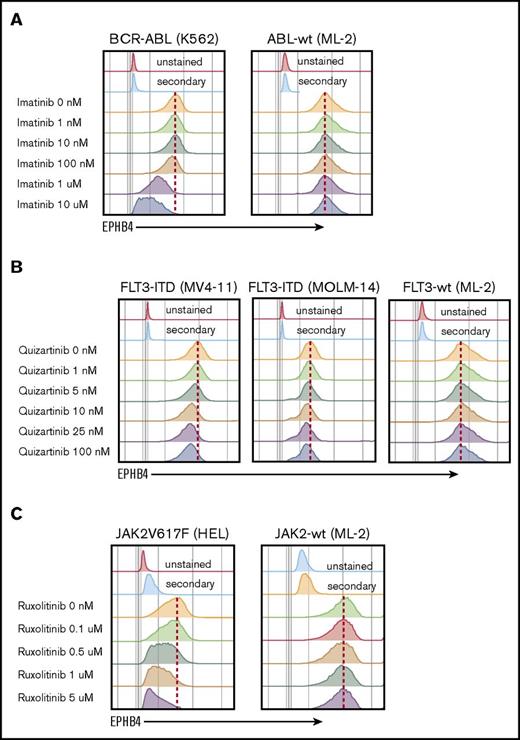
The mechanism of EPHB4 induction in leukemia cells is not understood. EPHB4 mutations and amplifications have been reported in some epithelial cancers, however, when we examined the The Cancer Genome Atlas AML dataset, we did not find evidence of mutations or copy number alterations of EPHB4.3 We therefore hypothesize that EPHB4 expression in AML may be induced by common driver mutations. To test this hypothesis, we studied leukemia cell lines with well-characterized mutations that can be targeted pharmacologically. K562 cells contain the BCR-ABL mutation, which can be inhibited by imatinib, and MV-4-11 and MOLM-14 cells bear the FLT3-ITD mutation, which can be inhibited by quizartinib. Finally, HEL cells contain the JAK2V617F mutation, which is sensitive to ruxolitinib. Treatment with imatinib, quizartinib, and ruxolitinib led to a dose-dependent decrease in EPHB4 in respective cell lines (Figure 5). The ML-2 cell line, which has wild-type ABL, FLT3, and JAK2 genes, was used as a control and showed no changed in EPHB4 expression after treatment with specific inhibitors.
EPHB4 expression is driven by mutations found in AML. (A) Treatment with inhibitor of BCR-ABL (imatinib) led to a decrease of EPHB4 expression on the surface of bcr-ABL–containing K562 cells lines, with no change in ML-2 cells, which had wild-type (wt) ABL. (B) Treatment with the specific FLT3 inhibitor, quizartinib led to a decrease of EPHB4 expression in FLT3-ITD–containing MV-4-11 and MOLM-14 cells, but no change in FLT3-wt cells. (C) Treatment with the JAK inhibitor ruxolitinib resulted in a decrease of EPHB4 expression on JAK2V617F-containing HEL cells, with no change in JAK2-wt controls.
EPHB4 expression is driven by mutations found in AML. (A) Treatment with inhibitor of BCR-ABL (imatinib) led to a decrease of EPHB4 expression on the surface of bcr-ABL–containing K562 cells lines, with no change in ML-2 cells, which had wild-type (wt) ABL. (B) Treatment with the specific FLT3 inhibitor, quizartinib led to a decrease of EPHB4 expression in FLT3-ITD–containing MV-4-11 and MOLM-14 cells, but no change in FLT3-wt cells. (C) Treatment with the JAK inhibitor ruxolitinib resulted in a decrease of EPHB4 expression on JAK2V617F-containing HEL cells, with no change in JAK2-wt controls.
Discussion
We show that EPHB4 is expressed at a high level in ∼30% of primary AML cells and is a driver in 6% of the cases. Steube and Wrobel21,22 have previously reported that EPHB4 is expressed in leukemia, but only at the messenger RNA level. We have demonstrated that EPHB4 is not expressed in normal bone marrow stem and progenitor cells. These data are similar to our previous studies in epithelial cancers and normal epithelial tissue and reinforce our previous findings that EPHB4 is an embryonic protein that is re-expressed only in certain tumors and tumor vessels. This restricted pattern of expression suggests that EPHB4 is an excellent therapeutic target in cancer, and that EPHB4 directed therapies will be safe with minimal effects on normal tissue. This is supported by initial data from clinical trials (identifiers NCT01642342, NCT02799485, and NCT02495896) targeting EPHB4/ephrinB2 with a soluble EPHB4 decoy receptor that have shown an excellent safety profile with no grade III or IV hematologic toxicity (El-Khoueiry et al28 and A. El-Khoueiry, USC Norris Comprehensive Cancer Center, oral communication, 14 December 2016).
EPHB4 signaling is activated by binding to its ligand, ephrinB2, which leads to homodimerization of the EPHB4 receptor and activation of downstream signaling.6 Like certain other receptor tyrosine kinases of its class, high expression of EPHB4 itself can lead to ligand-independent activation of downstream signaling.29 Because all of our in vitro experiments, including the RNAi screen, were done in the absence of ephrinB2-expressing stroma, it is expected that signaling in leukemia cells lines and primary cells can occur independent of the ligand. We believe this signaling would be further enhanced in an environment, such as bone marrow, where stroma expresses ephrinB2. Previous work has shown that tumor cells with EPHB4 expression can interact with ephrinB2 on stroma and vasculature to promote growth30 and confer resistance to therapy.31 Disrupting EPHB4/ephrinB2 interaction may, therefore, be a strategy to overcome treatment resistance.
We and others have observed that certain oncogenic pathways, such as EGFR, IGF-1, N-Ras, K-Ras, and loss of tumor suppressor genes, such as PTEN, can induce EPHB4 expression (Xia et al,8 Masood et al,17 and Huang et al,17 and P.S.G., unpublished observations). STAT proteins are commonly activated downstream of BCR-ABL, JAK2V617F, and FLT3-ITD mutations in leukemia, suggesting that oncogenic STAT activity might be driving EPHB4 expression in leukemia, and others have reported that imatinib treatment of BCR-ABL–mutated leukemia led to decreased EPHB4 expression.32 Studies to precisely determine how EPHB4 is induced in leukemia are currently underway. We believe that future patient selection strategies for EPHB4-targeted therapy will use both the cell surface expression of EPHB4 and genetic alterations in leukemia that may drive EPHB4 expression. Our data also suggest that monitoring PI3K/AKT activity could serve as a pharmacodynamic marker for assessing target response with EPHB4-targeted therapy. Finally, our data lay the ground work for additional translation opportunities for targeting EPHB4 beyond humanized antibody (MAb131) alone; efforts to develop antibody drug conjugates, chimeric antigen receptor T cells, and bispecific antibodies targeting EPHB4 are currently under development.
The full-text version of this article contains a data supplement.
Acknowledgments
This work was supported in part by award P30CA014089 and career development award K08 CA154975-01A1 (A.A.M.) from the National Institutes of Health, National Cancer Institute. J.W.T. was supported by the Leukemia and Lymphoma Society, the V Foundation for Cancer Research, the Gabrielle’s Angel Foundation for Cancer Research, and the National Institutes of Health, National Cancer Institute (grants 5R00CA151457-04 and 1R01CA183947-01).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Authorship
Contribution: A.A.M. designed and performed experiments, analyzed data, and prepared the manuscript; A.J., A.M., R.L., V.K., P.C., M.S., L.H., M.K., M.A., and T.J.T. performed experiments and analyzed data; and P.S.G., J.W.T., and B.J.D. designed experiments, analyzed data, and contributed to the manuscript.
Conflict-of-interest disclosure: A.A.M. received research funding and served as a consultant for Pfizer, Inc. J.W.T. received research support from Aptose, Array, AstraZeneca, Constellation, Genentech, Gilead, Incyte, Janssen, Seattle Genetics, Syros, and Takeda and is on the Scientific Advisory Board for Leap Oncology. The remaining authors declare no competing financial interests.
Correspondence: Akil A. Merchant, Division of Hematology, Department of Medicine, USC Norris Comprehensive Cancer Center, Keck School of Medicine, University of Southern California, 1450 Biggy St, Norris Research Tower 3501, Los Angeles, CA 90033; e-mail: akil.merchant@med.usc.edu.