Key Points
Transcriptome-wide differences exist between type 1 VWD and control-derived endothelial cells during basal and stimulated VWF release.
Dysregulation of specific miRNA-mRNA axes may contribute to the pathogenesis of type 1 VWD.
Abstract
Type 1 von Willebrand disease (VWD) is associated with a reduction in qualitatively normal von Willebrand factor (VWF). Current diagnostic guidelines only take into consideration the contribution of basal VWF levels, despite a lack of correlation with bleeding severity. Defects in stimulated VWF release, which occurs after hemostatic challenge, may contribute to bleeding in type 1 VWD, but the pathogenic mechanisms are poorly defined. In this study, a layered multiomic approach including messenger RNA (mRNA) and microRNA (miRNA) sequencing was used to evaluate transcriptome-wide differences between type 1 VWD- and control-derived endothelial colony forming cells (ECFCs) during basal and stimulated VWF release. ECFCs from 8 patients with type 1 VWD and 4 other patients were included in this study as controls. VWF protein analysis revealed heterogenous responses to stimulation among type 1 VWD and control ECFCs. During basal VWF release, 64 mRNAs and 7 miRNAs were differentially regulated between type 1 VWD and control ECFCs, and 65 putatively pathogenic miRNA-mRNA interactions were identified. During stimulated VWF release, 190 mRNAs and 5 mRNAs were differentially regulated between type 1 VWD and control ECFCs, and 110 putatively pathogenic miRNA-mRNA interactions were identified. Five gene ontology terms including coagulation, regulation of cell shape, and regulation of cell signaling were also differentially regulated in type 1 VWD ECFCs during stimulated release. To our knowledge, we have shown for the first time that transcriptome-wide differences exist between type 1 VWD and control ECFCs. These differences may contribute to bleeding in type 1 VWD, and further investigation may reveal novel biomarkers and therapeutic targets.
Introduction
von Willebrand disease (VWD) is the most common inherited bleeding disorder, with a symptomatic prevalence of ∼1 in 1000.1,2 VWD is caused by qualitative or quantitative defects in von Willebrand factor (VWF), a large plasma glycoprotein that tethers platelets to exposed subendothelial collagen after vascular injury.3 The International Society on Thrombosis and Haemostasis (ISTH) recognizes 3 types of VWD.4 The most common is type 1 VWD, which accounts for ∼80% of cases and is currently defined as a partial reduction in qualitatively normal VWF.4 Therapeutic management of type 1 VWD typically involves prophylactic and on-demand desmopressin (DDAVP) which actives endothelial vasopressin 2 receptors (V2R) to induce VWF release.5,6
The majority of VWF is produced by endothelial cells and stored in rod-shaped vesicles called Weibel-Palade bodies (WPBs).7 VWF is released into the circulation in both a basal and stimulated fashion.8 Basal release of VWF occurs constitutively in the absence of stimulation and determines an individual’s baseline VWF levels. Stimulated VWF release occurs after hemostatic challenge and results in rapid release of ultra-large molecular weight VWF, the most hemostatically active form of this protein. Two groups of agonists function on 2 pathways to release VWF. DDAVP and epinephrine act on cAMP to exocytose peripheral WPBs, whereas histamine, thrombin, and the secretagogue phorbol 12-myristate 13-acetate (PMA) increase intracellular Ca2+ to rapidly exocytose peripheral and perinuclear WPBs.9
Current diagnostic guidelines recommend that patients with VWF levels between 0.05 and 0.30 IU/mL, regardless of bleeding symptoms, are given a diagnosis of type 1 VWD.10 Patients with VWF levels below 0.50 IU/mL should receive a diagnosis of type 1 VWD if they present with abnormal bleeding. These guidelines, along with other published data,11-14 demonstrate a lack of correlation between bleeding severity and baseline VWF levels. This discrepancy complicates risk stratification of patients with type 1 VWD and often leads to homogeneous therapeutic management, creating potential for both over and under treatment of bleeding symptoms.
Notably, VWF levels assessed during the diagnostic workup are measured at baseline and, therefore, do not represent the contribution of stimulated VWF release. It is possible that the ability to elevate plasma VWF levels in response to hemostatic challenge is a better predictor of bleeding severity in type 1 VWD. A retrospective analysis of therapeutic DDAVP trials in type 1 VWD revealed a significant correlation between stimulated VWF levels and bleeding severity.15 Our lab has recently evaluated this association in a prospective clinical study which showed a reduced and less sustained response to DDAVP among patients with type 1 VWD, relative to controls, which correlated with bleeding severity (manuscript submitted for publication).
At this time, the pathological mechanisms underlying differences in stimulated VWF release among patients with type 1 VWD are poorly understood, therefore precluding large scale clinical studies required to inform evidence-based diagnostic guidelines. To address this lack of understanding, this study aims to use RNA sequencing (RNA-Seq) to evaluate differences in the transcriptome profiles of basal and stimulated VWF release between type 1 VWD- and control-derived endothelial colony forms cells (ECFCs). We integrate both messenger RNA-Seq (mRNA-Seq) and microRNA-Seq (miRNA-Seq) data to generate comprehensive transcriptome profiles. miRNAs are small noncoding RNAs that function by binding to complimentary mRNAs to reduce their expression. To our knowledge, this is the first study to evaluate VWF release in type 1 VWD using a layered multiomic approach. Along with improving our understanding of the pathogenesis of type 1 VWD, this study has the potential to identify novel biomarkers or therapeutic targets, for which antisense oligonucleotides can be developed.
Methods
Participant recruitment
Patients with type 1 VWD aged ≥18 years were recruited through the Inherited Bleeding Disorders Clinic of Southeastern Ontario in Kingston, Canada. Inclusion criteria for patients with type 1 VWD included abnormal bleeding as defined by an elevated ISTH-BAT bleeding score (BS), VWF antigen (VWF:Ag) and/or VWF activity (VWF:GPIbM or VWF:RCo) levels between 0.05 and 0.50 IU/mL, a VWF:GPIbM/VWF:Ag ratio >0.6, and normal VWF multimers. Exclusion criteria included patients with concomitant bleeding or clotting disorders, those taking medications known to influence hemostasis, being pregnant or breast-feeding, or those with a history of vascular disease. Controls were recruited from the general public and were subject to the exclusion criteria described above. All participants provided informed consent and this study was approved by the Queen’s University Health Sciences Ethics Review Board.
VWF and ABO genotyping
All participants had blood drawn and were isolated for sanger sequencing of the VWF gene and its promoter region. ABO genotyping was performed using the mutagenically separated polymerase chain reaction (MS-PCR) as described previously.16
Isolation and culture of ECFCs
ECFCs were isolated by drawing 48 mL of blood into BD Vacutainer Cell Preparation Tubes (BD Biosciences). The whole blood was centrifuged and the buffy coat containing peripheral blood mononuclear cells was retained. Peripheral blood mononuclear cells were washed in phosphate buffered saline (PBS)/10% fetal bovine serum (FBS; Wisent) and seeded to collagen coated 6-well cell culture plates at a density of 4×107 cells/well. Cell culture media was Endothelial Cell Growth Media 2 (ECGM-2) (PromoCell) supplemented with 10% FBS, Growth Media 2 Supplement Pack (PromoCell), and antibiotics (Invitrogen). Media was changed every day for the first 7 days and then every second day thereafter until ECFCs showing the typical cobblestone endothelial morphology appeared. ECFCs used for experimentation were between passages 4 and 9.
Endothelial cell phenotype was confirmed by the formation of confluent cell monolayers with characteristic cobblestone-like morphology and flow cytometry, as described previously.17 In cases in which cell numbers were limited, relative expression of endothelial-specific markers measured by mRNA-seq were used to confirm the cellular phenotype.
Stimulation of ECFCs
Because ECFCs do not respond to DDAVP,18 the PMA was used to induce VWF release. Dimethyl sulfoxide (DMSO) served as a control to mimic basal VWF release. By using measures of media and lysate VWF:Ag, confocal microscopy, and RNA-Seq, control and type 1 VWD ECFCs were compared during both basal and stimulated VWF release.
For all experiments, ECFCs in supplemented ECGM-2 were seeded at a density of 2 × 105 cells/well to collagen coated 24-well plates. After a 24-hour incubation, the plates were washed with Hanks Balanced Salt Solution (HBSS; Sigma) and replenished with serum free Opti-MEM media (Life Technologies). After an additional 24 hours, half of the wells were treated with DMSO and half were treated with 160 nM PMA for 1 hour at 37°C. Samples were subsequently prepared for analysis as described in the following subsections.
VWF:Ag secretion analysis
After the 1-hour incubation with either DMSO or PMA, media and lysate samples were collected to compare VWF secretion in type 1 VWD and control ECFCs during basal and stimulated release. Media was collected and then cells were washed with PBS and treated with NaCl lysis buffer. After 15 minutes, lysate samples were collected. VWF:Ag was measured using an enzyme-linked immunosorbent assay (ELISA) with DAKO antibodies A0082 and P0226, as described previously.17
Confocal immunofluorescence microscopy
For confocal immunofluorescence microscopy, the stimulation protocol was performed as described above with the exception that collagen coated coverslips were added to the wells and the cells seeded to these. VWF, VE-Cadherin, and nuclear staining was performed as described previously.19 The coverslips were then mounted to slides using antifade mounting medium (Vecta-Sheild) and imaged using a Leica TCS SP8 confocal fluorescence microscope and the LASX software (Leica Microsystems). Images were taken by a blinded microscopist. We measured the mean fluorescence intensity and normalized by the number of cells in the field. We quantified the number of the WPBs/cell using machine learning (identical settings, including intensity and minimum voxel size of 10). The WPBs were classified into 5 bins accordingly to their volume (bin 1, 0.1-0.5 μm; bin 2, 0.5-1 μm; bin 3, 1-2.5 μm; bin 4, 2.5-10 μm; and bin 5, 10-20 μm).
RNA isolation
After the 1-hour incubation with either DMSO or PMA, total RNA was isolated from ECFCs using an organic extraction protocol. Briefly, ECFCs were lysed with Trizol (Thermo Fisher) and chloroform was added to the lysates and mixed vigorously. RNA was precipitated from the supernatant using isopropanol and then washed with 80% ethanol. The pelleted RNA was dissolved in nuclease-free water and assessed for quality and concentration using the DropSense 16 (Trinean).
mRNA-seq and data analysis
Illumina compatible libraries were constructed using the QuantSeq 3’ mRNA-Seq Library Prep Kit (Lexogen, Austria) using 350 ng of total RNA. Independently indexed and purified libraries were combined at an equimolar concentration and the library pool was subject to bead purification, denaturation, and dilution. This pooled library was loaded onto a MidOutput v2 reagent cartridge at a concentration of 2.2 pM and subject to 75 cycles of single-ended sequencing to a depth exceeding 5 million clusters per sample on a NextSeq550 sequencer (Illumina, California). Raw data were transferred to the Queen’s Center for Advanced Computing and assessed for quality and trimmed using an established pipeline.20,21 Briefly, sequencing reads were aligned to the Ensembl_GRCh38 human genome using STAR aligner and counts were generated using HTSEQ-COUNT.22,23 Differential expression between unstimulated and PMA stimulated ECFCs was established using EdgR with adjusted P value cutoff of .1.24 Gene Ontology enrichment analysis was performed using the R package, Cluster Profiler and putative miRNA-mRNA interactions were evaluated using miRNET (https://www.mirnet.ca) which allowed for the identification of targets.25 miRNET queries differentially expressed mRNA and miRNA lists for target interactions that can be used to understand putative pathogenic mechanisms of differentially expressed miRNAs. Validation of the mRNA-seq data set was performed on a subset of differentially expressed genes (DEGs) using TaqMan real-time PCR assays (Thermo Fisher).
miRNA-seq and data analysis
miRNA profiling was performed using an established barcoded small RNA sequencing protocol.26 Briefly, 100 ng of total RNA was spiked with a set of synthetic calibration markers before ligation to a sample-specific 3’ oligonucleotide adaption specific for small RNA species. After sample pooling, 5’adapter ligation, and reverse transcription PCR, the cDNA was sent for Illumina deep sequencing using the HiSeq 2500 platform at the Ontario Institute of Cancer Research. After sequencing, .fastq files were uploaded through a web-accessible RNA sequencing pipeline (RNAworld.rockefeller.edu) hosted in the Tuschl Laboratory at The Rockefeller University. Data preprocessing, filtering, and subsequent analyses were performed in MATLAB (Mathworks) as described previously.27 To report miRNA abundance independent of sequencing depth, read counts were normalized against total sequence reads annotated as miRNAs. Sample outlier identification and batch effect assessment and removal were performed through correlation analyses of miRNA expression profiles as described previously.28 To exclude miRNAs with low expression across samples, a filtering threshold was applied as described previously.29 To prioritize high expression, we set the filtering threshold to the ninetieth percentile. miRNAs or miRNA clusters expressed above this threshold in more than 5% of the samples were retained. After preprocessing, all nonhuman miRNAs and human miRNA STAR sequences were excluded from further analyses. To identify miRNAs that accurately discriminated between control and type 1 VWD ECFCs, we used an established feature selection algorithm with leave out cross validation.28
Results
Patient characteristics
ECFCs were successfully isolated from 8 patients with type 1 VWD and 4 controls group. Demographic information, blood type, baseline VWF levels, and ISTH-BAT BS are provided in Table 1. Patients with type 1 VWD had significantly lower baseline VWF:Ag levels (0.43 vs 0.73 IU/mL; P = .008) and VWF activity levels (0.35 vs 0.73 IU/mL; P = .0078) compared with controls. The mean ISTH-BAT BS of the patients with type 1 VWD (BS = 9) was significantly higher than the controls (BS = 2; P = .0064). VWF sequencing was performed on all patients with type 1 VWD included in this study. Pathogenic VWF variants were identified in 6 out of 8 (75.0%) patients (Table 2).
Confirmation of ECFC phenotype
All ECFCs displayed cobblestone morphology. The endothelial cell phenotype was confirmed in 3 out of 4 (75.0%) control ECFCs and 5 out of 8 (62.5%) type 1 VWD ECFCs using flow cytometry (supplemental Table 1). Because of low cell yield, we could not confirm the phenotype of 4 ECFCs through flow cytometry, but high levels of CD31, CD144, and CD146 expression and low levels of CD14 and CD45 expression were observed through mRNA-seq across all type 1 VWD and control ECFCs (supplemental Table 2).
Analysis of VWF secretion from ECFCs
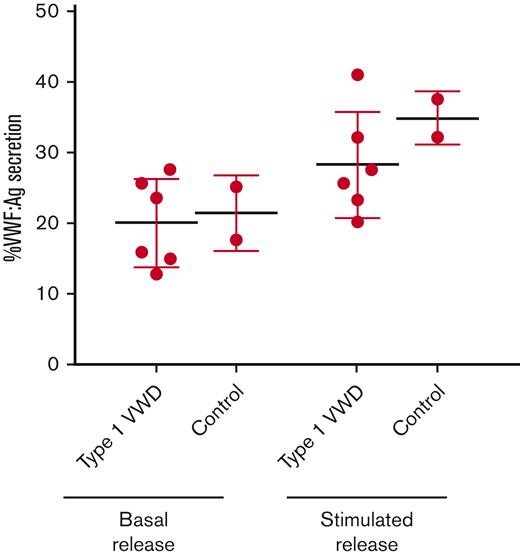
ECFCs were stimulated with a DMSO control or PMA to evaluate basal and stimulated VWF release, respectively. Because of low cell yield, data from only 6 type 1 VWD ECFCs (1P, 2P, 3P, 6P, 7P, and 8P) and 2 control ECFCs (1C and 4C) were included in this experiment. During basal VWF release, type 1 VWD and control ECFCs exhibited similar VWF secretion (20.1% and 21.4%, respectively; Figure 1). During stimulated VWF release, type 1 VWD ECFCs exhibited lower VWF secretion (28.3%) compared with control ECFCs (34.9%), but the sample size precluded statistical analysis.
%VWF secretion from type 1 VWD and control ECFCs after basal and stimulated VWF release. Mean %VWF secretion from type 1 VWD and control ECFCs during basal VWF release and stimulated VWF release. VWF:Ag was measured using an ELISA. %VWF secretion was calculated using the following formula: VWF:Ag in media / (VWF:Ag in media + VWF:Ag in lysate)∗100. Data points indicate mean %VWF:Ag secretion for each cell line (performed in triplicate). Error bars represent the mean ±SD. Because of limited cell yield, this experiment could only be performed on 2/4 control-derived ECFCs and 6/8 type 1 VWD-derived ECFCs. Low cell yield also precluded statistical analysis.
%VWF secretion from type 1 VWD and control ECFCs after basal and stimulated VWF release. Mean %VWF secretion from type 1 VWD and control ECFCs during basal VWF release and stimulated VWF release. VWF:Ag was measured using an ELISA. %VWF secretion was calculated using the following formula: VWF:Ag in media / (VWF:Ag in media + VWF:Ag in lysate)∗100. Data points indicate mean %VWF:Ag secretion for each cell line (performed in triplicate). Error bars represent the mean ±SD. Because of limited cell yield, this experiment could only be performed on 2/4 control-derived ECFCs and 6/8 type 1 VWD-derived ECFCs. Low cell yield also precluded statistical analysis.
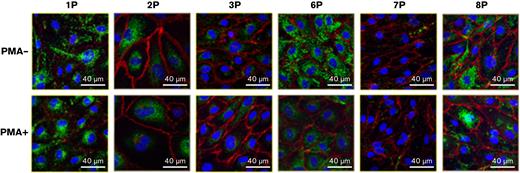
Confocal microscopy was used to qualitatively analyze the release of VWF during basal and stimulated release from the 6 type 1 VWD ECFCs and 2 control ECFCs mentioned above (Figure 2A,B). A variety of staining patterns were noted; however, upon quantification of these images, a statistically significant inverse correlation (r = −0.66; P = .0056) was observed with VWF secretion measured by ELISA (Figure 2C) and number of WPBs/cells. In addition, during basal VWF release, control ECFCs contained significantly more WPBs compared type 1 VWD ECFCs (111 vs 64 WPBs/cell; P = .0107). Upon stimulation, control ECFCs exocytosed an average of 82 WPBs, whereas type 1 VWD ECFCs exocytosed only 32 WPBs (Figure 2D).
Confocal immunofluorescence microscopy of ECFCs after basal and stimulated VWF release. ECFCs derived from healthy controls (A) and patients with type 1 VWD (B). Images in the top row were taken during basal VWF release (PMA-) and images in the bottom row were taken after stimulated VWF release (PMA+). ECFC identification labels are displayed across the top of the figure. ECFCs were stained for VWF (green), VE-cadherin (red) and DAPI (blue). Images are shown at 63× magnification with white scale lines representing 40 μm. Images were taken using a Lecia TCS SP8 Confocal Microscope. (C) Pearson r correlation between VWF section measured by ELISA and WPB count quantified from confocal microscopy images of control and type 1 VWD ECFCs during basal and stimulated release. (D) The number of WPBs contained in control and type 1 VWD ECFCs after basal and stimulated secretion. ∗P<.05.
Confocal immunofluorescence microscopy of ECFCs after basal and stimulated VWF release. ECFCs derived from healthy controls (A) and patients with type 1 VWD (B). Images in the top row were taken during basal VWF release (PMA-) and images in the bottom row were taken after stimulated VWF release (PMA+). ECFC identification labels are displayed across the top of the figure. ECFCs were stained for VWF (green), VE-cadherin (red) and DAPI (blue). Images are shown at 63× magnification with white scale lines representing 40 μm. Images were taken using a Lecia TCS SP8 Confocal Microscope. (C) Pearson r correlation between VWF section measured by ELISA and WPB count quantified from confocal microscopy images of control and type 1 VWD ECFCs during basal and stimulated release. (D) The number of WPBs contained in control and type 1 VWD ECFCs after basal and stimulated secretion. ∗P<.05.
ECFC transcriptomics
mRNA-Seq of all type 1 VWD (n = 8) and control (n = 4) ECFCs yielded high quality reads (supplemental Figure 1). A total of 173.6 million reads were sequenced and a mapping rate of 92.8% was achieved (supplemental Table 3). RNA-Seq data are available on Mendeley (doi. 10.17632/jc2vnrsccw.1). DEGs identified during basal and stimulated VWF release were validated using TaqMan assays (supplemental Table 4). Principal component analysis (PCA) plots for basal and stimulated release are shown in supplemental Figures 2 and 3, respectively. These plots show a considerable heterogeneity among the control groups, which likely blunted the number of DEGs identified between patients and controls during basal and stimulated VWF release.
Transcriptomics of basal VWF release
A total of 64 genes were differentially regulated in type 1 VWD ECFCs during basal VWF release: 60 genes were downregulated and 4 were upregulated, relative to controls (Figure 3A). The 10 most significantly DEGs during basal VWF release are outlined in Table 3. We were interested to determine if VWF or V2R were differentially expressed between control and type 1 VWD ECFCs during basal VWF release. VWF was highly expressed in both type 1 VWD and control ECFCs, but there was no difference between the 2 groups (Figure 3B). V2R encodes the V2 receptor, which has been shown to be absent on ECFCs18, therefore explaining the inability to respond to DDAVP. Relative expression of V2R in both controls and patients was near absent and there was no difference between the 2 groups (Figure 3C), therefore confirming prior findings. GO enrichment analysis was performed using the R Studio package, ClusterProfiler, but no molecular functions, biological processes, or cellular components were differentially regulated during basal VWF release.
Transcriptome analysis of basal VWF release. (A) Volcano plot outlining DEGs between type 1 VWD and control ECFCs during basal release. The horizontal dashed line represents an adjusted P value of 0.1 and the vertical dashed lines represent 1.5-fold change relative to control ECFCs. DEGs meeting the 0.1 corrected P value threshold and that exhibit a fold expression >1.5 are in red. Relative expression of VWF (B) and V2R (C). (D) Interacting miRNAs and mRNAs that were differentially regulated during basal VWF release are displayed in a chord diagram. Green indicates upregulation and red indicates downregulation relative to control ECFC expression. The outer ring indicates direction of transcript regulation, and the ribbon color represents specifically the direction of miRNA regulation.
Transcriptome analysis of basal VWF release. (A) Volcano plot outlining DEGs between type 1 VWD and control ECFCs during basal release. The horizontal dashed line represents an adjusted P value of 0.1 and the vertical dashed lines represent 1.5-fold change relative to control ECFCs. DEGs meeting the 0.1 corrected P value threshold and that exhibit a fold expression >1.5 are in red. Relative expression of VWF (B) and V2R (C). (D) Interacting miRNAs and mRNAs that were differentially regulated during basal VWF release are displayed in a chord diagram. Green indicates upregulation and red indicates downregulation relative to control ECFC expression. The outer ring indicates direction of transcript regulation, and the ribbon color represents specifically the direction of miRNA regulation.
During basal VWF release, 7 miRNAs were differentially expressed: 6 miRNAs were upregulated, and 1 miRNA was downregulated relative to controls (Table 4). We wanted to determine if these miRNAs targeted any of the differentially expressed mRNAs that we identified during mRNA-seq. To this end, we inputted the differentially expressed miRNA and mRNA datasets into miRNET to identify putative pathological interactions. During stimulated VWF release, 65 miRNA-mRNA interactions were identified (Figure 3D).30
Transcriptomics of stimulated VWF release
A total of 190 genes were differentially regulated in type 1 VWD ECFCs during stimulated VWF release; 127 genes were downregulated and 63 were upregulated, relative to controls (Figure 4A). The 10 most significantly DEGs are outlined in Table 5. Similar to basal VWF release, VWF was highly expressed in both type 1 VWD and control ECFCs, but there was no difference between the 2 groups (Figure 4B). Relative expression of V2R in both patients and controls was near absent and there was no difference between the 2 groups (Figure 4C). Through GO enrichment analysis, a total of 3 biological processes and 2 cellular components were identified that were differentially regulated between type 1 VWD and control ECFCs during stimulated VWF release (Table 6). These GO terms included coagulation and other cellular functions involved in cell shape, signaling, and intracellular trafficking.
Transcriptome analysis of stimulated VWF release. (A) Volcano plot outlining DEGs between type 1 VWD and control ECFCs during stimulated release. The horizontal dashed line represents an adjusted P value of 0.1 and the vertical dashed lines represent 1.5-fold change relative to control ECFCs. DEGs meeting the 0.1 corrected P value threshold and that exhibit a fold expression >1.5 are in red. Relative expression of VWF (B) and V2R (C). (D) Interacting miRNAs and mRNAs that were differentially regulated during stimulated VWF release are displayed in a chord diagram. Green indicates upregulation and red indicates downregulation relative to control ECFC expression. The outer ring indicates direction of transcript regulation, and the ribbon color represents specifically the direction of miRNA regulation.
Transcriptome analysis of stimulated VWF release. (A) Volcano plot outlining DEGs between type 1 VWD and control ECFCs during stimulated release. The horizontal dashed line represents an adjusted P value of 0.1 and the vertical dashed lines represent 1.5-fold change relative to control ECFCs. DEGs meeting the 0.1 corrected P value threshold and that exhibit a fold expression >1.5 are in red. Relative expression of VWF (B) and V2R (C). (D) Interacting miRNAs and mRNAs that were differentially regulated during stimulated VWF release are displayed in a chord diagram. Green indicates upregulation and red indicates downregulation relative to control ECFC expression. The outer ring indicates direction of transcript regulation, and the ribbon color represents specifically the direction of miRNA regulation.
Discussion
The pathogenesis of type 1 VWD is incompletely understood which complicates the development of evidence-based diagnostic and therapeutic guidelines. Current diagnostic guidelines only take into consideration baseline VWF levels despite a lack of correlation with bleeding severity. The ability to elevate VWF levels in response to hemostatic challenge may be a better predictor of bleeding severity, but the factors contributing to difference in stimulated VWF release between patients with type 1 VWD and healthy individuals are poorly defined. To our knowledge, for the first time, we used a layered multiomic approach to evaluate transcriptome-wide differences between ECFCs from patients with type 1 VWD and healthy controls during basal and stimulated VWF release.
We first evaluated basal and stimulated release of VWF from ECFCs quantitatively using secretion analysis and qualitatively using confocal IF microscopy. Although these experiments were underpowered to preserve ECFCs for mRNA- and miRNA-seq, general trends can be examined. During basal VWF release, type 1 VWD and control ECFCs released comparable amounts of VWF. Conversely, during stimulated release, the control ECFCs secreted greater proportions of their VWF stores. Confocal microscopy performed during basal and stimulated release revealed a reduction in WPBs/cell among the type 1 VWD ECFCs, compared with controls. Control ECFCs expelled twice as many WPBs/cell than controls upon stimulation with PMA. In addition, we found that the number of WBPs/cell inversely correlated with VWF secretion measured by ELISA during basal and stimulated release. Taken together, these findings further support the hypothesis that type 1 VWD is associated with a reduced response to stimulation.
Through mRNA- and miRNA-seq, transcriptome profiles of basal and stimulated VWF release were synthesized and compared between type 1 VWD and control ECFCs. A 1-hour stimulation period was chosen for these experiments to align with timepoints of therapeutic DDAVP trials. VWF levels typically peak at 1 to 2 hours post-DDAVP administration. Because PMA signaling is more rapid than DDAVP signaling,31,32 we speculated that a 1 hour stimulation period would capture the transcriptional changes most relevant to the response to PMA stimulation.
During simulated VWF release, 190 DEGs were identified between type 1 VWD and control ECFCs. One of the most notable DEGs was urokinase-type plasminogen activator (PLAU) which encodes urokinase (uPA) and was upregulated 3.3-fold in type 1 VWD ECFCs. Increased expression of PLAU upon stimulation/hemostatic challenge in type 1 VWD may be associated with accelerated fibrinolysis and, therefore, prolonged bleeding. We also found that 2 uPA inhibitors, plasminogen activator inhibitor-1 (SERPINE1; PAI-1) and plasminogen activator inhibitor-2 (SERPINB2; PAI-2), were upregulated in stimulated type 1 VWD ECFCs. A similar phenomenon is observed in Quebec platelet disorder (QPD), a rare bleeding disorder characterized by abnormally high levels of uPA in platelets due to a partial duplication of chromosome 10 which contains the PLAU gene. In QPD, bleeding symptoms persist despite a 2-fold increase in PAI-1, relative to healthy individuals.33 This is consistent with our observed transcriptional changes and indicates that dysregulation of fibrinolysis may also contribute, in part, to the pathogenesis of type 1 VWD. Several in vivo studies conducted in the 1990s have addressed this question by evaluating the relationship between uPA, tPA, and type 1 VWD after vascular occlusion or administration of DDAVP, but the results were conflicting.34-36 Future studies should investigate if this observed increase in PLAU, SERPINE1, and SERPINB2 expression translates to an increase in plasma levels of uPA, PAI-1, and PAI-2, respectively, upon stimulation.
Five GO terms were differentially regulated between type 1 VWD and control ECFCs during stimulated VWF release. The finding that coagulation is differentially regulated in type 1 VWD ECFCs during stimulated VWF release, but not basal VWF release, supports the notion that an insufficient response to stimulation may be an important contributor to bleeding in this patient population. Regulation of cell shape was another differentially regulated biological process. It is known that the endothelial cytoskeleton is a dynamic structure that is rearranged in response to stimulation to exocytose WPBs.37 WPBs are also trafficked through the cell along actin via various motor proteins. Dysregulation of this biological process in ECs would be expected to disrupt VWF release and may account for our observed reduction in WPB exocytosis among type 1 VWD ECFCs after stimulation.
Many studies have identified miRNAs that target mRNAs involved in coagulation,38 but no published studies have investigated miRNAs that are differentially regulated in type 1 VWD. Remarkably, during stimulated release, 5 miRNAs were differentially regulated between type 1 VWD and control ECFCs and 110 putative miRNA-mRNA interactions were identified. A particularly interesting finding was a 15.7-fold decrease in miR-23b expression in stimulated type 1 VWD ECFCs relative to controls. miR-23b targets with several differentially regulated coagulation- and cytoskeleton-related mRNAs including PLAU, SERPINE1, SERPINB2, profilin-2 (PFN2), transgelin (TAGLN), and versican (VCAN).39 In addition, it has been demonstrated that miR-23b is involved in cell migration, cytoskeletal remodeling, and metastasis in multiple forms of cancer.40-42 These findings support future studies analyzing miR-23b function in animal models of type 1 VWD. Interestingly, we found that some interacting miRNA-mRNA pairs exhibited the same direction of regulation, such as miR-19b and FAM102A which were both downregulated. In these cases, more potent upregulated miRNAs or other unknown factors likely contribute to this observation. These findings highlight the complexity of miRNA-mRNA interactions and, in some cases, require further functional studies to identify which miRNAs exhibit the strongest regulation of specific mRNAs.
The findings of this study are consistent with a previous report by Ng et al43 which used single-cell RNA-Seq to evaluate differential transcriptional regulation in ECFCs from individuals with low VWF. Although this study only analyzed ECFCs in the basal state, alterations in WPB metrics, VWF release, and transcriptional regulation were observed. We also identified reductions in WPBs among type 1 VWD ECFCs in the basal state, as well as a reduction in the number of expelled WPBs upon stimulation. In contrast to our study which showed only a numerical reduction in VWF mRNA in type 1 VWD ECFCs, Ng et al found a significant reduction in VWF expression in their low VWF ECFC cohort. Since the isolation of ECFCs used in this study and subsequent RNA-Seq, it has been shown that variations exist within ECFCs isolated from a single individual.44 These “clones” show variations in morphology, proliferation, and VWF production. As such, it is possible that clonal heterogeneity in our cohort blurred the ability to detect true differences in VWF expression between control and type 1 VWD ECFCs. This may also explain the heterogeneity observed in the PCA plots.
Another limitation of this study is the low number of replicates. Given the heterogenous nature of type 1 VWD, a larger and more homogeneous patient cohort could reveal additional DEGs. In addition, although the 1 hour PMA stimulation was strategically selected to align with the timing of therapeutic DDAVP trials, it is possible that other pathologically relevant genes are differentially expressed either before or after this 1 hour timepoint. As such, future studies could repeat this experiment using qPCR across several timepoints.
In summary, we have shown that distinct transcriptional differences exist between type 1 VWD and control ECFCs during basal and stimulated VWF release. Notably, these differences are more common during stimulated release which challenges current diagnostic guidelines for type 1 VWD that only consider baseline VWF levels. Although we have not discussed every DEG or miRNA-mRNA interaction identified in this study, we present the data and make it available to the community for further experimentation. These data may lay the groundwork for future functional, animal, and clinical studies, and may also reveal novel biomarkers and therapeutic targets.
Acknowledgment
The authors acknowledge Xiaojing Yang for assisting with RNA extraction.
Authorship
Contribution: R.K. performed research, analyzed data, and wrote the manuscript; M.Z.S. performed research; J.G. recruited participants and performed phlebotomy; L.T. performed phlebotomy; P.A.D.A. performed confocal microscopy and flow cytometry; M.B. performed research and contributed to the writing of the manuscript; K.T. analyzed transcriptomic data; C.C.T.H. performed mRNA-Seq, analyzed transcriptomic data, and prepared figures; N.R. performed miRNA-Seq and analyzed transcriptomic data; P.J. designed the study, supervised research, and contributed to the writing of the manuscript; and all authors reviewed and approved the final version of the manuscript.
Conflict-of-interest disclosure: P.J. receives funding from Bayer Pharmaceuticals, CSL Behring, and Takeda Pharmaceutical Company. The remaining authors declare no competing financial interests. This research was funded by a Translational Institute of Medicine (TIME) Incubator grant.
Correspondence: Paula James, Room 2025, Etherington Hall, Queen’s University Kingston, ON K7L 3N6 Canada; e-mail: jamesp@queensu.ca.
References
Author notes
Data are available on request from the corresponding author, Paula James (jamesp@queensu.ca).
The full-text version of this article contains a data supplement.