Abstract
The MALT1 gene was identified through its involvement in t(11;18)(q21;q21), seen in 30% of cases of mucosa-associated lymphoid tissue (MALT) lymphoma. Here, we show that deregulated MALT1 expression may occur in B-cell non-Hodgkin lymphoma (B-NHL) of various histologic subtypes either through translocation to the immunoglobulin heavy chain (IGH) locus or by genomic amplification. First, 2 cases, one case of MALT lymphoma and another of aggressive marginal zone lymphoma (MZL) with t(14;18)(q32;q21), cytogenetically identical to the translocation involving BCL2, were shown by fluorescence in situ hybridization (FISH) to involve MALT1, which lies about 5 Mb centromeric of BCL2. Molecular cloning of both by long-distance inverse polymerase chain reaction showed breakpoints lying 1 to 2 kilobase (kb) centromeric of the first 5′ MALT1 exon; both cases showed MALT1 overexpression at either RNA or protein levels. Second, we examined the structure and gene expression profile of genomic amplifications involving 18q21 in a panel of 40 B-NHL cell lines using comparative genomic hybridization to microarrays (array CGH) and gene expression profiling techniques. Using array CGH, 2 peaks of genomic amplification were observed, one centered around BCL2 and the other around MALT1.Ofthe 3 cell lines with MALT1 amplification, 2 showed MALT1 overexpression as assessed by gene profiling, quantitative reverse transcription–polymerase chain reaction (QRT-PCR), and Western blotting. To determine if comparable events occurred in primary MALT and splenic MZL tumors, 40 cases were analyzed by FISH or QRT-PCR; genomic amplification and MALT1 overexpression were seen in 2 cases. Together, these data implicate MALT1 as a dominant oncogene that may play a role in the pathogenesis of B-NHL.
Introduction
The chromosome band 18q21 is a frequent site of rearrangement in B-cell non-Hodgkin lymphoma (B-NHL). In the t(14;18)(q32; q21), the BCL2 gene located in 18q21.3 becomes juxtaposed to the immunoglobulin heavy chain (IGH) locus in about 80% of follicle center lymphomas (FCLs) and in 20% to 30% of diffuse large B-cell lymphomas (DLBCLs).1 Most BCL2 breakpoints are clustered within the 3′ end of the BCL2 gene or more rarely in the 5′ region.2 Besides translocation, chromosomal amplification of band 18q21 is observed in up to 20% of B-NHL cases and may be the cause of BCL2 overexpression seen in both FCLs and DLBCLs that appear to lack BCL2 translocation.3,4 In chronic lymphocytic leukemia, deregulated BCL2 expression may also occur as a consequence of promoter hypomethylation.5
Some evidence, however, suggests that BCL2 is not the only target gene for some abnormalities of chromosome 18q21. First, an alternative breakpoint telomeric (5′) of BCL2 involving a gene termed FVT1 has been reported, although such translocations appear to be infrequent.6 Chromosomal amplifications involving 18q21 have been reported in primary central nervous system B-NHL with 18q21 amplicon, but these do not seem to involve the BCL2 gene.7,8 The role of BCL2 in 18q21 amplifications seen in mantle cell lymphomas is also controversial; in one study, BCL2 gene was the target of the 18q21 amplicon, whereas Bentz et al9 and Bea and coworkers10 did not confirm such involvement in similar genomic rearrangements. These studies suggest that other genes may be deregulated as a consequence of alterations within 18q21.
Another frequent translocation of band 18q21 is the t(11;18)(q21; q21), which characterizes about one third of extranodal marginal zone lymphomas (MZLs) of the mucosa-associated lymphoid tissue (MALT) type.11, 12, 13 This translocation fuses MALT1, located about 5 Mb centromeric of BCL2 in chromosome 18q21, with API2 on 11q21, resulting in a chimeric API2-MALT1 fusion.14, 15, 16 MALT1 is a paracaspase that possesses a death domain, 2 adjacent immunoglobulin-like domains, and a caspaselike domain.17 The functions of MALT1 remain unknown. MALT1 overexpression in fibroblasts does not induce either nuclear factorκB (NF-κB) activation or apoptosis.17,18 However, MALT1 interacts directly with BCL10, another protein implicated in MALT lymphoma pathogenesis through its involvement in t(1;14)(p22;q32) and synergistically induces NF-κB.17,18 These data along with those from the BCL10 knockout mice suggest that both BCL10 and MALT1 link the antigen receptor signaling to the NF-κB pathway.19 Unlike wild-type MALT1, the API2-MALT1 fusion protein activates NF-κB presumably through homodimerization mediated via the API2 component of the fusion protein.17,18
In this report, we have determined the consequences of abnormalities involving 18q21 but not involving BCL2. First, using fluorescence in situ hybridization (FISH), we delineated and cloned the translocation breakpoints of 2 patients with a t(14;18)(q32;q21) with an 18q21 breakpoint centromeric of BCL2. In addition, the structural analysis of an 18q21 amplicon was determined in a panel of B-NHL cell lines using parallel comparative gene hybridization to microarrays (array CGH) and cDNA microarray analyses. Our data demonstrate the deregulated expression of MALT1 in both instances and therefore implicate MALT1 as a dominant oncogene in the pathogenesis of B-NHL.
Patients, materials, and methods
Patient material
Two patients with t(14;18)(q32;q21) not involving the BCL2 locus were studied. Case 1 was observed in a previously reported series of MALT lymphomas.12 This low-grade MALT lymphoma affecting the orbit showed a t(14;18)(q32;q21) as well as a trisomy of chromosome 3 in cytogenetic analysis. Case 2 was observed in a series of patients with splenic MZLs.20 This tumor also displayed t(14;18)(q32;q21) and trisomy of chromosome 3 but exhibited a complex karyotype. Abbreviated clinical details are shown in Table 1.
To screen for MALT1 gene alterations in MZL, samples from a series of 40 patients diagnosed with either splenic MZL (n = 35) or MALT lymphoma (n = 5), diagnosed using accepted criteria,21 were studied using quantitative real-time polymerase chain reaction (QRT-PCR) and FISH.
B-NHL cell lines
To characterize the genomic changes in B-NHL, 40 derived cell lines were studied by CGH, as previously described.22 Eleven displayed amplification (ranging from 4- to 6-fold) of band 18q21. These cell lines comprised Granta 519, HBL-2, and Z-138 (derived from mantle cell lymphoma); Karpas 1718 and SSK-41 (MZL); PR-1, SC-1, and ROS-50 (lymphoma with t(14;18)(q32;q21) and BCL2-IGH gene rearrangement); RIVA and MD-901 (DLBCL); and KHM-10B (Burkitt lymphoma with t(8;22)(q24; q11)). In addition, 4 cell lines with t(14;18)(q32;q21) and BCL2-IGH gene rearrangement (Karpas 422, RL, VAL, and SUDHL-6), and the mediastinal B-cell lymphoma cell line Karpas 1106, showed low-level gain (3 DNA copies) of 18q21 band in CGH analysis. A description of derivation and characteristics of the cell lines is available in Supplemental Table S1 on the Blood website; see the Supplemental Material link at the top of the online article.
Molecular cytogenetic techniques
Patients and cell lines were karyotyped according to standard methods. FISH was performed on fixed cells from cytogenetic samples as reported,8,22 using the following probes: LSI-BCL2-IGH and LSI-IGH (Vysis, Downers Grove, IL), BAC 248E24 containing BCL2; MALT1 PAC clones either flanking (119K19, 83A16, 628B12, and 59N7) or containing the MALT1 gene (60F18 and 152M15) and PAC 166G16 containing API2 gene (as described in Dierlamm et al23 ). To search for genomic abnormalities of the MALT1 gene, 2 FISH assays were applied to the series of 40 patients. First, clones 59N7 and 119K19 flanking and labeled in different colors (Spectrum-Green-dUTP and Spectrum-Red-dUTP, Vysis) were used to evaluate a chromosomal translocation on interphase nuclei. Second, PAC clone 60F18 containing the MALT1 gene in combination with a centromeric chromosome 18 probe (Vysis) were used to assess chromosome 18 copy number on interphase nuclei. Normal FISH patterns of both assays were initially evaluated on 3 samples from healthy donors (data not shown).
Long-distance inverse PCR
Long-distance inverse PCR (LDI-PCR) for detection of IGHJ rearrangements was performed in the 2 cases with t(14;18)(q32;q21), as previously described.24 In both cases, DNA was digested with BglII and ligated at low concentration, followed by nested PCR using the following primers: J6E 5′ CCCACAGGCAGTAGCAGAAAACAA 3′, JBE 5′ GAAGCAGGTCACCGCGAGAGT 3′ and nested primers J6I 5′ TCTGGGCTCGAGTCGACGCAGAAAACAAAGGCCCTAGAGGG 3′, JBI 5′ CTTCTGGTTGTGAAGAGGTGGTTTTG 3′. PCR products were cloned and sequenced as previously reported.24 Sequences of both cloned breakpoints have been submitted to GenBank (accession nos. AJ514428 and AJ512985).
Array CGH
Genome-wide analysis of DNA-copy number variation was performed using array CGH on a microchip with 2460 BAC and P1 clones in triplicate (HumArray 1.14). Fabrication and validation of the array, hybridization methods, analytic procedures, and clone content have been described elsewhere in detail.25,26 For the purpose of this study, we analyzed 55 BAC/PAC clones covering the chromosome 18 from pter to qter. A full list of these clones is available in Supplemental Table S2. Fluorescent Cy3 (tumor)/Cy5 (normal) log2 ratios were defined as follows: between -0.5 and +0.4, normal range; between 0.41 and 0.95, 3-fold genomic gain; between 0.96 and 1.6, 4- to 6-fold amplification; more than 1.61 corresponds to 7-fold or greater amplification; genomic deletion was defined by values lower than -0.5.
Gene expression analysis
“Lymphochip” cDNA microarrays were used for the analysis. These microarrays were constructed from 12 196 clones of cDNA and were used to quantify the expression of mRNA in the cell lines. Isolation of polyA+ mRNA, hybridization procedures, and evaluation of global-wide gene expression profiles were performed as described.27 For this study, we focused on the expression values of the clones mapped to the long arm of chromosome 18 that included 37 different sequence-verified genes/expressed sequence tags (ESTs). A full list of the clones studied and their localization on chromosome 18 is provided in Supplemental Table S3.
QRT-PCR assay for MALT1 expression
Total RNA was extracted from the cell lines, one patient with t(14;18)(q32; q21) with available material from peripheral blood (case 2), the blood lymphocytes from 16 healthy donors, 2 reactive lymph nodes, and 5 normal spleens, using the RNeasy kit (Qiagen, Valencia, CA). First-strand cDNA synthesis was performed using the GeneAmp kit with random hexamer primers (RNA PCR system; PE Applied Biosystems, Foster City, CA). On the basis of the published sequence of MALT1 gene (GenBank accession no. AF130356), a set of gene-specific primers to produce a product of 124 base pair (bp) was designed using the Primer Express program (PE Applied Biosystems), as follows: forward 5′ TGAGATGCGTAATGCTGTGG 3′ at exon 10, and reverse 5′ TATGGATTTGGAGCATCAACG 3′ at exon 11. These primers include an intron of 11 kilobase (kb), thus avoiding gDNA contamination. Real-time monitoring of PCR reactions was performed using the ABIPrism 7700 (PE Applied Biosystems) and the SYBR Green I dye, which binds preferentially to double-stranded (ds) DNA. The reactions were carried out in a total volume of 50 μL according to the manufacturer's instructions. MALT1 mRNA expression was normalized by using GADPH and TATA-binding protein (TBD) housekeeping gene products as endogenous control of expression.28 Each cDNA sample was analyzed in triplicate with appropriate negative controls. MALT1/TBP expression ratios were calculated in the normal samples. The median value of all these measurements was 1.08 (± 0.07), which was assigned the value of 1. After the MALT1 gene standardization and normalization, MALT1 mRNA expression was quantified in the 40 patient samples; measurements in the patients were expressed with respect to the value of 1.
Immunohistochemistry
Paraffin section and cytospin immunohistochemistry were performed using routine methods with a wide panel of monoclonal antibodies (MAbs). Flow cytometric immunophenotypic analysis was performed in case 2. For BCL10 detection, a mouse MAb (clone 151) that recognizes an epitope in the carboxy terminus of BCL10 was used.29 For MALT1 staining, a mouse MAb raised against the amino-terminal portion of the protein was kindly provided by Dr V. Dixit (Genentech, South San Francisco, CA).17
Western blotting analysis
Cell pellets and frozen tissue sections were homogenized in 50 mM Tris (tris-(hydroxymethyl)aminomethane-HCl) (pH 8.0), 150 mM NaCl, 0.02% sodium azide, 0.1% sodium dodecyl sulfate (SDS), 1% Nonidet P-40, 0.5% sodium deoxycholate, 100 mg/mL phenylmethylsulfonyl fluoride, and 1 mg/mL leupeptin. The protein concentration of each extract was quantified and equal amounts of protein from each sample were mixed with SDS loading buffer, separated on 8% polyacrylamide gels, and eletrotransfered onto nitrocellulose membranes. The membrane was sequentially incubated with a MALT1 N-terminal antibody, biotinylated rabbit antimouse immunoglobulin, and alkaline phosphatase–conjugated avidin and was finally visualized with 5-bromo-4chloro-3-indolyl phosphate and nitroblue tetrazolium. For more information, see “Methods” in the data supplement on the Blood website.
Results
t(14;18)(q32;q21) may involve MALT1 rather than BCL2 in both MZLs and MALTs
The t(14;18)(q32;q21) occurs frequently in follicular and diffuse B-NHL, but is seen rarely, if at all, in other histologic subtypes of mature B-cell malignancy. The detection of 2 cases, one with MALT lymphoma and the other with MZL with this translocation, was therefore unexpected and prompted our investigation of the breakpoints. Clinical characteristics and karyotypes on the 2 patients with t(14;18)(q32;q21) are shown in Table 1. The t(14; 18)(q32;q21) was indistinguishable cytogenetically from the one characteristic of FCL. Case 1 was a histologically typical orbital MALT lymphoma, which has remained in remission since treatment with local radiotherapy (Figure 1A). In contrast, case 2 was an aggressive lymphoma that presented with pulmonary infiltration, pleural effusion, massive splenomegaly, and lymphocytosis of mature B cells (CD20+, CD5-, CD10-, CD3-, CD43-, FMC7+, CD23-). A diagnosis of MZL in leukemic phase was made on the basis of both the histologic appearances and the immunophenotype of the peripheral blood cells (Figure 1B-C). However, the possibility that this case represented a case of transformed MALT lymphoma cannot be entirely excluded. Sections of the spleen showed that the malignant cells expressed IgD, but failed to express DBA44 and appeared to coexpress CD43, both unusual findings in splenic MZL29 (data not shown). Despite initially achieving complete remission with combination chemotherapy, the patient had a relapse within 1 year (Figure 1D).
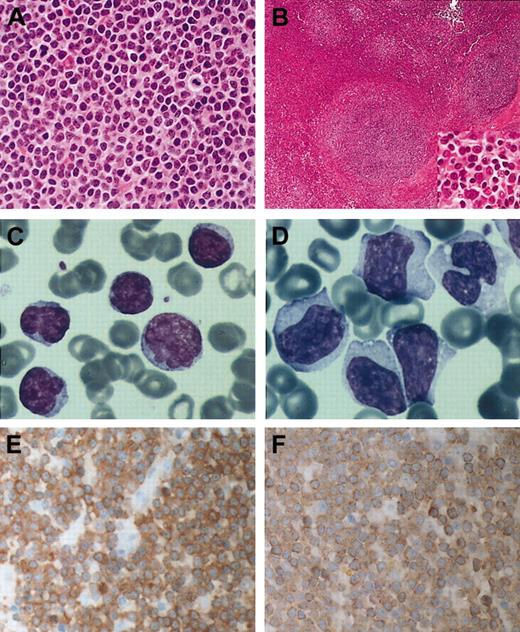
Histology, cytology, MALT1, and BCL10 expression in B-cell lymphomas with t(14;18)(q32;q21) involvingMALT1 (A) Hematoxylin and eosin–stained section of case 1 showing typical centrocyte-like cells of MALT lymphoma. (B) Hematoxylin and eosin–stained splenic section of case 2; inset shows the morphology of tumor cells in the marginal zone. (C) Peripheral blood smear of case 2 at diagnosis. May-Grünewald-Giemsa stain. (D) Peripheral blood smear at transformation from case 2 shows blastic cells with abundant cytoplasm, irregular nuclei with nucleoli. May-Grünewald-Giemsa stain. (E) Immunohistochemstry for MALT1 in case 1 showing high levels of the protein in the cytoplasm of all tumor cells. Stained with hematoxylin. (F) Case no. 1 stained with BCL10 Mab 151 showing increased cytoplasmic BCL10 expression. Original magnifications: × 400 (A, B inset, E-F); × 100 (B); and × 1000 (C-D).
Histology, cytology, MALT1, and BCL10 expression in B-cell lymphomas with t(14;18)(q32;q21) involvingMALT1 (A) Hematoxylin and eosin–stained section of case 1 showing typical centrocyte-like cells of MALT lymphoma. (B) Hematoxylin and eosin–stained splenic section of case 2; inset shows the morphology of tumor cells in the marginal zone. (C) Peripheral blood smear of case 2 at diagnosis. May-Grünewald-Giemsa stain. (D) Peripheral blood smear at transformation from case 2 shows blastic cells with abundant cytoplasm, irregular nuclei with nucleoli. May-Grünewald-Giemsa stain. (E) Immunohistochemstry for MALT1 in case 1 showing high levels of the protein in the cytoplasm of all tumor cells. Stained with hematoxylin. (F) Case no. 1 stained with BCL10 Mab 151 showing increased cytoplasmic BCL10 expression. Original magnifications: × 400 (A, B inset, E-F); × 100 (B); and × 1000 (C-D).
Both patients exhibited trisomy of chromosome 3, an alteration seen commonly in both MALT and MZL, but rarely in FCL.1,11, 12, 13,30 These were the sole abnormalities seen in case 1, whereas case 2 exhibited a complex karyotype with multiple other numeric and structural abnormalities (Table 1; Figure 2A).
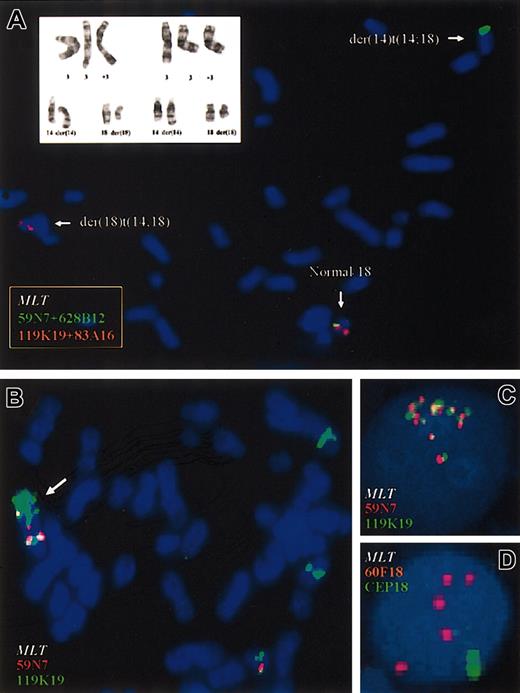
Cytogenetic and FISH analyses of patients with t(14;18)(q32;q21). (A) FISH study using flanking probes for MALT1 gene in peripheral blood cells from case 2. PACs 119K17 and 83A16 centromeric to MALT1 locus are shown in red; PACs 59N7 and 628B12, telomeric to MALT1 are shown in green. The pooled telomeric probes (green) hybridized to the derivative chromosome 14, whereas the pooled centromeric probes (red) hybridized to the derivative 18. A partial karyotype from case 1 including the t(14;18)(q32;q21) and the trisomy of chromosome 3 is shown in the top left corner. (B) In the SSK-41 cell line, selective amplification of MALT1 but not of BCL2 was observed. Cytogenetic analysis showed der(18)trp(18)(q11q21), which is marked with an arrow, as well as several insertions of chromosome material from 18q21 in different chromosomes. FISH analysis showed amplification of PAC 119K19 flanking 5′ region of MALT1 gene locus but not of PAC 59N7 located 3′ of MALT1, indicating that a chromosome break occurred before amplification. (C) Genomic amplification of MALT1 gene in KHM-10B cell line using MALT1 flanking probes. Eight copies of MALT1 were detected, confirming the array CGH data. (D) Genomic amplification of MALT1 in one case of gastric MALT lymphoma with bulky retroperitoneal disease at diagnosis. Five copies of MALT1 were seen.
Cytogenetic and FISH analyses of patients with t(14;18)(q32;q21). (A) FISH study using flanking probes for MALT1 gene in peripheral blood cells from case 2. PACs 119K17 and 83A16 centromeric to MALT1 locus are shown in red; PACs 59N7 and 628B12, telomeric to MALT1 are shown in green. The pooled telomeric probes (green) hybridized to the derivative chromosome 14, whereas the pooled centromeric probes (red) hybridized to the derivative 18. A partial karyotype from case 1 including the t(14;18)(q32;q21) and the trisomy of chromosome 3 is shown in the top left corner. (B) In the SSK-41 cell line, selective amplification of MALT1 but not of BCL2 was observed. Cytogenetic analysis showed der(18)trp(18)(q11q21), which is marked with an arrow, as well as several insertions of chromosome material from 18q21 in different chromosomes. FISH analysis showed amplification of PAC 119K19 flanking 5′ region of MALT1 gene locus but not of PAC 59N7 located 3′ of MALT1, indicating that a chromosome break occurred before amplification. (C) Genomic amplification of MALT1 gene in KHM-10B cell line using MALT1 flanking probes. Eight copies of MALT1 were detected, confirming the array CGH data. (D) Genomic amplification of MALT1 in one case of gastric MALT lymphoma with bulky retroperitoneal disease at diagnosis. Five copies of MALT1 were seen.
Using metaphase FISH and 2 differentially labeled probes mapping to IGHV and centromeric to IGHC, a breakpoint within IGH was observed in both cases. The 2 IGH probes were split, with the IGHV probe hybridizing to chromosome 18q21, with IGHC retained on the derivative chromosome 14. In contrast, the BCL2/FVT1 locus was not involved because probes containing BCL2 gene completely hybridized to the derivative chromosome 14, thus indicating a more centromeric break on chromosome 18q21. To determine the possible 18q21 breakpoint a number of PAC clones mapping to the region were hybridized to metaphases from both cases (Figure 2A). Clone 60F18, which contains MALT1, was split with partial hybridization to the derivative chromosome 14. Additional FISH mapping using probes covering and flanking the MALT1 locus, indicated a recurrent breakpoint in both cases 1 and 2, resulting in fusion of MALT1 and IGH gene probes. Involvement of the API2 gene in a cytogenetically cryptic t(11;18)(q21;q21) or insertion of API2 sequences31 was excluded by using FISH probes for this gene, which showed retention of both signals on chromosome 11 (data not shown).
Molecular cloning of variant t(14;18)(q32;q21) translocation breakpoints identifies MALT1 as the targeted gene
The FISH studies therefore suggested MALT1 as a potential target gene for the variant t(14;18)(q32;q21). To confirm this directly, molecular cloning of the translocation breakpoints using LDI-PCR from IGHJ was performed.24 In case 1, the sequence of a 1.45-kb LDI-PCR product showed loss of homology with IGH beyond IGHJ5, indicating that this clone contained the translocation breakpoint. Comparison of the non-IGH sequence with the human genome sequence (http://www.ncbi.nlm.nih.gov/BLAST/) showed that the non-IGH sequence derived from chromosome 18q21.3 close to the MALT1 gene, although initially it was not possible to link this sequence directly to MALT1 due to “gaps” in the genome sequence. In case 2, sequence analysis of a 700-bp LDI-PCR product showed loss of homology with IGH sequences beyond DH7-JH5, indicating that the breakpoint occurred after a presumably nonproductive IGH rearrangement. Further sequencing of the BAC clone RP11-126O1 showed the 2 cloned breakpoints to be 1.1 kb and 1.7 kb upstream of the first 5′ noncoding exon of MALT1 in cases 1 and 2, respectively (Figure 3). It should be noted that there might be alternative 5′ noncoding exons for MALT1; 2 more centromeric 5′ exons have been described by one group15 (and Figure 3). Nevertheless, from the consensus sequence, the open-reading frame of MALT1 (http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=NP_006776.1) would not appear to be involved by either translocation breakpoint. This translocation is therefore similar in structure to other IGH translocations involving several genes such as MYC, BCL6, BCL10, and BCL11A with breakpoints that fall immediately 5′ to the first exons or within the 5′ noncoding regions, but leaving the coding regions of the genes intact.32
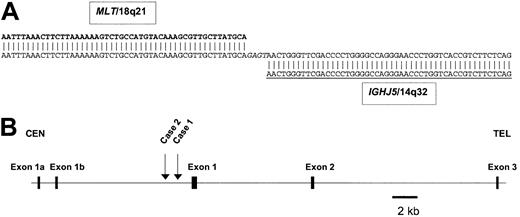
Location of the breakpoints withinMALT1and theIGHlocus. (A) Sequence of the der(14)t(14;18)(q32;q21) breakpoint from case 1 (GenBank accession no. pending). Chromosome 18q21 sequences shown in bold, IGH sequences underlined, inserted nontemplated nucleotides in italics. DNA sequence analysis of cloned LDI-PCR product from case 1 showed loss of homology with IGH sequences beyond IGHJ5. The sequence beyond IGHJ5 was identical to sequence from BAC clone RP11-126O1, which contained not only both breakpoints but also the 5′ end of the MALT1 gene as well. (B) Ideogram to show the localization of chromosome 18q21 breakpoints. Both cloned breakpoints fell 5′ (centromeric) of the most 5′ exon of the database consensus MALT1 gene sequence (http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=NP_006776.1). However, it should be noted that one report has indicated the presence of alternative 5′ MALT1 exons15 ; these are depicted as exons 1a and 1b. Neither of these exons is coding and therefore the open-reading frame of MALT1 is maintained in both translocations. Within the BAC clone RP11-126O1, the breakpoints were located at nucleotides 27 558 (case 1), and 28 209 (case 2), thus placing the breakpoints 1.1 and 1.7 kb upstream, respectively, of the first 5′ MALT1 exon.
Location of the breakpoints withinMALT1and theIGHlocus. (A) Sequence of the der(14)t(14;18)(q32;q21) breakpoint from case 1 (GenBank accession no. pending). Chromosome 18q21 sequences shown in bold, IGH sequences underlined, inserted nontemplated nucleotides in italics. DNA sequence analysis of cloned LDI-PCR product from case 1 showed loss of homology with IGH sequences beyond IGHJ5. The sequence beyond IGHJ5 was identical to sequence from BAC clone RP11-126O1, which contained not only both breakpoints but also the 5′ end of the MALT1 gene as well. (B) Ideogram to show the localization of chromosome 18q21 breakpoints. Both cloned breakpoints fell 5′ (centromeric) of the most 5′ exon of the database consensus MALT1 gene sequence (http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?val=NP_006776.1). However, it should be noted that one report has indicated the presence of alternative 5′ MALT1 exons15 ; these are depicted as exons 1a and 1b. Neither of these exons is coding and therefore the open-reading frame of MALT1 is maintained in both translocations. Within the BAC clone RP11-126O1, the breakpoints were located at nucleotides 27 558 (case 1), and 28 209 (case 2), thus placing the breakpoints 1.1 and 1.7 kb upstream, respectively, of the first 5′ MALT1 exon.
To determine whether MALT1 was deregulated as a consequence of the translocation, QRT-PCR for MALT1 was performed in case 2, where RNA was available. Higher-level expression of MALT1 was observed in this specimen compared with normal samples; levels of expression were in the same range as seen in the 2 cell lines with MALT1 amplification (Figure 4). In case 1, expression of MALT1 protein was determined by immunohistochemistry. High levels of MALT1 expression were detected in the malignant cells (Figure 1E). In contrast, the protein was either not expressed or expressed at a low level in lymphomas lacking MALT1 chromosomal translocation. Fixation of material of case 2 was not optimal and reproducible results with the MALT1 MAb could not be obtained. In addition, because MALT lymphomas with t(11;18)(q21;q21) are usually associated with aberrant nuclear BCL10 expression,29,33 BCL10 expression was evaluated in case 1 and showed increased cytoplasmic (but not nuclear) staining in the tumor cells (Figure 1F).
Quantification ofMALT1gene expression by real-time PCR. Logarithmic plot of fluorescence signal (y-axis) versus number of PCR cycles (x-axis). Samples D1, D2, D3, and D4 represent results obtained with RNA extracted from peripheral blood mononuclear cells from 4 healthy donor samples. MALT1 curves in duplicate for case 2 (red and green lines) and SSK-41 cell line (blue and yellow lines), all with similar MALT1 expression levels with respect to healthy donors, are shown.
Quantification ofMALT1gene expression by real-time PCR. Logarithmic plot of fluorescence signal (y-axis) versus number of PCR cycles (x-axis). Samples D1, D2, D3, and D4 represent results obtained with RNA extracted from peripheral blood mononuclear cells from 4 healthy donor samples. MALT1 curves in duplicate for case 2 (red and green lines) and SSK-41 cell line (blue and yellow lines), all with similar MALT1 expression levels with respect to healthy donors, are shown.
MALT1 is a target for amplification and overexpression in the 18q21 amplicon of B-NHL
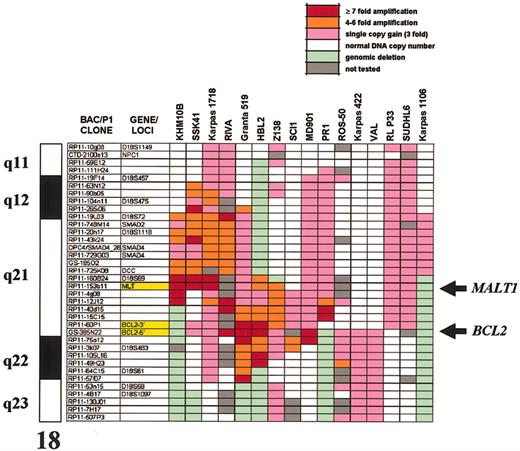
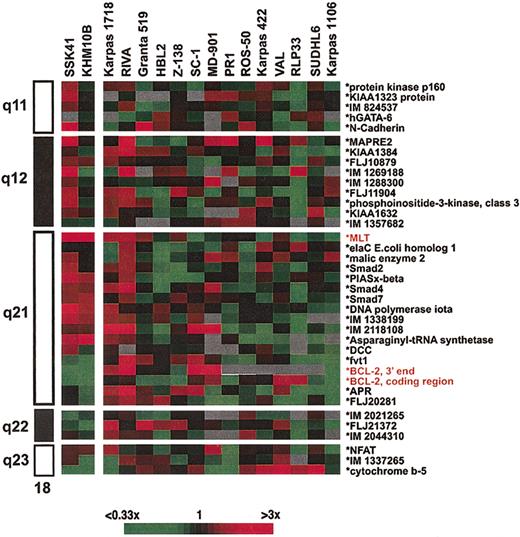
Genomic amplification of 18q21 has been noted to occur frequently in B-cell malignancies and has been suggested in FCLs and DLBCLs to account for BCL2 overexpression in the absence of t(14;18)(q32;q21).3,4 However, the possibility that other genes may also be deregulated as a consequence of such amplifications has not been examined. To determine more precisely the boundaries and the pathologic consequences of the 18q21 amplicon, we have performed both array CGH and gene profiling on a panel of well-characterized B-NHL cell lines. Array CGH defined genomic aberrations affecting chromosome 18q21 in 16 of 40 B-NHL cell lines studied, delineated amplification borders and peaks, and measured the number of DNA copies within the amplified area on 18q21 (Figure 5). A recurrently amplified/gained segment of about 9 Mb encompassing 18q21.31 to 18q21.33 was defined. This interval contained 2 separate peaks of amplification, one containing the MALT1 locus (ranging from 3- to 21-fold amplification), and a more telomeric amplicon, centered around the BCL2 locus (ranging from 3- to 15-fold amplification). The only cell line not showing involvement of this interval was the mediastinal B-cell lymphoma-derived cell line Karpas 1106,34 which showed gain of chromosome band 18q21.1 (3-fold). The DNA copy number variation observed in array CGH analysis was confirmed by FISH analysis using the individual probes containing MALT1 and BCL2 genes on interphase and metaphase spreads from selected cell lines (Figure 2B-C).
Genomic structure of the 18q amplicon shown by comparative genomic hybridization to microarrays in B-NHL cell lines. Only the 18q clones from the HumArray 1.14 CGH microarray are shown; a full list of these clones and the numeric results of array CGH are provided in Supplemental Table S2. Each clone contains at least one sequenced tagged site (STS), allowing linkage to the genome sequence (http://genome.cse.ucsc.edu). The order of the clones was arranged according to the April 2002 version of the draft. The array provides an average resolution of 1.4 Mb across chromosome 18. Both BCL2 and MALT1 peaks of amplification are indicated by arrows.
Genomic structure of the 18q amplicon shown by comparative genomic hybridization to microarrays in B-NHL cell lines. Only the 18q clones from the HumArray 1.14 CGH microarray are shown; a full list of these clones and the numeric results of array CGH are provided in Supplemental Table S2. Each clone contains at least one sequenced tagged site (STS), allowing linkage to the genome sequence (http://genome.cse.ucsc.edu). The order of the clones was arranged according to the April 2002 version of the draft. The array provides an average resolution of 1.4 Mb across chromosome 18. Both BCL2 and MALT1 peaks of amplification are indicated by arrows.
The cytogenetic mechanisms leading to amplification and gain of 18q21 were also investigated. In most DLBCL and mantle cell lymphoma cell lines, the amplification peak occurred predominantly in the BCL2 gene locus but spared the MALT1 locus. In contrast, in the 2 MZL-derived cell lines, SSK-4135 and Karpas 1718 (MJSD; A. Karpas, unpublished observations, November 2001) displayed the amplification maxima centered around the MALT1 locus. In SSK-41, as well as in the Burkitt cell line KHM-10B,36 exclusive amplification of MALT1 not affecting BCL2 was detected.
These data indicated that the MALT1 and BCL2 loci were independent targets of amplification in B-NHL. To search for putative genes with deregulated expression as a consequence of the amplification, we measured gene expression of 37 different genes/ESTs along chromosome 18q in the cell lines by gene expression profiling (Figure 6). In all but 3, BCL2 mRNA overexpression was identified. The 3 cell lines with lower BCL2 expression included the 2 cell lines with selective amplification of MALT1 (SSK-41 and KHM-10B), as well as Karpas 1106 that displayed gain of 18q21.1; low-level BCL2 expression has been previously reported in this cell line.34 SSK-41 and KHM-10B cell lines displayed high-level MALT1 expression. In the case of MALT1, the gene profiling data were validated using the QRT-PCR assay and confirmed elevated MALT1 expression only in SSK-41 and KHM-10B cell lines at a level comparable to those observed in case 2 with t(14;18)(q32; q21). No other gene/EST mapping within 18q on the lymphochip showed consistently altered expression in the cell line panel, excluding other putative genes as targets of amplification.
Gene expression profiling of 18q in B-NHL cell lines with genomic amplification of 18q21. The ratios are a measure of relative gene expression of 37 genes/ESTs in each experimental sample and are depicted according to the color scale shown at the bottom. Only sequence-verified clones are shown. A full list of these clones with their respective expression measurements in the cell lines is provided in Supplemental Table S3. For genes/ESTs with more than one clone into the microarray, the average values from the different clone measurements are depicted. Clones are ordered according to their cytogenetic position in chromosome 18, which is shown on the left. Both SSK-41 and KHM-10B cell lines show MALT1 overexpression but normal BCL2 expression values.
Gene expression profiling of 18q in B-NHL cell lines with genomic amplification of 18q21. The ratios are a measure of relative gene expression of 37 genes/ESTs in each experimental sample and are depicted according to the color scale shown at the bottom. Only sequence-verified clones are shown. A full list of these clones with their respective expression measurements in the cell lines is provided in Supplemental Table S3. For genes/ESTs with more than one clone into the microarray, the average values from the different clone measurements are depicted. Clones are ordered according to their cytogenetic position in chromosome 18, which is shown on the left. Both SSK-41 and KHM-10B cell lines show MALT1 overexpression but normal BCL2 expression values.
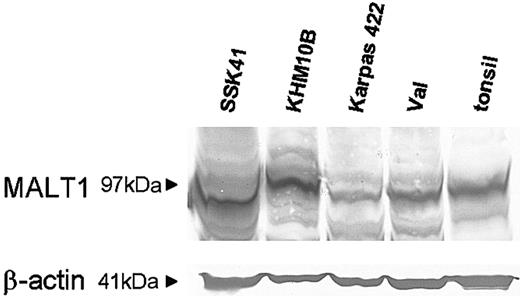
To examine MALT1 protein expression, we performed Western blotting analysis in selected cell lines with 18q21 amplification. MALT1 was expressed at a much higher level in both SSK-41 and KHM-10B that had MALT1 gene amplification, compared with the remaining cell lines (Figure 7).
Western blotting analysis for MALT1 in cell lines with 18q amplification. Equal amounts of protein extracts from various cell lines and a tonsil were subjected to Western blotting analysis for MALT1. The protein is expressed at a much higher level in SSK-41 and KHM-10B that have MALT1 gene amplification, than in Karpas 422, VAL, and tonsil.
Western blotting analysis for MALT1 in cell lines with 18q amplification. Equal amounts of protein extracts from various cell lines and a tonsil were subjected to Western blotting analysis for MALT1. The protein is expressed at a much higher level in SSK-41 and KHM-10B that have MALT1 gene amplification, than in Karpas 422, VAL, and tonsil.
Screening for MALT1 alterations in patients with MZL and MALT lymphoma
To extend these observations from cell lines, we sought amplification or overexpression of MALT1 using FISH and QRT-PCR assays in a panel of primary cases consisting of 35 cases of splenic MZLs and 5 MALT lymphomas. Among the patients studied by FISH, only one with gastric MALT lymphoma showed amplification of MALT1, including 5 copies of MALT1 gene probes as well as coamplification of the BCL2 (Figure 2D). Genomic gain of 18q21 was confirmed using standard CGH (data not shown). No mRNA measurements could be performed in this case due to lack of suitable material. No other translocation or amplification event was detected by FISH. Using QRT-PCR we measured MALT1 gene expression in the series of patients. Only one case with typical splenic MZL presented elevated MALT1 gene expression level. In this patient, neither cytogenetic nor genomic abnormalities affecting MALT1 gene were identified in bone marrow or spleen specimens. Unfortunately, lack of well-preserved material from this case precluded further studies. No other MALT1 gene expression alteration was detected in any other cases (data not shown).
Discussion
MALT1 was isolated through its direct involvement in the chromosomal translocation t(11;18)(q21;q21), which characterizes about 30% of cases of MALT lymphoma and results in API2-MALT1 fusion.11, 12, 13, 14, 15, 16 The functions of MALT1 are not known, but the protein contains several recognized domains including a death domain (DD), 2 immunoglobulin domains, and a caspaselike domain; the last is maintained in all API2-MALT1 fusions reported to date.14, 15, 16, 17, 18,37 MALT1 has been implicated in NF-κB signaling from the B-cell receptor for antigen through its direct interaction with BCL10; MALT1 itself does not induce NF-κB signaling, and neither does it homodimerize, but synergizes markedly with BCL10.17,18 This is of some interest, because BCL10 is deregulated in a subset of MALT lymphomas as a consequence of t(1;14)(p22;q32).38,39 The present study implicates MALT1 as the target not only of a variant t(14;18)(q32;q21) of MZL and MALT lymphoma but also in some cases with an 18q21 amplicon. From the in vitro data mentioned, it is likely that MALT1-induced transformation is dependent on the formation of MALT1-BCL10 heterodimers; this hypothesis is currently being tested.
From the preliminary data reported here, deregulation of MALT1 seems to be a relatively uncommon event. However, it would appear that MALT1 deregulation might occur in B-cell malignancies of various histologic subtypes. First, the variant t(14;18)(q32;q21) was seen not only in one patient with orbital MALT lymphoma (case 1) but also in another patient with an aggressive MZL of possible splenic type (case 2). This finding is noteworthy because splenic MZL (SMZL) and MALT lymphoma are 2 distinct diseases, defined by characteristic histologic and cytogenetic appearances.40,41 Thus, for example, SMZL may typically exhibit cytogenetically deletion of chromosome 7q31-q32 and trisomy of chromosomes 3q and 18 as the more common events and IG chromosomal translocations involving either CDK6 or CCND3, whereas MALT lymphomas are characterized by t(11;18)(q21;q21) or more rarely t(1;14)(p22;q32).11, 12, 13,38,39,42, 43, 44 Trisomy 3 may also be found frequently in MALT lymphoma45 and because both cases reported here exhibited this abnormality, it is possible that case 2 classified here as MZL, in fact represented leukemic transformation of an undetected pulmonary low-grade MALT lymphoma. However, it is also possible that the variant t(14;18)(q32;q21) involving MALT1 occurs at low frequency in both lymphoma subtypes, indicating that at least a small fraction of cases share a common genetic background.46
During the preparation of this manuscript, 2 other reports were published documenting by FISH the occurrence of the same chromosomal translocation in patients with nongastric MALT lymphoma.47,48 In the first, the cytogenetic detection of t(14;18)(q32; q21) in MALT lymphoma of the liver and skin prompted the identical FISH investigations to those reported here. The authors subsequently showed that 12 (18%) of 66 MALT lymphomas harbored the IGH/MALT1 translocation.47 However, the frequency at which this translocation occurred varied significantly with primary location of disease. Thus, the t(14;18)(q32;21) was seen in all 4 hepatic MALT lymphomas, 3 of 11 primary cutaneous, 3 of 8 orbital, and 2 of 11 salivary MALT lymphomas but not in any of the 17 gastrointestinal MALT lymphomas studied. Similarly, the second group showed the IGH/MALT1 translocation in 3 of 27 primary pulmonary MALT lymphomas; in this series, all 3 cases also exhibited trisomy 3, whereas 2 exhibited trisomy 12 and 1 trisomy 11.48 Confirming these observations, we have failed to find any evidence for FISH in a large series of primary gastrointestinal MALT lymphomas.49 Together, these data would indicate that the genetic abnormalities in MALT lymphomas reflect their site of origin and possibly either environmental or autoimmune causes.
However, deregulation of MALT1 may also follow genomic amplification. We correlated the expression of BCL2 and MALT1 with the corresponding DNA copy number increase in a panel of derived B-NHL cell lines with amplification of 18q21. Gene expression profile analysis excluded various genes on 18q as targets of the amplification, such as FVT1 and APR, and highlighted BCL2 and MALT1 as the activated genes influenced by the respective DNA gains. In most of the B-NHL cell lines, BCL2 was targeted by the amplifications. Two cell lines, however, showed exclusive involvement of MALT1, one derived from a patient with MZL and one from a patient with Burkitt lymphoma. Gene amplification of MALT1 may therefore represent another mechanism of MALT1 activation in B-NHL. In support of this hypothesis, it has recently been shown that in primary B-NHL of the central nervous system, gains of 18q21 included the MALT1 gene locus.8 Similarly, other oncogenes deregulated by translocation to IG genes, such as BCL1, BCL2, MYC, and BCL11A, have also been shown to be altered by gene amplification in B-NHL.4,9,10,50, 51, 52, 53
In summary, our data indicate that the MALT1 may act as a dominant oncogene in the pathogenesis of some B-cell malignancies. The mechanisms through which MALT1 contributes to lymphoma pathogenesis remain to be elucidated. Additional genetic and functional analyses are currently under investigation to address these questions. Activation of MALT1, a paracaspase with unknown target proteins, may represent a novel therapeutic target.
Prepublished online as Blood First Edition Paper, January 30, 2003; DOI 10.1182/blood-2002-10-3236.
Supported by grants from Spanish Ministries of Health and Science and Technology, Union Internationale contra le Cancer, Kay Kendall Leukaemia Fund, University Hospitals Leicester, Leukaemia Research Fund, Interdisziplinäres Zentrum für klinische Krebsforschung (IZKF) Kiel, Deutsche Krebshilfe/Dr Mildred Scheel Stiftung für Krebsforschung (10-1641-De1), and National Institutes of Health grant U01-CA84967-1.
M.J.S.D. and J.A.M.-C. contributed equally to this study.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Drs Jacqueline Batanian, Isabel Benet, Miguel Angel Piris, Elías Campo, Maria Angeles Ruiz, Isabel Navarro, Jesus Maria Hernandez, Francesc Sole, Antonio Ferrandez, Isabel Marugan, Maria Jose Terol, Jack Rootman, and Joseph Connors for kindly providing patient material and clinical data; Dr Abraham Karpas (University of Cambridge, United Kingdom) for kindly providing the Karpas 1718 cell line; and Dr V. Dixit for kindly providing MALT1 MAb.