TO THE EDITOR:
Introduction
Past and current tobacco smoking is significantly associated with higher incidence and worse survival among patients with acute myeloid leukemia (AML) than among nonsmokers; however, there are no tailored treatment strategies available for these individuals.1-3 For patients with AML with the fms-like tyrosine kinase 3-internal tandem duplication (FLT3-ITD) mutation, clinical outcomes are compromised because of aggressive disease features, and the additional impact of smoking in this molecular subtype of AML has not been well studied. DNA methylation signatures persist for decades in smokers but have not been applied to patients with AML with exposure to cigarette smoke.4,5 Characterization of molecular changes attributable to cigarette smoke has potential to uncover targets for therapy that could be applied to improve treatment outcomes.
Methods
Review of patients with newly diagnosed, treatment-naive FLT3-ITD+ AML was conducted after approval by the University of Texas MD Anderson Cancer Center Institutional Review Board (protocols DR09-0223 and PA12-0395), and informed consent was obtained in compliance with the Declaration of Helsinki. Raw bisulfite sequencing and patient survival data were evaluated based on those reported by Ley et al.6
Mice were placed in whole-body smoke exposure chambers of a cigarette smoking robot SciReq InExpose, 5 days per week, with an intake of 24 cigarettes per day. Nonsmoking (NS) mice were removed from normal housing during the exposure of smoking mice. Calbiotech Mouse/Rat Cotinine enzyme-linked immunosorbent assay kit was used in mouse urine. MOLM13-luc, MOLM14-luc, or MV411-luc cells were introduced via tail-vein injection into mice and monitored by in vivo imaging system. Daunorubicin was administered thrice weekly at 2 mg/kg via tail-vein. All mouse experiments were approved by the University of Texas MD Anderson Cancer Center Institutional Animal Care and Use Committee under protocol 638 (principal investigator, J.C.).
Reduced representation bisulfite sequencing7,8 was applied to AML-bearing mouse spleens to compare global changes in DNA methylation. Genomic DNA was obtained with PureLink Genomic DNA Mini Kit. Samples passing quality control were subsequently sequenced on an Illumina HiSeq 3000, with between 49 and 85 million reads generated per sample. Western blots, immunohistochemistry, flow cytometry, and mass cytometry were used for protein analyses of spleen and liver tissues from mice.
Results and discussion
Poor prognosis after AML diagnosis for past and current smokers has been documented,3 but it has not been examined in a uniform cohort of patients with FLT3-ITD mutation. Overall response rates in treatment-naive patients with AML, bearing the FLT3-ITD mutation, who have never smoked (never smokers; n = 55) or have a smoking history (ever smokers; n = 29) were 93% and 73% (P = .005), respectively. Complete response rates were significantly different in never smokers (75%; n = 75) compared with ever smokers (43%; n = 17; P = .001; Figure 1A). Smokers also had worse survival rates than nonsmokers (18 vs 58 months for never smokers; P = .0092; Figure 1B). No significant differences in cytogenetics, ELN 2017 risk classification, blast, blood or platelet counts, age, or sex were noted in smokers (supplemental Table 1).
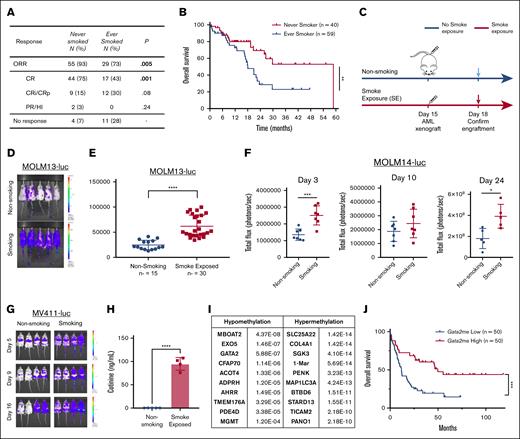
Effects of cigarette smoke exposure in patients and mouse models with FLT3-mutant AML. (A) Table with response rates of patients shown in Figure 1B. Values are n (%). χ2 test analysis was used for categorical variables. (B) Overall survival curve of patients (from panel A) (P = .0092). (C) Schematic representation of SE and AML introduction timeline for mouse modeling of smoking. (D) Representative MOLM13-luc bearing mice 3 days after being NS or SE. (E) Bioluminescent flux shown of MOLM13 bearing mice (n = 15 for NS and n = 30 for SE mice). (F) Leukemic burden of MOLM14-bearing mice 3, 10, and 24 days after injection (n = 7 mice per group for days 3 and 10; n = 5 for NS; and n = 4 for SE on day 24). (G) Representative mice exposed to NS or SE bearing the MV411-luc cells after days 5, 9, or 16 of engraftment. (n = 4 per group). (H) Cotinine levels in urine from NS or SE mice in ng/mL (n = 4). (I) Table with the top hypermethylated and hypomethylated genes from SE in AML cells from mouse spleens as compared with NS mice. (J) The Cancer Genome Atlas (TCGA) data from NEJM 2013 study.6 Patients with quartile of high or low Gata2 methylation survival depicted. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. CR, complete remission; CRi, complete remission with incomplete count recovery; FLT3i, FLT3 inhibitor; HI, hematologic improvement; N, number; ORR, overall response rate; PR, partial response.
Effects of cigarette smoke exposure in patients and mouse models with FLT3-mutant AML. (A) Table with response rates of patients shown in Figure 1B. Values are n (%). χ2 test analysis was used for categorical variables. (B) Overall survival curve of patients (from panel A) (P = .0092). (C) Schematic representation of SE and AML introduction timeline for mouse modeling of smoking. (D) Representative MOLM13-luc bearing mice 3 days after being NS or SE. (E) Bioluminescent flux shown of MOLM13 bearing mice (n = 15 for NS and n = 30 for SE mice). (F) Leukemic burden of MOLM14-bearing mice 3, 10, and 24 days after injection (n = 7 mice per group for days 3 and 10; n = 5 for NS; and n = 4 for SE on day 24). (G) Representative mice exposed to NS or SE bearing the MV411-luc cells after days 5, 9, or 16 of engraftment. (n = 4 per group). (H) Cotinine levels in urine from NS or SE mice in ng/mL (n = 4). (I) Table with the top hypermethylated and hypomethylated genes from SE in AML cells from mouse spleens as compared with NS mice. (J) The Cancer Genome Atlas (TCGA) data from NEJM 2013 study.6 Patients with quartile of high or low Gata2 methylation survival depicted. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. CR, complete remission; CRi, complete remission with incomplete count recovery; FLT3i, FLT3 inhibitor; HI, hematologic improvement; N, number; ORR, overall response rate; PR, partial response.
To characterize molecular mediators of this poor treatment outcome, we used leukemia-bearing NS or smoke-exposed (SE) mice for 2 weeks before orthotopic implantation of luciferase-tagged human FLT3-ITD+ AML lines (Figure 1C) and continued exposing them to smoking throughout the experiment. This experimental design was chosen to mimic leukemia occurring in nonsmokers vs smokers, without smoking cessation (SC) upon diagnosis. Leukemic burden through noninvasive imaging was followed, and a significantly higher leukemic burden 3 days after engraftment was seen in SE mice (n = 30; Figure 1D-E) using the MOLM13 line (P < .0001). The same experiment in MOLM14-luc (n = 7 mice per group) or MV4-11-luc-bearing SE mice (n = 4 mice per group) (Figure 1F-G) showed enhanced leukemic burden on days 3 and 24 (Figure 1F; P < .05). Because leukemic cells were engrafted 2 weeks after smoke exposure, these results suggest that the microenvironment contributes to the accelerated leukemia growth in vivo. To ensure that the 2-week SE schedule used in these experiments was relevant to human exposures, we measured cotinine, the major metabolite of nicotine, in urine and found a significant increase (Figure 2H; P < .0001) in SE mice. In addition to cotinine levels, hypomethylation of the Ahrr promoter is an established biomarker of cigarette smoking.9 We conducted DNA methylation analyses in spleens from SE and NS mice. Among the top genes with significantly altered methylation in human cells were hypomethylated Ahrr and Gata2 (supplemental Table 2; Figure 1I). In a cohort of patients with AML from The Cancer Genome Atlas, low Gata2 methylation was also tracked with significantly worse survival (P = .0 001 383; Figure 1J). Median survival of patients with low Gata2 methylation was significantly (P < .001) shorter (11.8 months) than that of patients with high Gata2 methylation (49.8 months). Interestingly, the methylation status of the other genes shown in Figure 1I did not correlate with survival.
Reduced treatment efficacy, SC effects, and molecular insights into smoking induced proliferative signaling in vivo. (A) Leukemic burden as measured via IVIS on days 3, 7, 15, and 17 after engraftment in NS mice bearing MOLM13-luc cells treated with vehicle (dimethyl sulfoxide [DMSO]) or daunorubicin, 2 mg/kg delivered 3 times weekly IV using 8 mice per group. Days 14 and 17 show a significant difference in leukemia burden in daunorubicin-treated mice compared with that in diluent-treated mice (P < .05). (B) Leukemia burden as measured by IVIS on days 3, 7, 15, and 17 after engraftment in SE mice bearing MOLM13-luc cells treated with vehicle (DMSO) or daunorubicin 2 mg/kg delivered 3 times weekly IV using 8 mice per group. No significant difference (n.s.) was seen in daunorubicin-treated mice at any of the time points. (C) Representative mice from day 14 of experiments depicted in panels A-B. (D) SC upon engraftment significantly slows leukemia progression in MV411-luc bearing mouse models (4 mice per group) and MOLM13-luc bearing mouse models (10 mice per group) compared with mice that continue smoking. P < .01 or .05, respectively. (E) HO-1 protein expression in MOLM13 bearing spleen lysates from NS, SC, and CSE mice. Ratio to tubulin is shown between westerns. (F) Densitometry of western blots from spleens of MOLM13 bearing NS, CSE, or SC mice for HO-1, BCL-2, and AHRR. (G) Immunohistochemistry for HO-1 in spleens and livers from MOLM13-luc bearing NS and SE mice (n = 6 mice per group with 5 slides per mouse depicted graphically and representative sections). (H) Mass cytometry of HO-1 of spleens from MOLM13-bearing mice. (I) Percent frequency of hematopoietic populations of whole bone marrow of NS or SE MOLM14-bearing mice. (J) Tsne plots of CD45+ or CD45− cells from spleens of NS or SE MOLM13-bearing mice. Two mouse samples were combined for each group in the analysis. (K) Heat maps of percentages of CD45− (left) or CD45+ (right) cells present in clusters depicted in panel J. (L) Tsne plots with expression of p21, p-FLT3, MCL-1, and RUNX1 in CD45+ samples from NS or SE mice, as depicted in panel J. (M) Overall median expression of RUNX1, MCL1, DNMT3B, P21, and P-FLT3 in CD45+ cells NS or SE samples. IVIS, in vivo imaging system; MCL1, myeloid cell leukemia 1.
Reduced treatment efficacy, SC effects, and molecular insights into smoking induced proliferative signaling in vivo. (A) Leukemic burden as measured via IVIS on days 3, 7, 15, and 17 after engraftment in NS mice bearing MOLM13-luc cells treated with vehicle (dimethyl sulfoxide [DMSO]) or daunorubicin, 2 mg/kg delivered 3 times weekly IV using 8 mice per group. Days 14 and 17 show a significant difference in leukemia burden in daunorubicin-treated mice compared with that in diluent-treated mice (P < .05). (B) Leukemia burden as measured by IVIS on days 3, 7, 15, and 17 after engraftment in SE mice bearing MOLM13-luc cells treated with vehicle (DMSO) or daunorubicin 2 mg/kg delivered 3 times weekly IV using 8 mice per group. No significant difference (n.s.) was seen in daunorubicin-treated mice at any of the time points. (C) Representative mice from day 14 of experiments depicted in panels A-B. (D) SC upon engraftment significantly slows leukemia progression in MV411-luc bearing mouse models (4 mice per group) and MOLM13-luc bearing mouse models (10 mice per group) compared with mice that continue smoking. P < .01 or .05, respectively. (E) HO-1 protein expression in MOLM13 bearing spleen lysates from NS, SC, and CSE mice. Ratio to tubulin is shown between westerns. (F) Densitometry of western blots from spleens of MOLM13 bearing NS, CSE, or SC mice for HO-1, BCL-2, and AHRR. (G) Immunohistochemistry for HO-1 in spleens and livers from MOLM13-luc bearing NS and SE mice (n = 6 mice per group with 5 slides per mouse depicted graphically and representative sections). (H) Mass cytometry of HO-1 of spleens from MOLM13-bearing mice. (I) Percent frequency of hematopoietic populations of whole bone marrow of NS or SE MOLM14-bearing mice. (J) Tsne plots of CD45+ or CD45− cells from spleens of NS or SE MOLM13-bearing mice. Two mouse samples were combined for each group in the analysis. (K) Heat maps of percentages of CD45− (left) or CD45+ (right) cells present in clusters depicted in panel J. (L) Tsne plots with expression of p21, p-FLT3, MCL-1, and RUNX1 in CD45+ samples from NS or SE mice, as depicted in panel J. (M) Overall median expression of RUNX1, MCL1, DNMT3B, P21, and P-FLT3 in CD45+ cells NS or SE samples. IVIS, in vivo imaging system; MCL1, myeloid cell leukemia 1.
Because treatment outcomes are worse in patients with AML with a smoking history,2 chemotherapy was tested in mouse models. In NS mice, daunorubicin reduced leukemia burden over time compared with that in untreated mice (P < .05; n = 8 mice per group; Figure 2A,C), whereas in smoking mice, treatment did not significantly reduce the leukemic burden (Figure 2B-C). To determine whether SC influenced AML progression, smoke exposure was halted after engraftment and showed reduced tumor burden compared with mice that continued to be SE in 2 mouse models MV411 (n = 4 mice per group) and MOLM13 (n = 10 mice per group; Figure 2D), suggesting that halting smoking upon AML diagnosis in patients could offer clinical benefit. To gain insight into the molecular signals emanating from smoking, immunoblotting was carried out in spleen samples from NS, continued SE (CSE), and SC mice. Heme-oxygenase-1 (HO-1) is an antioxidant elevated in FLT3-ITD+ AML and associated with therapy resistance.10,11 HO-1 expression in spleens from NS vs CSE mice was elevated (Figure 2E-F), and this increase was lessened in SC mice. A similar pattern was seen for BCL-2 expression. AHRR protein expression in CSE mice was decreased, consistent with gene hypomethylation (Figure 2F). Additional proapoptotic and antiapoptotic proteins were probed in lysates from NS, CSE, and SC spleens, and MCL-1 and phosphorylated extracellular signal-regulated kinase were increased by CSE; however, this increase was blunted in cessation models (supplemental Figure 2). HO-1 expression was further examined via immunohistochemistry in spleen and liver sections and via mass cytometry in splenocytes, and it was elevated in the SE mice (Figure 2G-H).
Potential explanations for enhanced leukemia growth in vivo include methylation events in leukemic cells or DNA damage associated mutations acting as drivers of proliferation and will be explored in future studies. However, because mice were exposed to cigarette smoke for 2 weeks before the engraftment of AML cells, we hypothesized that the microenvironment could also contribute to the enhanced leukemia growth (Figure 1E-F). In order to address microenvironmental vs direct leukemia cell effects of smoking, hematopoietic stem cell populations from the bone marrow of SE MOLM14-bearing mice were characterized via flow cytometry, as described in the supplemental Methods; however, no significant changes in progenitor populations were seen in SE mice compared with that in NS mice (n = 6; Figure 2I). These data indicate that immune cells retained in NOD-SCID mice or other differentiated cells in the microenvironment rather than progenitors may contribute to the proliferative effect seen with SE mice. Additional explanations for the enhanced proliferation include a direct stimulatory effect on human leukemia cells. To further address this question, using mass cytometry analyses of SE or NS mice, we separated human CD45+ AML cells from CD45− cells in spleen samples (Figure 2J). NS and SE mice had differences in the percent of cells found within clusters based on the following expression of proteins: CD34, p-ERK1/2, RUNX1, MCL-1, DNMT3B, HIF-1α, p21, and p-FLT3 (Figure 2J-K); however, differences in cluster populations were distinct in human CD45+ AML cells vs CD45− mouse cells. This indicates that there are selective protein changes seen in leukemia cells relative to the microenvironment upon in vivo smoke exposure. In CD45+ human AML cells from the spleens of NS and SE mice, SE samples expressed higher levels of p21, p-FLT3, MCL-1, and RUNX1 (Figure 2L). The overall expressions of LSD1, interleukin-10, p-ERK1/2, RUNX1, MCL-1, DNMT3B, p21, and p-FLT3 were elevated in AML cells from spleens of SE mice (Figure 2M).
Past work reports smokers with AML have worse survival outcomes than nonsmokers2; however, to the best of our knowledge, this is the first study to model cigarette smoke exposure in FLT3-mutant AML–bearing mice to examine potential molecular mediators of leukemia progression and chemotherapy resistance. SE accelerated disease progression in 3 FLT3-ITD AML mouse models. SC upon leukemia engraftment slowed acceleration, providing the first evidence that smoking and cessation deliver “go” and “no-go” signals to FLT3-ITD AML cells. Because many patients with cancer continue smoking after their diagnosis,12,13 these data from xenograft models provide evidence for cessation recommendations, but it will require further validation in primary AML samples. Additionally, mass cytometry revealed that SE increased protein expression of MCL-1, DNMT3B, and RUNX1. MCL-1 inhibitors are currently being investigated for AML treatment, especially in association with resistance to venetoclax,14 and RUNX1 mutations occur in 10% of patients with AML and are associated with inferior prognosis.15 Cumulatively, our data provide novel insights into previously undescribed molecular regulators of aggressive disease seen in patients with AML with histories of smoking.
Acknowledgments: This research was supported by funds from the Texas Tobacco Settlement–Molecular Mechanisms of Tobacco Carcinogenesis (J.C.), MD Anderson Center for Cancer Epigenetics Scholar award and the Schissler Foundation Fellowship (M.F.), Cancer Prevention Research Institute of Texas grant RP170002 (Y.L. and M.R.E.), National Institutes of Health grant R35 HL155672 (K.Y.K.), unrelated to this work, and the HHMI Gilliam Fellowship (D.E.M.-M.). Core facilities associated with MD Anderson's Cancer Center support grant CA016672 were used for this work, including the Flow Cytometry & Cellular Imaging Core Facility, which is supported, in part, by the National Cancer Institute’s Research Specialist Grant R50 CA243707 through the use of Division of Veterinary Medicine and Surgery, and a Shared Instrumentation Award from the Cancer Prevention and Research Institute of Texas (RP121010).
Contribution: M.F., H.M., D.E.M.-M., and F.W. performed research; M.F., J.C., M. Alfayez, Y.L., and M.R.E. analyzed data; K.Y.K., E.K., S.J.M., N.D., M. Andreeff, M.K., and C.D. contributed vital reagents or analytical tools; and M.F. and J.C. designed research and wrote the paper.
Conflict-of-interest disclosure: M.K. has received research funding and/or has served as a consultant for AbbVie, Allogene, Ablynx, Agios, Cellectis, Kisoji, Forty Seven, Daiichi, Precision BioSciences, Gilead, Genentech, Ascentage, Calithera, Eli Lilly, Amgen, Janssen, Sanofi, Hoffman-La Roche, AstraZeneca, MEI Pharma, Menarini-Stemline Therapeutics, Rafael Pharmaceutical, Stemline Therapeutics, and Reata Pharmaceutical. C.D. has received funding from AbbVie, Servier, Astellas, Bristol Myers Squibb (BMS), Celgene, Cleave, Foghorn, Genentech, Gilead, Kura, Novartis, Notable Labs, and Takeda, which are unrelated to the submitted work. N.D. has received research funding, distinct from this work, from Daiichi-Sankyo, BMS, Pfizer, Gilead, Sevier, Genentech, Astellas, Daiichi-Sankyo, AbbVie, Hanmi, Trovagene, FATE Therapeutics, Amgen, Novimmune, GlycoMimetics, Trillium, and ImmunoGen, and has served in a consulting or advisory role for Daiichi-Sankyo, BMS, Arog, Pfizer, Novartis, Jazz, Celgene, AbbVie, Astellas, Genentech, ImmunoGen, Servier, Syndax, Trillium, Gilead, Amgen, Shattuck Labs, and Agios. S.J.M. reports funding from Arrowhead Pharma and Boehringer Ingelheim, outside the scope of the submitted work. The remaining authors declare no competing financial interests.
Correspondence: Joya Chandra, The University of Texas MD Anderson Cancer Center, Box 853, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: jchandra@mdanderson.org.
References
Author notes
Data are available on request from the corresponding author, Joya Chandra (jchandra@mdanderson.org).
The full-text version of this article contains a data supplement.

![Reduced treatment efficacy, SC effects, and molecular insights into smoking induced proliferative signaling in vivo. (A) Leukemic burden as measured via IVIS on days 3, 7, 15, and 17 after engraftment in NS mice bearing MOLM13-luc cells treated with vehicle (dimethyl sulfoxide [DMSO]) or daunorubicin, 2 mg/kg delivered 3 times weekly IV using 8 mice per group. Days 14 and 17 show a significant difference in leukemia burden in daunorubicin-treated mice compared with that in diluent-treated mice (P < .05). (B) Leukemia burden as measured by IVIS on days 3, 7, 15, and 17 after engraftment in SE mice bearing MOLM13-luc cells treated with vehicle (DMSO) or daunorubicin 2 mg/kg delivered 3 times weekly IV using 8 mice per group. No significant difference (n.s.) was seen in daunorubicin-treated mice at any of the time points. (C) Representative mice from day 14 of experiments depicted in panels A-B. (D) SC upon engraftment significantly slows leukemia progression in MV411-luc bearing mouse models (4 mice per group) and MOLM13-luc bearing mouse models (10 mice per group) compared with mice that continue smoking. P < .01 or .05, respectively. (E) HO-1 protein expression in MOLM13 bearing spleen lysates from NS, SC, and CSE mice. Ratio to tubulin is shown between westerns. (F) Densitometry of western blots from spleens of MOLM13 bearing NS, CSE, or SC mice for HO-1, BCL-2, and AHRR. (G) Immunohistochemistry for HO-1 in spleens and livers from MOLM13-luc bearing NS and SE mice (n = 6 mice per group with 5 slides per mouse depicted graphically and representative sections). (H) Mass cytometry of HO-1 of spleens from MOLM13-bearing mice. (I) Percent frequency of hematopoietic populations of whole bone marrow of NS or SE MOLM14-bearing mice. (J) Tsne plots of CD45+ or CD45− cells from spleens of NS or SE MOLM13-bearing mice. Two mouse samples were combined for each group in the analysis. (K) Heat maps of percentages of CD45− (left) or CD45+ (right) cells present in clusters depicted in panel J. (L) Tsne plots with expression of p21, p-FLT3, MCL-1, and RUNX1 in CD45+ samples from NS or SE mice, as depicted in panel J. (M) Overall median expression of RUNX1, MCL1, DNMT3B, P21, and P-FLT3 in CD45+ cells NS or SE samples. IVIS, in vivo imaging system; MCL1, myeloid cell leukemia 1.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/21/10.1182_bloodadvances.2023010111/3/m_blooda_adv-2023-010111-gr2.jpeg?Expires=1765884820&Signature=cXEivKldpkOkGmpqYhgHhLVqq6U8ktC5~NJ4HQD6foQuNE1fWQ9Ts3~Gza3RVcior-FCnn4KTJh5CLmC9TThukOoL~yTIgQ0UylVwU4pPFvwiMygABNL~Ifd-YVmJ1kc~fD0Fj14EplOgkGSAAmn4CkLJkyVRok0qnlz-j7OhE2D0WDbpuKD6Yyh25CsGSFCtM-jdg3x-SQ0qiWRpGfxkkWS1TSU3vhQBCQOjT~KnMgqF9f1oNawKvsOm6Ai14hX7DaQL3oHCUw3bTxUcaI4gtuREXkZMJfaa9oWzKxDtHCt29vwSKSE39CNvf3X6j4fTBopOX3CrIb9WtUwt729mA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)