Abstract
Transcription factor GATA-1 is essential for the development of erythroid cells and megakaryocytes. Each of its 2 zinc fingers is critical for normal function. The C-terminal finger is necessary for DNA binding. The N finger mediates interaction with FOG-1, a cofactor for GATA-1, and also modulates DNA-binding affinity, notably at complex or palindromic GATA sites. Residues of the N finger–mediating interaction with FOG-1 lie on the surface of the N finger facing away from DNA. Strong sequence conservation of residues facing DNA suggests that this other surface may also have an important role. We report here that a syndrome of X-linked thrombocytopenia with thalassemia in humans is caused by a missense mutation (Arg216Gln) in the GATA-1 N finger. To investigate the functional consequences of this substitution, we used site-directed mutagenesis to alter the corresponding residue in GATA-1. Compared with wild-type GATA-1, Arg216Gln GATA-1 shows comparable affinity to single GATA sites but decreased affinity to palindromic sites. Arg216Gln GATA-1 interacts with FOG-1 similarly with wild-type GATA-1. Arg216Gln GATA-1 supports erythroid maturation of GATA-1 erythroid cells, albeit at reduced efficiency compared with wild-type GATA-1. Together, these findings suggest that residues of the N finger of GATA-1–facing DNA contribute to GATA-1 function apart from interaction with the cofactor FOG-1. This is also the first example of β-thalassemia in humans caused by a mutation in an erythroid transcription factor.
Introduction
The zinc finger transcription factor GATA-1 is expressed in erythroid, megakaryocytic, eosinophilic, and mast cells and plays multiple roles in hematopoiesis (reviewed in Orkin1). For instance, GATA-1 is essential for the differentiation of erythroid and megakaryocytic precursors, as demonstrated by gene targeting in the mouse.2-4
Members of the GATA family are notable for the presence of 2 closely spaced, homologous zinc fingers of the C2-C2 class.5 Although highly similar to each other, the 2 fingers have different functions, both with respect to DNA binding and interaction with other proteins.1 In vitro DNA-binding experiments demonstrate that the C-terminal zinc finger (C finger) is required for binding to WGATAR GATA consensus sites.6 Whereas the N-terminal zinc finger (N finger) is not required for DNA binding per se, its presence increases the stability of the interaction between GATA-1 and specific DNA sequences, particularly complex or palindromic GATA sites.6,7 Functional studies show that the zinc fingers of GATA-1 are individually required for normal erythroid development. For example, an erythroid cell line (G1E) that is developmentally arrested because of the lack of GATA-1 differentiates on the introduction of wild-type GATA-1, but not GATA-1 lacking either finger.8Similar findings have been reported recently in transgenic mice.9
Several protein interactions of GATA-1 are mediated through the zinc finger domain. Such interactions are mediated by the C finger or by both zinc fingers. However, the N finger specifically mediates association with a zinc finger protein cofactor, FOG-1 (friend of GATA-1).10,11 Structural predictions guided by nuclear magnetic resonance (NMR) studies suggest that the N finger structure may participate simultaneously in interactions with DNA and with FOG-1.12 Site-specific mutagenesis has defined several amino acid residues of the N finger that are critical to the interaction of GATA-1 with FOG-1. These residues lie on the same face of the predicted structure of the N finger, suggesting that the N finger may be viewed as having a FOG-1 surface and another potentially in apposition to DNA. Indeed, amino acid substitutions of residues on the FOG-1 surface of the finger have been identified in patients with X-linked thrombocytopenia with (or without) anemia.13 14These mutations perturb the association of GATA-1 with its cofactor, FOG-1. Sequence comparisons across species reveal that the entire N finger of GATA-1 is highly conserved with respect to GATA-1 and other GATA factors. Despite the high degree of conservation of residues on the presumptive DNA face of GATA-1, there are no findings to date that address the biologic relevance of the DNA-binding contribution of its N finger.
We show here that a previously reported family with X-linked thrombocytopenia and β-thalassemia (XLTT; Online Mendelian Inheritance in Man [OMIM] accession number 314050)15carries a single amino acid substitution in the N finger of GATA-1 that destabilizes binding of GATA-1 to complex DNA sites. Mutant GATA-1 harboring this substitution (Arg216Gln) interacts normally with FOG-1 and programs erythroid differentiation, though somewhat less efficiently than wild-type protein. The identification of this mutation in GATA-1 establishes a critical in vivo requirement for the DNA-binding contribution of the N finger of GATA-1, as distinguished from its involvement in FOG-1 interaction. In addition, our findings reveal the genetic basis of the rare syndrome of X-linked thrombocytopenia with β-thalassemia.
Patients, materials, and methods
Patients and genetic mapping
Under a protocol approved by the Human Subjects Committee of the University of Washington, blood samples were obtained from 13 members in 4 generations of the XLTT family as previously described.15,16 DNA was extracted from leukocytes or Epstein-Barr virus (EBV)–transformed cell lines and was polymerase chain reaction (PCR)–amplified as previously described.17We sequenced 830 base pairs in GATA-1 coding exons, across splice junctions, and in a portion of the promoter region with the following primer sets: P1F, 5′CTTTCCTACCCTATCCCACT-3′; P1R, 5′-ATGACTTCACTGGGCTTTT-3′; P2F, 5′-CGCACATACACAGGAGTCTA-3′; P2R, 5′-TGACCTGGGCTGGTG-3′; 2.1F, GGAAGGATTTCTGTGTCTGA-3′; 2.1R, 5′-CCACTCAATGGAGTTACCTG-3′; 2.2F, TACTACAGGGACGCTGAGG-3′; 2.2R, 5′-CAGCCGGCATATGGTGAG-3′; 3F, 5′-GTGCGCTGACCCTAGACT-3′; 3R, 5′-GGTGGAGAGGAGAAGAGG-3; 4F, 5-CCCTGACTTTTCCAGTACCT-3′; 4R, 5′-CCTATATAATGGGAAGATGTGG-3′; 5.1F, 5′-AGGCCACTACCTATGCAACG-3′; 5.1R, 5′-ATCCCACTGGCATTTCTC-3′; 5.2F, 5′-CCTGTAGTACCCTAGATTGTCAG-3′; 5.2R, GAGCCCCGTTTCTTTTC-3′; 6F, 5′-AAAGAAGTGGGGTAGAGAGG-3′; 6R, GCTACAAGAGGAGAAGGACA-3′. Genomic DNA was PCR-amplified using Promega Taq polymerase (Promega, Madison, WI) and purified with the QIAquick PCR purification kit (QIAGEN, Valencia, CA). Sequencing was performed on [γ32]P or [γ33]P-labeled DNA using the AmpliCycle kit or on fluorescently labeled DNA using the ABI PRISM DyeTerminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA).
Transfections
Site-directed mutagenesis was performed on the GATA-1 expression vector pXM–GATA-16 with the Quik-Change Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) using the oligonucleotide 5′-CAACGGCTACTCCACTGTGGCAGAGGGACAGGACAGGTCACTA-3′ and its complement to create pXM-Arg216Gln–GATA-1. COS cells were transfected with pXM–GATA-1, pXM-Arg216Gln–GATA-1, or pXM vector alone using Fugene (Roche Molecular Biochemicals, Indianapolis, IN), according to the manufacturer's instructions. Nuclear extracts were prepared 60 hours following transfection, as described.10
Electrophoretic mobility shift analyses
Double-stranded probes containing either a palindromic GATA site present in the chicken GATA-1 promoter (5′-GCGCTATCAGATAAGGCCTTG -3′ and 5′-CAAGGCCTTATCTGATAGCGC-3′) or a single GATA site patterned from this site (5′-GCGCTCAGAGATAAGGCCTTG-3′ and 5′-CAAGGCCTTATCTCTGAGCGC-3′) were used as previously described7,10 to study GATA-1 dissociation from DNA in electrophoretic mobility shift analyses. Briefly, nuclear extracts of COS cells transfected with pXM–GATA-1, pXM-Arg216Gln–GATA-1, or pXM vector alone were incubated with32P-labeled double-stranded probe (palindromic GATA site or single GATA site) for 20 minutes. After 100-fold molar excess, unlabeled competitor probe was added (t = 0 minute), and reaction mixtures were loaded onto polyacrylamide gels and run under established conditions.6 Quantities of bound probe and free probe were measured by Storm PhosphorImager analysis using ImageQuant software (Molecular Dynamics, Piscataway, NJ).
Coimmunoprecipitation assays
COS cells were cotransfected with pXM–GATA-1, pXM-Arg216Gln–GATA, or pXM vector alone and either FLAG–FOG-1 (a gift from S. G. Katz) or FLAG vector alone (pEFrFLAGPGKpuropAV18, a gift from D. Huang). Immunoprecipitation with anti-FLAG antibody and detection of FLAG proteins and GATA-1 by Western blot were performed as described earlier,10 except that the ECL+ Kit (Amersham Pharmacia Biotech, Piscataway, NJ) was used for final visualization in Western blots.
G1E-ER2 clones
G1E cells were cultivated as described.8 Using the vector pGD-G1/ER-puro, we created pGD-Arg216Gln-G1/ER-puro using site-directed mutagenesis as described above. The wild-type or mutant vector was used to retrovirally infect G1E cells as described.10 GATA-1 was detected by the N6 monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) on Western blots as described.8
Northern blot analyses
Results
Syndrome of X-linked thrombocytopenia with thalassemia maps to the GATA-1 locus
We previously determined16 that a syndrome of X-linked thrombocytopenia with thalassemia maps to band p11-12 on the human X chromosome, a region that includes a number of genes involved in hematopoiesis, including GATA-1. Noting defects in the megakaryocytic and erythroid lineages, 2 lineages that require GATA-1 for normal development, we investigated whether the GATA-1 locus was altered in affected males and obligate carrier females.
Sequencing of the GATA-1 exons, splice junctions, and promoter showed that affected males and obligate carrier females had a GATA-1 allele with a transition mutation in codon 216 (CGG>CAG), recoding arginine (Arg) as glutamine (Gln; Figure 1A). The missense mutation segregated with disease in the XLTT family and was not found in 100 control X chromosomes studied. Arg216 is within the N finger of GATA-1 and is strictly conserved in GATA-1 orthologs in various species (Figure 1B). The NMR structure of the GATA-1 N finger (National Center for Biotechnology Information [NCBI] structure 1GNF12) indicates that this residue lies on the face of the N finger that faces DNA—that is, it lies on the surface that does not contact the GATA-1 cofactor FOG-1. Consistent with this location, Arg216 was not altered in an unbiased mutagenesis screen10designed to identify GATA-1 N finger mutants unable to bind to FOG-1. Thus the N finger has a “FOG face” and a “DNA face” whose approximate “border” is represented in Figure 1C. The locations of Arg216, on the DNA face, and Val205, on the FOG face, that are replaced by methionine in a syndrome of X-linked dyserythropoietic anemia and thrombocytopenia14 are also shown.
X-linked thrombocytopenia with thalassemia maps to a mutation (Arg216Gln) in GATA-1.
(A) Pedigree of a family with X-linked thrombocytopenia with thalassemia.15,16 Black squares represent affected males; circles with black discs represent obligate carrier females. Differences in GATA-1 DNA sequence at codon 216 are indicated below individuals: CGG (arginine, wild-type); CAG (glutamine, mutant). Patients who were evaluated for α-globin/β-globin chain imbalance15,16 are designated with + (elevated α/β ratio) or − (within normal limits). (B) Partial sequence of the N finger of GATA-1. Arg216 is conserved across GATA-1 orthologs of various species. Here, amino acid numbering relates to the human, rat, and mouse polypeptides. Val205, mutated to methionine in familial dyserythropoietic anemia with thrombocytopenia,14 is also highlighted here. Polypeptide sequences are shown as single-letter code. (C) The N finger of GATA-1 has 2 faces, a FOG face and a DNA face.10,12 Approximate division of these faces is superimposed on the NMR structure.12 Val205 (green) is highlighted on the FOG face, and Arg216 (blue) is highlighted on the DNA face.
X-linked thrombocytopenia with thalassemia maps to a mutation (Arg216Gln) in GATA-1.
(A) Pedigree of a family with X-linked thrombocytopenia with thalassemia.15,16 Black squares represent affected males; circles with black discs represent obligate carrier females. Differences in GATA-1 DNA sequence at codon 216 are indicated below individuals: CGG (arginine, wild-type); CAG (glutamine, mutant). Patients who were evaluated for α-globin/β-globin chain imbalance15,16 are designated with + (elevated α/β ratio) or − (within normal limits). (B) Partial sequence of the N finger of GATA-1. Arg216 is conserved across GATA-1 orthologs of various species. Here, amino acid numbering relates to the human, rat, and mouse polypeptides. Val205, mutated to methionine in familial dyserythropoietic anemia with thrombocytopenia,14 is also highlighted here. Polypeptide sequences are shown as single-letter code. (C) The N finger of GATA-1 has 2 faces, a FOG face and a DNA face.10,12 Approximate division of these faces is superimposed on the NMR structure.12 Val205 (green) is highlighted on the FOG face, and Arg216 (blue) is highlighted on the DNA face.
Arg216Gln GATA-1, like wild-type GATA-1, binds to FOG-1
To determine whether Arg216Gln mutation affects protein interaction with FOG-1, we performed coimmunoprecipitation experiments with different GATA-1::FOG-1 complexes. We have shown previously that a shortened and FLAG-tagged version of FOG-1, which spans zinc fingers 5 and 6, interacts with wild-type GATA-1 on the overexpression of both proteins in COS cells.10Interaction is markedly reduced, however, with the Val205Gly mutation on the FOG face.10 Based on the structure model, we predicted that Arg216Gln GATA-1 would associate normally with FOG-1. To assess this hypothesis, we tested the interaction of full-length, rather than shortened, versions of these proteins.
Wild-type murine GATA-1 or GATA-1 mutants were coexpressed with FLAG–FOG-1 (full-length murine FOG-1) in COS cells. Immunoprecipitation of nuclear extracts with anti-FLAG antibody demonstrates that Arg216Gln GATA-1, like wild-type GATA-1, forms a complex with FLAG–FOG-1 (Figure 2, lanes 5 and 4, respectively). As expected, interaction is not observed with Δ200 to 248 GATA-1, in which the entire N finger is deleted (lane 6). No GATA-1 was immunoprecipitated in various control experiments (lanes 1, 2, 3, 7).
Arg216Gln GATA-1, like wild-type GATA-1, binds to FOG-1.
A FLAG-tagged FOG-1 expression construct was cotransfected with various GATA-1 expression constructs into COS cells as indicated. Western blots (top panels) confirm expression from FOG and GATA constructs. (bottom panel) After immunoprecipitation (IP) with anti-FLAG antibody (or anti-myc control antibody), Western blot detection of precipitated GATA-1 shows that Arg216Gln GATA-1 (lane 5), like wild-type GATA-1, binds to FOG-1 (lane 4), whereas Δ200 to 248 GATA-1 (N finger–deleted; lane 6) do not. No immunoprecipitation of GATA-1 is observed in control experiments (remaining lanes).
Arg216Gln GATA-1, like wild-type GATA-1, binds to FOG-1.
A FLAG-tagged FOG-1 expression construct was cotransfected with various GATA-1 expression constructs into COS cells as indicated. Western blots (top panels) confirm expression from FOG and GATA constructs. (bottom panel) After immunoprecipitation (IP) with anti-FLAG antibody (or anti-myc control antibody), Western blot detection of precipitated GATA-1 shows that Arg216Gln GATA-1 (lane 5), like wild-type GATA-1, binds to FOG-1 (lane 4), whereas Δ200 to 248 GATA-1 (N finger–deleted; lane 6) do not. No immunoprecipitation of GATA-1 is observed in control experiments (remaining lanes).
DNA binding is altered in Arg216Gln GATA-1
Because Arg216 lies on the predicted DNA-facing side of the N finger, we characterized the DNA-binding properties of Arg216Gln GATA-1. Previous studies6,7,18 show that the C finger is required for GATA-1 to bind DNA, whereas the N finger stabilizes the interaction between GATA-1 and palindromic GATA sites. Accordingly, a mutation in the DNA face of the N finger could be expected to dissociate more easily from a palindromic site than would wild-type GATA-1. In contrast, as suggested from binding studies in which the entire N finger is deleted,7 this mutation would not be expected to show a difference in dissociation from a single GATA site.
Using electrophoretic mobility shift assays, we assessed the dissociation profiles of wild-type and Arg216Gln GATA-1 proteins with single and palindromic GATA-site targets. We used site-directed mutagenesis to introduce the Arg216Gln mutation into pXM–GATA-1, an expression vector containing the complete cDNA of murine GATA-1. Nuclear extracts were prepared from COS cells transfected with expression vectors encoding wild-type or Arg216Gln GATA-1. Using radioactively labeled, double-stranded DNA probes encoding either the palindromic GATA site from the chicken GATA-1 promoter or a single GATA site modeled after this site (Figure 3, top), we assembled GATA-1::DNA complexes in vitro.
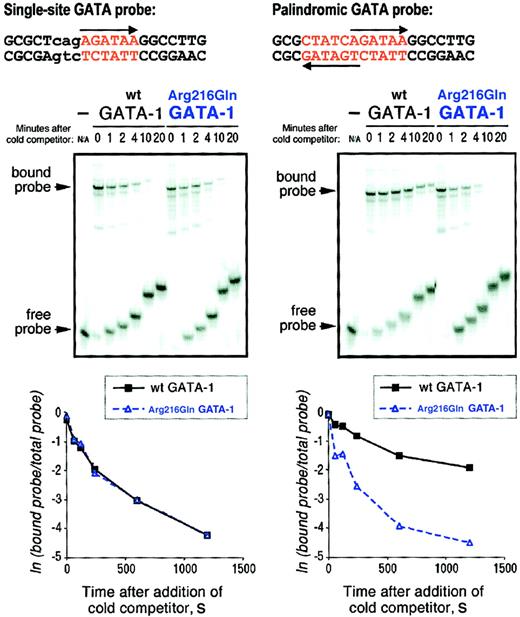
Arg216Gln GATA-1 dissociates from a palindromic GATA site more easily than wild-type GATA-1.
GATA-1::DNA complexes were formed by incubating either wild-type or Arg216Gln GATA-1 with 32P-labeled single GATA site or palindromic GATA site probes (top). One hundred–fold molar excess of corresponding unlabeled probe was then added att = 0 minute, and reaction mixtures were loaded onto gels at the indicated times (middle panels). The first lane of each set shows migration of the free probe alone. (bottom panels) PhosphorImager quantitation of percentage of bound probe over time (here, in seconds).
Arg216Gln GATA-1 dissociates from a palindromic GATA site more easily than wild-type GATA-1.
GATA-1::DNA complexes were formed by incubating either wild-type or Arg216Gln GATA-1 with 32P-labeled single GATA site or palindromic GATA site probes (top). One hundred–fold molar excess of corresponding unlabeled probe was then added att = 0 minute, and reaction mixtures were loaded onto gels at the indicated times (middle panels). The first lane of each set shows migration of the free probe alone. (bottom panels) PhosphorImager quantitation of percentage of bound probe over time (here, in seconds).
Following the addition of the respective unlabeled (cold) competitor probe, we observed the dissociation of GATA-1::DNA complexes over time by following the electrophoretic mobility shift of bound probe to free probe (Figure 3, middle panels). (Note: the “slant” in the migration of either the bound or free probe across time points is due merely to the different times at which the samples are loaded onto the gel, which is running continuously.) PhosphorImager quantitation (Figure 3, bottom panels) demonstrates that wild-type and Arg216Gln GATA-1 dissociate at similar rates from a single GATA site (left graph), whereas only wild-type GATA-1 and not Arg216Gln shows a slower rate of dissociation from a palindromic GATA site (right). Indeed, it seems as though Arg216Gln GATA-1 shows similar binding characteristics on the palindromic site and the single site, as predicted. Experiments with other point mutants in this region (Pro213Gly, Leu214Phe, Arg216Ala, Arg216Glu, Leu230Glu, Leu230Phe) also demonstrate this altered DNA-binding profile without disruption of FOG-1 binding (not shown). Of note, the dissociation rates seen in these experiments are similar to those previously reported.7 10 Extracts from COS cells transfected with pXM vector alone did not bind palindromic or single GATA site probes (not shown).
Arg216Gln GATA-1 allows erythroid maturation to proceed in a GATA-1 null cell line
We next examined the biologic implications of the Arg216Gln mutation in G1E cells, which lack GATA-1 and undergo late erythroid maturation on restoration of GATA-1 function. We have shown previously in these cells that an intact N finger is required for erythroid maturation,8 probably because of recruitment by GATA-1 of FOG-1,10,11 which, like GATA-1, is required for normal erythropoiesis.11
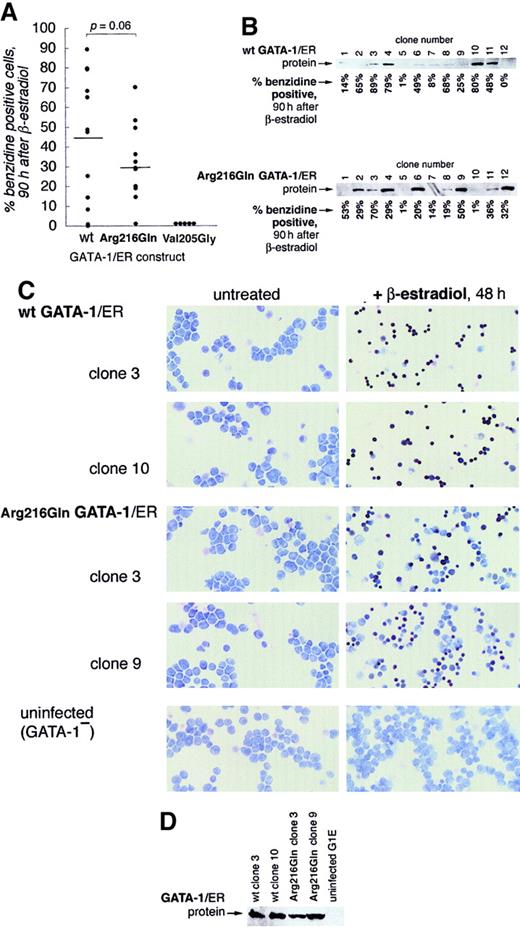
Wild-type GATA-1 or Arg216Gln GATA-1 was stably introduced into G1E cells as a GATA-1/ER fusion (murine GATA-1 cDNA fused in frame to the human estrogen receptor ligand-binding domain).11,19Synchronous GATA-1–dependent maturation can then be induced by the addition of hormone (β-estradiol). Twelve puromycin-resistant clones of each type were selected for further analysis. At 90 hours after induction, wild-type GATA-1/ER G1E clones (hereafter, wt G1E-ER2 clones) were benzidine positive at a slightly higher mean percentage (t test, P = .06; Figure4A) than Arg216Gln GATA-1/ER clones (hereafter, Arg216Gln G1E-ER2 clones), albeit with a range of overlap. Clones from both groups displayed a range of GATA-1/ER protein expression levels (Figure 4B) that approximated the degree of benzidine positivity (not statistically significant). Meanwhile, 5 Val205Gly G1E-ER2 clones were generated in parallel; none of these showed benzidine-positive cells 90 hours after induction, as observed in previous studies.10
Arg216Gln GATA-1 allows erythroid maturation in G1E cells.
G1E cells, which lack GATA-1, undergo erythroid differentiation after retroviral infection with GATA-1.8 Stable introduction of estrogen-receptor fusion versions of GATA-1 (GATA-1/ER) allows this differentiation to be controlled by the addition of β-estradiol, which activates the GATA-1/ER fusion protein.19 29 (A) Wild-type and Arg216Gln versions of GATA-1/ER fusion proteins allow G1E cells to undergo erythroid differentiation, evidenced here by quantitation of benzidine-positive cells 90 hours after induction by β-estradiol of 12 independent clones from each group. The mean percentage of benzidine-positive cells is slightly lower in the Arg216Gln clones (t test, P = .06). No benzidine-positive cells are observed in the Val205Gly clones. (B) GATA-1/ER protein is expressed at varying levels in wild-type (top) and Arg216 (bottom) clones. Corresponding percentages of benzidine-positive cells 90 hours after induction by β-estradiol (from panel A) in individual clones are indicated below the blots. (C) Benzidine staining of clones of G1E cells following stable introduction of wild-type GATA-1/ER (the ensuing clones are named G1E-ER2) or Arg216Gln GATA-1/ER, or G1E cells alone. Benzidine-positive cells (black) are visible after β-estradiol treatment (right panels) when wt GATA-1/ER or Arg216Gln GATA-1/ER is present. Origninal magnification, × 200. (D) GATA-1/ER protein levels of clones pictured in panel C.
Arg216Gln GATA-1 allows erythroid maturation in G1E cells.
G1E cells, which lack GATA-1, undergo erythroid differentiation after retroviral infection with GATA-1.8 Stable introduction of estrogen-receptor fusion versions of GATA-1 (GATA-1/ER) allows this differentiation to be controlled by the addition of β-estradiol, which activates the GATA-1/ER fusion protein.19 29 (A) Wild-type and Arg216Gln versions of GATA-1/ER fusion proteins allow G1E cells to undergo erythroid differentiation, evidenced here by quantitation of benzidine-positive cells 90 hours after induction by β-estradiol of 12 independent clones from each group. The mean percentage of benzidine-positive cells is slightly lower in the Arg216Gln clones (t test, P = .06). No benzidine-positive cells are observed in the Val205Gly clones. (B) GATA-1/ER protein is expressed at varying levels in wild-type (top) and Arg216 (bottom) clones. Corresponding percentages of benzidine-positive cells 90 hours after induction by β-estradiol (from panel A) in individual clones are indicated below the blots. (C) Benzidine staining of clones of G1E cells following stable introduction of wild-type GATA-1/ER (the ensuing clones are named G1E-ER2) or Arg216Gln GATA-1/ER, or G1E cells alone. Benzidine-positive cells (black) are visible after β-estradiol treatment (right panels) when wt GATA-1/ER or Arg216Gln GATA-1/ER is present. Origninal magnification, × 200. (D) GATA-1/ER protein levels of clones pictured in panel C.
To examine the qualitative differences between wt G1E-ER2 and Arg216Gln G1E-ER2, we selected for further analysis 2 clones of each group that showed higher levels of differentiation. Arg216Gln clones exhibited morphologic changes similar to those observed in wild-type clones and showed a comparable degree of benzidine staining in individual benzidine-positive cells (Figure 4C). No benzidine-positive cells were observed in parental G1E cells. The 4 clones studied here expressed GATA-1/ER protein at similar magnitudes (Figure 4D).
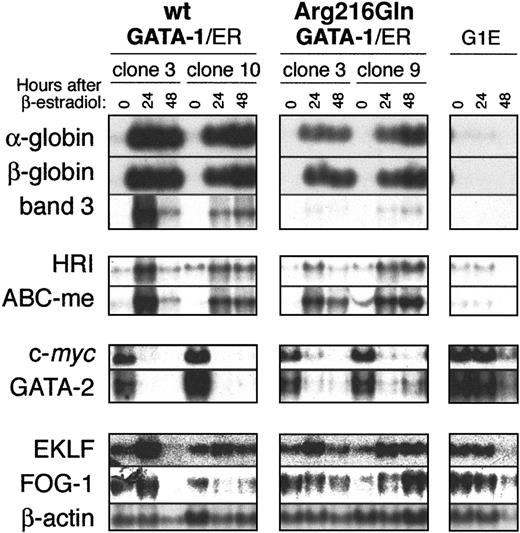
Northern blot analysis of a panel of transcription factors and erythroid markers (as performed similarly in Crispino et al10) shows that Arg216Gln G1E-ER2 clones display changes in gene expression similar to wt G1E-ER2 clones following activation of GATA-1 (Figure 5). Transcripts of adult globins, the erythroid transporter band 3, the heme-regulated eukaryotic initiation factor-2α kinase (HRI), and the mitochondrial transporter ABC-me are induced by Arg216Gln GATA-1/ER but to a somewhat lesser extent than wt GATA-1/ER (upper panels). This blunted level of induction is especially apparent in band 3 transcript levels. The genesc-myc and GATA-2, which promote proliferation in erythroid precursors,10,20 21 are down-regulated in Arg216Gln G1E-ER2 clones, albeit to a lesser degree as well (middle panels). Transcript levels of the transcription factors EKLF and FOG-1 are modestly induced in wt G1E-ER2 or Arg216Gln G1E-ER2 clones relative to the β-actin loading control. As expected, transcripts of none of these genes were altered relative to β-actin after induction of the parental G1E cells, which lack any GATA-1/ER protein (bottom panels).
Changes in gene expression following activation of wild-type or Arg216Gln G1E-ER2 by β-estradiol.
Two clones of wild-type or Arg216Gln G1E-ER2 cells were induced with β-estradiol, and transcripts of erythroid genes were assayed by Northern blotting. Like wild-type G1E-ER2 clones, Arg216Gln G1E-ER2 clones showed changes in erythroid gene expression. However, in some instances (eg, the erythroid anion transporter band 3; see “Results” for details), the level of induction was reduced relative to wild-type controls. Parental G1E cells, which lack GATA-1, do not display changes in expression of these markers following β-estradiol induction.
Changes in gene expression following activation of wild-type or Arg216Gln G1E-ER2 by β-estradiol.
Two clones of wild-type or Arg216Gln G1E-ER2 cells were induced with β-estradiol, and transcripts of erythroid genes were assayed by Northern blotting. Like wild-type G1E-ER2 clones, Arg216Gln G1E-ER2 clones showed changes in erythroid gene expression. However, in some instances (eg, the erythroid anion transporter band 3; see “Results” for details), the level of induction was reduced relative to wild-type controls. Parental G1E cells, which lack GATA-1, do not display changes in expression of these markers following β-estradiol induction.
Discussion
Here we demonstrate that a human syndrome of X-linked thrombocytopenia with thalassemia maps to a missense mutation (Arg216Gln) in the transcription factor GATA-1, a factor required for the normal development of erythroid cells and platelets. Affected males, who have a single mutant allele, have a reduction in platelet levels with disproportionately increased bleeding times and an increase in the α/β globin ratio in reticulocytes.
Our work is informative with respect to the function of GATA-1 in 2 respects. First, it reveals a physiologic role for the DNA face of the N finger of the protein. Second, it reveals a novel mechanism for β-thalassemia in humans: the mutation of an erythroid transcription factor.
GATA-1 N finger has a role in binding DNA
We have shown that the Arg216Gln mutation perturbs the DNA face of the N-terminal zinc finger, thereby reducing the affinity of GATA-1 for palindromic GATA sites. The opposite FOG-binding face of the N finger is unaffected as Arg216Gln GATA-1 binds normally to FOG-1. Several human mutations identified in X-linked thrombocytopenic syndromes, some with concomitant anemia, map to the FOG face and show no alteration in DNA-binding properties. Therefore, the mutation seen in this family provides the first evidence in humans of an anticipated, but undocumented, role for the N finger in DNA-binding and function in vivo.
Determining the function of the DNA face of the N finger has broader implications for understanding the mechanism of GATA-1 action. The strong conservation of the residues on this face across organisms suggests that this region is likely to be important. To date, a number of “complex” sites that depend on N-finger binding have been identified in vertebrate globin genes, including the chicken αD-globin promoter, the human Aγ-globin promoter, and the human ε-globin gene silencer.6,7,22,23 Whether palindromic or double sites are important in GATA-1–dependent transcription or whether the N finger modulates transcription through other motifs in isolated circumstances24 awaits further study. A recent report25 suggested a possible transcriptional role for the N finger at GATC sites within a nonhematopoietic environment. Given the importance of tissue context for GATA-1 function (eg, Weiss et al8), the significance of such findings remains unclear. Our analysis of Arg216Gln in a GATA-1− erythroid cell line (G1E) shows that Arg216Gln GATA-1 directs erythroid differentiation but at a slightly weaker extent relative to that of wild-type GATA-1. Thus, we observe how a specific mutation on the DNA face of the N finger demonstrates an effect on erythropoiesis. Unlike mutation of a residue on the FOG face (Val205Met), which leads to dyserythropoietic anemia with thrombocytopenia in humans and poor induction of G1E cell differentiation,14 the Arg216Gln mutation leads to a mild form of β-thalassemia in humans and considerable ability to promote G1E differentiation.
The bleeding disorders that brought patients in this family to clinical attention underscore the importance of the N finger in normal platelet development. We have found previously that GATA-1 is critical for the creation of functional platelets.3 4 Unfortunately, evaluation of the consequences of the Arg216Gln mutation on megakaryocyte development and platelet biogenesis is precluded by the lack of a suitable experimental system for in vitro study. Furthermore, loss of these patients to follow-up prevents more detailed analysis of platelet function with the Arg216Gln mutation.
Of note, the mild thalassemia seen in this family is not reproduced in the G1E differentiation model. Ribonuclease protection assays examining the ratio between α- and β-globin transcript levels in Arg216Gln GATA-1/ER clones reveal no significant difference from ratios observed in wt G1E-ER2 clones (data not shown). The failure to observe globin chain imbalance is most likely attributed to the limitations of the model system. Supranormal levels of overexpressed Arg216Gln GATA-1/ER may overcome differences in lowered binding affinity. Alternatively, the thalassemic effect of mutant GATA-1 may be masked in the milieu of late-stage erythropoiesis represented by the G1E cell line. In these cells, GATA-2 levels are significantly elevated relative to normal proerythroblasts and may have already played a compensatory role in elaborating low levels of α- and β-globin in the absence of GATA-1.8 26 In addition, the organization of the globin gene loci differs in mice and humans. Nonetheless, we infer also from initial studies of an anemic and thrombocytopenic “knock-in” mouse harboring a different point mutation (Pro213Gly) in the N finger DNA face (data not shown) that this face is indeed important in erythroid and megakaryocytic development.
With the identification of naturally occurring mutations on both sides of the N finger that result in distinct biochemical alterations and distinguishable phenotypes, it is now possible to distinguish the FOG-binding and DNA-binding functions of the N finger. This is important in assigning targets of GATA-1 that depend on interaction with FOG-1 and those independent of FOG-1. As we have previously shown, slightly different phenotypes are observed in GATA-12versus FOG-111 knockout mice. Like GATA-1−embryos, FOG-1−/− embryos also die of severe anemia at days 11.5 to 12.5 of gestation. FOG-1−/− erythroid precursors are arrested at the proerythroblast stage as are GATA-1− erythroid precursors but do not undergo apoptosis. However, FOG-1−/− embryos show a more extreme effect in megakaryocyte development. Although GATA-1− embryos produce immature megakaryocytes that have defects in granule production and disturbances in nuclear structure, FOG-1−/− embryos are unable to produce megakaryocytes at all. Together these observations suggest that detailed analyses of gene expression with specific missense GATA-1 proteins may lead to the identification of functionally distinct classes of gene expression that result from varying combinations of transcription factors acting with GATA-1.
Phenotype resulting from Arg216Gln GATA-1 reveals a new mechanism of β-thalassemia in humans: mutation of an erythroid transcription factor
The thalassemia syndromes have been ascribed to numerous and diverse mutations, acting principally in cis to the affected globin chain.27 Our study of the Arg216Gln mutation in GATA-1 represents the first example of mutation in an erythroid transcription factor as a trans-mechanism for β-thalassemia in humans. While this work was in preparation, an example of β-thalassemia caused by the mutation of a general transcription factor, TFIIH, was reported.28
We thank John Crispino for the Val205Gly/ER expression construct.
Prepublished online as Blood First Edition Paper, May 17, 2002; DOI 10.1182/blood-2002-02-0387.
Supported in part by grants from the National Institutes of Health and by a grant from NASA to the Washington Space Grant program. S.H.O. is an Investigator of the Howard Hughes Medical Institute.
C.Y. and K.K.N. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Stuart H. Orkin, Children's Hospital, Division of Hematology/Oncology, 300 Longwood Ave, Boston, MA 02115; e-mail: stuart_orkin@dfci.harvard.edu.