Abstract
HOX genes, notably members of the HOXA cluster, and HOX cofactors have increasingly been linked to human leukemia. Intriguingly, HOXD13, a member of the HOXD cluster not normally expressed in hematopoietic cells, was recently identified as a partner of NUP98 in a t(2;11) translocation associated with t-AML/MDS. We have now tested directly the leukemogenic potential of the NUP98-HOXD13 t(2; 11) fusion gene in the murine hematopoietic model. NUP98-HOXD13 strongly promoted growth and impaired differentiation of early hematopoietic progenitor cells in vitro; this effect was dependent on the NUP98 portion and an intact HOXD13 homeodomain. Expression of the NUP98-HOXD13 fusion gene in vivo resulted in a partial impairment of lymphopoiesis but did not induce evident hematologic disease until late after transplantation (more than 5 months), when some mice developed a myeloproliferative-like disease. In contrast, mice transplanted with bone marrow (BM) cells cotransduced with NUP98-HOXD13 and the HOX cofactor Meis1 rapidly developed lethal and transplantable acute myeloid leukemia (AML), with a median disease onset of 75 days. In summary, this study demonstrates that NUP98-HOXD13 can be directly implicated in the molecular process leading to leukemic transformation, and it supports a model in which the transforming properties of NUP98-HOXD13 are mediated through HOX-dependent pathways.
Introduction
Therapy-related myelodysplastic syndrome (t-MDS) and acute myeloid leukemia (t-AML) have emerged as serious long-term complications, denoting poor prognosis, of genotoxic high-dose chemotherapy or radiotherapy in patients with cancer.1,2 Recent data have described a novel group of t-MDS/t-AML with mostly myelomonocytic features that are characterized by balanced translocations involving the nucleoporin NUP98 gene on chromosome 11p15. In all cases, the chromosomal translocations involving 11p15 create a chimeric gene fusing the same N-terminal portion of NUP98 in-frame to one of multiple potential partner genes.
The most frequently observed NUP98 fusion partners belong to the clustered homeobox gene family previously implicated in normal regulation of hematopoiesis and leukemia (for reviews, see van Oostveen et al,3 Buske and Humphries,4 and Owens and Hawley5 ). Since the first identification of the NUP98-HOXA9 fusion gene,6,7 HOXD13 (t(2;11),8 HOXA11, HOXA13,9 HOXC11,10 HOXD11,11 and the nonclustered homeobox-containing gene PMX112 have all been identified as NUP98 fusion partners in de novo AML, t-AML, or both. The involvement of homeobox genes as partner genes of NUP98 is of particular interest given the growing evidence linking HOX genes, particularly members of the 5′-located members of the HOXA cluster, such as HOXA9 and HOXA10, to leukemia.13, 14, 15, 16
The involvement of 2 members of the HOXD cluster in NUP98 translocation-associated leukemia is intriguing. In contrast to members of the A cluster,17 HOXD cluster genes are not normally expressed in hematopoietic cells but rather are recognized as key regulators in the morphogenesis of vertebrate limbs.18,19 The NUP98-HOXD13 (ND13) chimeric protein thus defines a novel class of an NUP98 fusion gene involving a nonhematopoietic clustered HOX gene and resulting in its de novo and aberrant expression in human hematopoietic cells.8,20,21 Moreover, as a 13-paralog member, HOXD13, unlike HOXA9, lacks the binding domain for PBX, a known HOX cofactor also implicated in normal hematopoiesis and leukemia.22, 23, 24 It is unclear how and to what extent ND13 alone may perturb hematopoiesis, in contrast to several other HOX genes so far characterized. Also of interest is the nature of other potential collaborating genes for the leukemogenic activity of ND13. One provocative candidate is the TALE homeobox protein Meis1, previously identified as a gene frequently coactivated with Hoxa9 or Hoxa7 in the BXH2 leukemia model25,26 and able to accelerate HOX gene-induced leukemogenesis for several HOX genes, including NUP98-HOXA9.27,28 The mechanisms responsible for the acceleration of NUP98-HOXA9–induced leukemogenesis by Meis1 remain elusive because the putative Meis1-interacting domain, which is located N-terminal of the homeodomain, is lost in the fusion gene.29 Interestingly, this domain is also absent in the ND13 fusion.
NUP98 is the second member of the nucleoporin gene family implicated in human leukemogenesis in addition to NUP214 (or CAN), which is found in the t(6;9)(p23;q34) in patients with AML.30 NUP98 regulates the unidirectional and the bidirectional transport of proteins and RNA–protein complexes between cytoplasm and nucleus. It belongs to a subgroup of nuclear pore complex proteins (NPCs) characterized by a common sequence motif, the phenylalanine–glycine (FG) repeat, that functions as a putative docking site for imported substrates.31,32 So far, the role of the nucleoporins in the malignant transformation of hematopoietic cells is only partly understood, but, importantly, all chimeric genes preserve the FG repeats of the N-terminal half of NUP98.
We now have directly tested the impact of the human leukemia–specific fusion gene ND13 on hematopoietic development. Here, we report that ND13 has potent myeloproliferative activity in vitro and causes myeloproliferative disease in vivo but is insufficient to induce AML in mice. However, ND13, together with the deregulated expression of Meis1, induced AML in all transplanted mice with a short latency time. The hematopoietic effects of ND13 were shown to be dependent on a functional homeodomain and the presence of the NUP98 portion of the fusion gene. These findings support a model in which aberrant activation of HOXD13-dependent pathways might be critical for ND13-associated leukemogenesis.
Materials and methods
cDNA constructs and retroviral vectors
The ND13 fusion gene was constructed by ligation of the 5′ end of the NUP98 cDNA (kindly provided by J. Borrow, Massachusetts Institute of Technology, Cambridge, MA) to the ApaI site upstream of the breakpoint region of the ND13 cDNA fragment isolated from a patient with t-MDS.8 The 1.2-kb coding sequence of Meis1a was kindly provided by G. Sauvageau (Clinical Research Institute of Montreal, Canada). A flagged-tagged version of ND13 was engineered by ligating a polymerase chain reaction (PCR) product of the fusion gene in-frame to the 3′ end of the FLAG epitope in the pSC plasmid (Clontech, Palo Alto, CA). The N51S-ND13 dead homeodomain mutant was constructed using the Quick-Change site-directed mutagenesis kit (Stratagene, La Jolla, CA) and the pSC-ND13 plasmid as a template. The TR-HOXD13 and TR-NUP98 constructs, consisting of the conserved HOXD13 and NUP98 portion of the ND13 fusion gene, respectively, were obtained by PCR and subcloned in-frame into the pSC plasmid. The cDNAs were subcloned into the murine stem-cell virus (MSCV) 2.1 vector (kindly provided by R. Hawley, American Red Cross, Rockville, MD) upstream of the internal ribosomal entry site (IRES) sequence linked to the gene encoding the enhanced green fluorescence protein (EGFP; Clontech) or to the enhanced yellow fluorescence protein (YFP) in the Meis1a construct (Clontech) (kindly provided by P. Leboulch, Harvard–Massachusetts Institute of Technology Division of Health Sciences and Technology, Cambridge, MA). In some experiments, a hemagglutinin (HA)–epitope tagged version of the Meis1a cDNA was used (fused in N-terminus). As a control, the MSCV vector carrying only the IRES-GFP cassette (GFP virus) was used. Production of high-titer, helper-free retrovirus was carried out by standard procedures.33 The presence of full-length provirus was determined by cutting the provirus twice with NheI and using Southern blot analysis with standard techniques.34
Mice and retroviral infection of primary bone marrow cells
Parental strain mice were bred and maintained at the British Columbia Cancer Research Centre animal facility. Donors of primary bone marrow (BM) cells were older than 12-week (C57Bl/6Ly-Pep3b × C3H/HeJ) F1 (PepC3) mice, and recipients were older than 8- to 12-week (C57Bl/6J × C3H/HeJ) F1 (B6C3) mice. Primary mouse BM cells were transduced as previously described.35 Briefly, BM cells were harvested from mice treated 4 days previously with 150 mg/kg 5-fluorouracil (Faulding, Underdaler, Australia) and prestimulated for 48 hours in Dulbecco modified Eagle medium (DMEM) supplemented with 15% fetal bovine serum (FBS), 10 ng/mL human interleukin-6 (hIL-6), 6 ng/mL murine interleukin-3 (IL-3), and 100 ng/mL murine Steel factor (mSF) (StemCell Technologies, Vancouver, BC, Canada). Cells were infected by cocultivation with irradiated (4000 cGy x-ray) GP+E86 viral producer cells with the addition of 5 μg/mL protamine sulfate (Sigma, Oakville, ON, Canada). A 1:1 mixture of ND13/GFP or Meis1/YFP producers was used to coinfect BM cells. Loosely adherent and nonadherent cells were harvested from the cocultures after 2 days and were cultured for 48 hours in the same medium without protamine sulfate.
BM transplantation and assessment of mice
Retrovirally transduced BM cells were highly purified based on the expression of GFP or YFP using a FACS Vantage (Becton Dickinson, Mississauga, ON, Canada). GFP, YFP, or both were excited at 488 nm, and fluorescence emission was detected using a 530/30BP filter (Omega Optical, Brattleboro, VT) with a 560SP dichroic mirror (Omega Optical) if only GFP or YFP was present. It was detected using 510/20BP and 550/30BP optical filters (Omega Optical) and a 525-nm short-pass dichroic mirror (Omega Optical) to separate emission wavelengths if both proteins were present.
Primary and secondary F1 (B6C3) mice were irradiated with 900 cGy of cesium Cs 137 γ-radiation. Irradiated mice underwent transplantation with sorted or unsorted transduced BM cells into the tail vein. Gene transfer efficiency to BM cells ranged from 50% to 90% for GFP, 25% to 40% for Meis1a, 10% to 30% for ND13, 20% to 35% for TR-HOXD13, 10% to 25% for N51S-ND13, and 0.3% to 10% (GFP+/YFP+) for double transduction with ND13 and Meis1a (Tables 1, 2). Peripheral blood (PB) cell progeny of transduced cells was tracked by the expression of GFP, YFP, or both. Engraftment and lineage differentiation were analyzed by the aspiration of cells from the femurs of the mice under light anesthesia or after killing. Lineage distribution was determined by fluorescence-activated cell sorter (FACS) analysis as previously described.36 Monoclonal antibodies (mAbs) were all purchased from PharMingen (San Diego, CA) (phycoerythrin [PE]–labeled Gr-1, Mac-1, B220, Ter-119, CD4, CD8, and c-Kit). Morphologic analysis of PB, BM, and spleen cells was determined from modified Wright-Giemsa–stained cytospin preparations, and nonspecific esterase activity was tested using alpha naphthyl butyrate. For the histologic analysis, selected organs were fixed in a buffered 4% paraformaldehyde solution, dehydrated in ethanol, and embedded in paraffin for sectioning. Sections were prepared and hematoxylin and eosin (H&E)–stained at the Academic Pathology Laboratory (University of British Columbia, Vancouver, BC, Canada) using standard protocols.
Absolute numbers of BM cells transplanted per ND13, Meis1, and GFP mouse
Mouse . | GFP, % . | No. cells/mouse . | No. transplants . |
|---|---|---|---|
| Experiments 1-4 | |||
| GFP | 95 | 1.6-2.4 × 105 | 5 |
| GFP* | 50 | 2.4 × 105 | 2 |
| ND13 | 95 | 2.1 × 105 | 7 |
| ND13* | 25 | 3.5 × 105 | 8 |
| Meis1 | 95 | 1.25 × 105 | 2 |
| Experiments 5-7 | |||
| GFP* | 60-70 | 1.0 × 106 | 5 |
| ND13 | 50 | 1.6 × 105 | 5 |
| Meis1 | 95 | 4.0 × 105 | 2 |
| Meis1* | 35 | 1.0 × 106 | 3 |
Mouse . | GFP, % . | No. cells/mouse . | No. transplants . |
|---|---|---|---|
| Experiments 1-4 | |||
| GFP | 95 | 1.6-2.4 × 105 | 5 |
| GFP* | 50 | 2.4 × 105 | 2 |
| ND13 | 95 | 2.1 × 105 | 7 |
| ND13* | 25 | 3.5 × 105 | 8 |
| Meis1 | 95 | 1.25 × 105 | 2 |
| Experiments 5-7 | |||
| GFP* | 60-70 | 1.0 × 106 | 5 |
| ND13 | 50 | 1.6 × 105 | 5 |
| Meis1 | 95 | 4.0 × 105 | 2 |
| Meis1* | 35 | 1.0 × 106 | 3 |
Unsorted BM cells injected.
Absolute numbers of ND13, Meis1, and ND13-Meis1 cells transplanted per ND13-Meis1 mouse
No. BM cells transplanted . | . | . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Experiment . | ND13 . | Meis1 . | ND13-Meis1 . | Untransduced . | No. transplants . | Median survival, d . | ||||
| 3* | 2.3 × 104 | 2.15 × 104 | 3 × 103 | 9.5 × 105 | 5 | 110 | ||||
| 4 | < 2000 | < 2000 | 1 × 105 | 2.5 × 104 | 4 | 66 | ||||
| 4† | NA | NA | NA | < 2000 | 4 | 88 | ||||
| 5 | < 2000 | < 2000 | 1 × 105 | 2.5 × 104 | 3 | 62 | ||||
| 6 | < 2000 | < 2000 | 1 × 105 | 2.6 × 104 | 3 | 76 | ||||
| 6* | 2 × 105 | 1.1 × 105 | 1 × 105 | 5.9 × 104 | 4 | 72 | ||||
No. BM cells transplanted . | . | . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Experiment . | ND13 . | Meis1 . | ND13-Meis1 . | Untransduced . | No. transplants . | Median survival, d . | ||||
| 3* | 2.3 × 104 | 2.15 × 104 | 3 × 103 | 9.5 × 105 | 5 | 110 | ||||
| 4 | < 2000 | < 2000 | 1 × 105 | 2.5 × 104 | 4 | 66 | ||||
| 4† | NA | NA | NA | < 2000 | 4 | 88 | ||||
| 5 | < 2000 | < 2000 | 1 × 105 | 2.5 × 104 | 3 | 62 | ||||
| 6 | < 2000 | < 2000 | 1 × 105 | 2.6 × 104 | 3 | 76 | ||||
| 6* | 2 × 105 | 1.1 × 105 | 1 × 105 | 5.9 × 104 | 4 | 72 | ||||
Unsorted BM cells injected
Each mice received 1.5 × 105 cells sorted for GFP or YFP expression (inoculum consisted of ND13, Meis1, and ND13- Meis1 cells).
In vitro assays
Cell proliferation was assessed in DMEM supplemented with 15% FBS, 10 ng/mL hIL-6, 6 ng/mL mIL-3, and 100 ng/mL mSF by viable cell counting. Differentiation of clonogenic progenitors was analyzed by plating cells in methylcellulose culture media (Methocult M3434; StemCell Technologies), containing 3 U/mL recombinant human erythropoietin (rhEpo), 10 ng/mL rmIL-3, 10 ng/mL rhIL-6, and 50 ng/mL rmSF. Pre-B progenitors were detected by culture in methylcellulose media containing 10 ng/mL IL-7 for 5 to 7 days (Methocult M3630; StemCell Technologies). All colonies were scored microscopically using standard criteria.
IL-3–dependent cell lines from ND13 or ND13/Meis1-infected bone marrow cells were established in vitro directly after sorting in DMEM/15% FBS with IL-3 alone (6 ng/mL). The differentiation capacity of cell lines was tested in DMEM/15% FBS supplemented with 100 ng/mL granulocyte–colony-stimulating factor (G-CSF) or 10 ng/mL macrophage-CSF (M-CSF) (R&D Systems, Wiesbaden, Germany) and after 7-day analysis of Wright-Giemsa–stained cytospin preparations.
CFU-S assay
BM cells from day 4 (d4) 5-fluorouracil (5-FU)–treated PepC3 donor mice were infected with the different viruses, and transduced BM cells were purified by FACS. To assess initial (d0) colony-forming spleen unit (CFU-S) numbers, 5000 cells were injected per lethally irradiated mouse 48 hours after infection. CFU-S numbers after 7 days of culture in DMEM, supplemented with 15% FBS, 10 ng/mL hIL-6, 6 ng/mL mIL-3, and 100 ng/mL mSF, were measured by injection of the total cell progeny derived from 25 to 1 × 106 input cells. Macroscopic colonies on the spleen at day 12 after injection were counted after fixation in Telleyesnickzky solution.
Semiquantitative RT-PCR
Expression of selected Hox genes and Meis1 was assayed by reverse transcription–polymerase chain reaction (RT-PCR) of RNA recovered from bone marrow cells harvested from sick or control mice or from GFP control, ND13, or ND13+Meis1–transduced cells isolated by FACS 48 hours after infection and maintained in extended liquid culture for at least 28 days in DMEM supplemented with 15% FBS, 10 ng/mL hIL-6, 6 ng/mL mIL-3, and 100 ng/mL mSF. RNA was isolated using RNAzol reagent (all reagents for RT-PCR were from Invitrogen, Burlington, ON, Canada) and were treated with DNase I (amp grade) to remove contaminating genomic DNA. cDNA was synthesized from 500 ng total RNA by random priming using SuperScript II reverse transcriptase, as recommended by the manufacturer. PCR conditions were identical for all genes (annealing temperature of 60°C) using the cDNA amount originating from 5 ng starting RNA. Conditions used were those recommended by the manufacturer of Platinum Taq Polymerase. The number of PCR cycles for each gene was chosen to stop the reaction in the linear phase of the amplification (20 cycles for glyceraldehyde-3-phosphate dehydrogenase [GAPDH] and total Meis1; 30 for Hoxa7 and Hoxa9; 20 and 30 for endogenous Meis1 for in vitro and in vivo samples, respectively). No significant expression was detected for Hoxd13 using up to 40 PCR cycles. Amplified products were separated on agarose gel and transferred to Zeta probe membrane (Bio-Rad, Hercules, CA) and were hybridized to randomly primed radioactive gene-specific DNA (p32-α–adenosine triphosphate [ATP]) fragments. The intensity for each signal was measured using a PhosphorImager STORM 860 and ImageQuant software from Molecular Dynamics (Sunnyvale, CA). The expression for each gene was normalized first to that of GAPDH and then to the value expressed relative to control BM (from a healthy mouse), which was given an arbitrary value of 1. Primer sequences are available on request.
Western blot analysis
Nuclear extract from 293T cells was prepared as follows: cells were washed twice in cold phosphate-buffered saline (PBS), then resuspended in hypotonic solution (10 mM N-2-hydroxyethylpiperazine-N'-2-ethanesulfonic acid [HEPES] pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 10% glycerol) for 10 minutes on ice. Cells were then spun down and resuspended in 1 mL hypotonic solution and homogenized using a Dounce tissue grinder. The cells were spun down and resuspended in a lysis buffer (20 mM HEPES pH 7.9, 400 mM KCl, 5 mM MgCl2, 25% glycerol); after 40 minutes of incubation, the supernatant was harvested (nuclear extract). The nuclear extract was electrophoresed in sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) 12.5% or a gradient gel (4%-12.5%) (Nu-PAGE; Invitrogene, Burlington, ON, Canada) and was blotted to nitrocellulose membranes (Millipore, Bedford, MA) as previously described.37 Antibodies used consisted of monoclonal anti-FLAG (M2; Sigma), an anti-HA (16B12; Babco, Richmond, CA), and donkey horseradish peroxidase–conjugated antimouse antibody (Jackson ImmunoResearch Lab, West Grove, PA). Protein expression was detected using an enhanced luminol reagent (Renaissance, Boston, MA).
Statistical analysis
Data were statistically tested using the t test for dependent or independent samples (Microsoft Excel). Differences with P < .05 were considered statistically significant.
Results
Retroviral vectors
To engineer and study the effect of expression of the ND13 fusion gene in hematopoietic cells, we generated the MSCV-based retroviral vector depicted in Figure 1A in which the ND13 cDNA was linked to the EGFP marker gene by an IRES element. In some experiments, an analogous vector carrying the Meis1a cDNA linked to the YFP marker gene was used (Figure 1A). Transmission of full-length provirus to target cells was demonstrated by Southern blot analysis of infected BM (data not shown) for each vector and expression of ND13 or Meis1 was verified by Western blot analysis of a FLAG- or an HA-tagged version of each vector, respectively (Figure 1B). As a control, an MSCV virus carrying GFP alone (GFP virus) was used.
ND13 expression in BM confers a growth advantage to transduced BM cells. (A) Retroviral vectors used to express ND13 and Meis1 in murine BM. The FG repeats of the NUP98 gene and the homeodomain of HOXD13 are illustrated as hatched boxes and black boxes, respectively. (B) Western blot analysis of nuclear extracts from 293T cells transfected with FLAG-ND13 and HA-tagged Meis1 as detected with an anti-FLAG and an anti-HA monoclonal antibody, respectively. Molecular mass markers are indicated. (C) Cell expansion of transduced (GFP+) and nontransduced (GFP-) cells in cultures initiated with BM infected with GFP control or ND13 virus (mean ± SD). Results of a representative experiment (n = 3) are shown; each experiment was performed in triplicate cultures. (D) Wright-Giemsa (original magnification, × 400) staining of a representative ND13-transduced, IL-3–dependent cell line that underwent monocytic differentiation with M-CSF stimulation but remained predominantly blastic without granulocytic differentiation with G-CSF stimulation. ND13 indicates NUP98-HOXD13; FS, fusion site; LTR, long terminal repeat; and CTL, control.
ND13 expression in BM confers a growth advantage to transduced BM cells. (A) Retroviral vectors used to express ND13 and Meis1 in murine BM. The FG repeats of the NUP98 gene and the homeodomain of HOXD13 are illustrated as hatched boxes and black boxes, respectively. (B) Western blot analysis of nuclear extracts from 293T cells transfected with FLAG-ND13 and HA-tagged Meis1 as detected with an anti-FLAG and an anti-HA monoclonal antibody, respectively. Molecular mass markers are indicated. (C) Cell expansion of transduced (GFP+) and nontransduced (GFP-) cells in cultures initiated with BM infected with GFP control or ND13 virus (mean ± SD). Results of a representative experiment (n = 3) are shown; each experiment was performed in triplicate cultures. (D) Wright-Giemsa (original magnification, × 400) staining of a representative ND13-transduced, IL-3–dependent cell line that underwent monocytic differentiation with M-CSF stimulation but remained predominantly blastic without granulocytic differentiation with G-CSF stimulation. ND13 indicates NUP98-HOXD13; FS, fusion site; LTR, long terminal repeat; and CTL, control.
ND13 enhances growth of early hematopoietic progenitor cells in vitro
To study the direct impact of the ND13 on hematopoietic differentiation and proliferation, we introduced the fusion gene into primary murine BM cells by retroviral gene transfer and assessed various growth properties in vitro in comparison to nontransduced or control GFP transduced BM cells.
In liquid cultures containing IL-3, IL-6, and SF, the proportion of GFP+ cells in the control culture remained constant at input levels (approximately 42.5%) over 2 weeks. In contrast, in cultures initiated with cells infected with the ND13 virus, the proportion of transduced ND13-GFP–expressing cells expanded from input levels of 35% to 94.5% ± 1.9% after 2 weeks. This competitive outgrowth of ND13-GFP+–transduced cells was further evidenced by their 40-fold greater increase in total cell number compared with GFP+ control cells (Figure 1C; total net fold expansion of 2.5 × 104; P < .003). Moreover, IL-3–dependent cell lines were readily established from expansion cultures of ND13-transduced cells that showed blast morphology on Wright-Giemsa–stained cytospin preparations. Interestingly, these cell lines retained limited capacity to undergo macrophage differentiation after stimulation with M-CSF, but most cells failed to differentiate into granulocytes following G-CSF stimulation (Figure 1D).
The growth-promoting effect of ND13 was further seen in clonogenic progenitor assays in methylcellulose culture. Although the frequency of clonogenic progenitors detected in FACS-purified GFP+ cells was essentially identical for control and ND13 cells (data not shown, n = 7), ND13-transduced progenitors formed much larger granulo-monocytic (GM) colonies (an average of 6.4 × 104 ± 1.9 × 104 cells per ND13 colony vs 3.8 × 103 ± 1.8 × 103 per control GFP colony). Moreover, ND13 GM progenitors cells were capable of forming equivalent large secondary colonies in elevated numbers on serial replatings (4 or more), whereas GFP+ control cells produced only mast cell colonies after one replating.
Growth-promoting effect of ND13 is dependent on an intact HOXD13 homeodomain and the NUP98 portion
To investigate further the effect of ND13 on more primitive hematopoietic cells, we used the colony-forming spleen assay (CFU-S). ND13-GFP+- and control GFP+-transduced BM cells isolated by FACS were injected into lethally irradiated mice 48 hours after infection. Although the d12 CFU-S frequency was comparable between the control GFP and ND13-transduced cells (Figure 2A), the colonies arising from ND13-transduced cells were significantly larger. Moreover, closer investigation of the ND13 d12 CFU-S progeny cells indicated a marked reduction in the proportion of Ter-119+ erythroid cells (9% ± 1% vs 56% ± 11% for controls) and a concomitant increase in the proportion of Gr-1+ myeloid cells (70% ± 12% versus 12% ± 2% for controls) (Figure 2B). The impact of the ND13 fusion gene on primitive hematopoietic progenitor cells was further evident following 1 week of liquid culture, which led to more than a 1300-fold increase in the yield of d12 CFU-S compared with untransduced BM cells or more than 5000-fold when compared with GFP+ control transduced cells (Figure 2D).
Fusion gene ND13 perturbs the differentiation and proliferative potential of primitive myeloid progenitors, and its activity is dependent on the HOXD13 homeodomain and NUP98 portion. (A) Total number of d12 CFU-S derived from 5000 FACS-purified control GFP or ND13-transduced injected BM cells (mean ± SD, n = 2). (B) Lineage distribution of GFP+ d12 CFU-S–derived transduced cells. 2 × 104 cells recovered after infection were injected without selection into irradiated mice, and single spleen cell suspensions (2 spleens/construct) were analyzed by FACS 12 days later for GFP and lineage-specific marker expression (n = 2; representative experiment shown). □ indicates GFP; ▦, ND13. (C) Expression of FLAG-tagged ND13 wild-type and mutant constructs in nuclear extracts of transfected 293T cells detected by Western blot using an anti-FLAG monoclonal antibody. (D) Total number of d12 CFU-S colonies derived per culture initiated with 1 × 106 cells transduced with the GFP, ND13, and mutant viruses after 1 week in liquid culture. Yields for TR-HOXD13 and ND13-N51S were significantly different from those for ND13 (P < .03) (mean ± SD; n = 3 for all except TR-HOXD13 [n = 2]).
Fusion gene ND13 perturbs the differentiation and proliferative potential of primitive myeloid progenitors, and its activity is dependent on the HOXD13 homeodomain and NUP98 portion. (A) Total number of d12 CFU-S derived from 5000 FACS-purified control GFP or ND13-transduced injected BM cells (mean ± SD, n = 2). (B) Lineage distribution of GFP+ d12 CFU-S–derived transduced cells. 2 × 104 cells recovered after infection were injected without selection into irradiated mice, and single spleen cell suspensions (2 spleens/construct) were analyzed by FACS 12 days later for GFP and lineage-specific marker expression (n = 2; representative experiment shown). □ indicates GFP; ▦, ND13. (C) Expression of FLAG-tagged ND13 wild-type and mutant constructs in nuclear extracts of transfected 293T cells detected by Western blot using an anti-FLAG monoclonal antibody. (D) Total number of d12 CFU-S colonies derived per culture initiated with 1 × 106 cells transduced with the GFP, ND13, and mutant viruses after 1 week in liquid culture. Yields for TR-HOXD13 and ND13-N51S were significantly different from those for ND13 (P < .03) (mean ± SD; n = 3 for all except TR-HOXD13 [n = 2]).
In an effort to functionally characterize the contribution of the HOXD13 and NUP98 portion to the hematopoietic effects of the ND13 fusion gene, 2 mutants were designed. The first mutant consisted of a truncated version of the fusion gene, which deleted the entire NUP98 portion (TR-HOXD13). In the second case, a dead-homeodomain ND13 mutant was created in which the highly conserved asparagine residue at position 51 in the homeodomain of HOXD13 was changed to serine (N51S-ND13). This mutation has been shown to eliminate the DNA-binding activity of various homeobox genes.38,39 Western blot analysis of 293T transfected cells confirmed that all mutant proteins were expressed (Figure 2C). The various mutants were directly tested in BM cells using the d12 CFU-S assay after 7 days of liquid culture and the growth competition assay in liquid culture.
In contrast to the marked increase in d12 CFU-S output seen in culture of ND13-transduced cells, the TR-HOXD13 and the ND13-N51S mutants yielded CFU-S numbers equivalent to those observed with the nontransduced or GFP-transduced controls (Figure 2D). Consistent with this loss of proliferative effect, none of the mutants conferred a proliferative advantage on transduced cells, as tested in liquid expansion competition culture or by colony replating (data not shown). In agreement with these results, a third mutant, consisting of the NUP98 portion only, also failed to reproduce any of the ND13 effects (data not shown). Collectively, these results demonstrate that the hematopoietic stimulatory activity of the chimeric protein is derived from both the NUP98 and the HOXD13 portion. Furthermore, the requirement of an intact HOXD13 homeodomain for functional activity indicates a potential role of the fusion gene as an aberrant transcription factor, likely involving HOX-dependent pathways.
Expression of ND13 perturbs myeloid and lymphoid development in vivo and induces myeloproliferative diseases
To assess the long-term effect of ND13 on hematopoiesis, we exploited the murine BM transplantation model. BM cells were harvested 48 hours following infection with ND13 or control GFP virus and then transplanted with or without FACS selection for transduced (GFP+) cells into lethally irradiated recipients (Table 1).
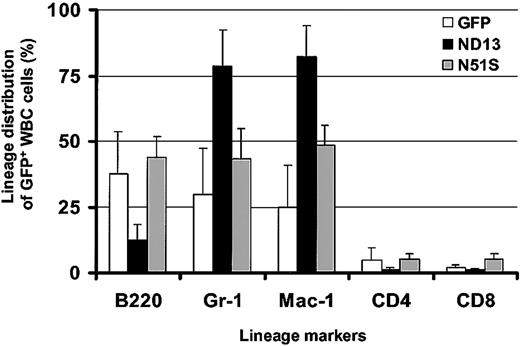
Remarkably, in view of the growth-promoting effect of ND13 seen in vitro, analysis of the peripheral blood 10 weeks after transplantation revealed a lower level of engraftment with transduced GFP+ cells in ND13 recipient mice (mean, 40.9% GFP+ white blood cells (WBCs); range, 22%-60%) compared with control GFP mice (mean, 55.5% GFP+ WBCs; range, 44%-66%) and a significant decrease in WBC number (3.7 ± 0.7 × 106/mL for ND13 vs 6.7 ± 1.1 × 106 for GFP [8 mice/arm; P < .0003]). Flow cytometry analysis indicated that the reduction of WBCs in ND13 recipients was the result of a substantial decrease in the level of B220+GFP+ B-lymphocytes and of CD4/CD8+GFP+ T cells (Figure 3). Most GFP+ WBCs in ND13 mice was myeloid, as evidenced by the high proportion of Gr-1+ and Mac-1+ cells. This impairment in lymphopoiesis by ND13-transduced cells was evident as early as 4 weeks after transplantation and was not seen in mice that received transplants of the homeodomain mutant (ND13-N51S) version of the fusion gene (Figure 3).
Expression of ND13 in vivo leads to impaired lymphopoiesis. Lineage distribution of transduced (GFP+) WBCs in recipient reconstituted with GFP-, ND13-, and ND13-N51S–transduced BM cells 4 weeks after transplantation (mean ± SD for 3-5 animals).
Expression of ND13 in vivo leads to impaired lymphopoiesis. Lineage distribution of transduced (GFP+) WBCs in recipient reconstituted with GFP-, ND13-, and ND13-N51S–transduced BM cells 4 weeks after transplantation (mean ± SD for 3-5 animals).
The impact of the ND13 fusion gene on lymphoid development was underlined by analyses of BM aspirates obtained from mice 8 weeks after transplantation. Quantification of IL-7–sensitive clonogenic B-cell precursors in the BM aspirates using the pre-B–cell colony assay showed a 10-fold reduction in ND13 transplanted mice compared with control transplanted mice (6.7 ± 0.7 vs 70 ± 1 per 1 × 106 BM cells, respectively [2 mice/arm]). In contrast, an ND13 growth-promoting of 16-fold effect was evident for myeloid cells in ex vivo liquid culture supplemented with IL-3 after 14 days, and it induced the outgrowth of a monoblastic cell population with partial differentiation into macrophages after 3 weeks.
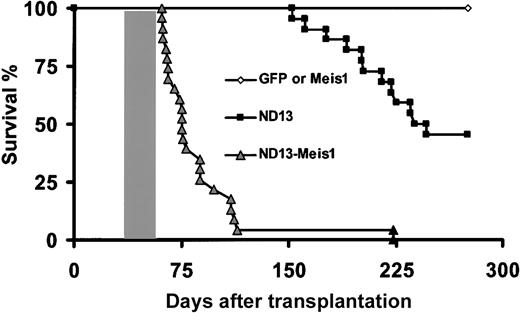
One ND13 mouse was dead 152 days after transplantation with splenomegaly (0.66 g vs 0.10 g for CTL), and stable ND13 provirus integration was confirmed. Further analysis of the remaining cohort at 22 weeks revealed 3 of 23 ND13 recipients with signs of myeloproliferation, as evidenced by elevated WBC counts (range, 20-90 × 106/mL). Two of those mice developed anemia (2.7-3.2 × 109/mL), and 1 was killed for additional analysis; it had an enlarged spleen (0.28 g), elevated WBC count (70 × 106/mL), and BM cells that stained positive for Gr-1 and Mac-1 (82% and 98% of GFP+ cells in BM, respectively). The accumulation of mature neutrophils in the PB, BM, and spleen confirmed the myeloproliferative disease. By 250 days after transplantation, 7 of 23 ND13 recipients had died (Figure 4), and 5 additional diseased primary recipients were killed and analyzed. These mice had anemia (3.2 ± 1.1 × 109/mL), splenomegaly (0.15-0.31 g), and, with one exception, normal or inferior WBC levels (mean, 7.9 ± 6.1 × 106 WBC/mL). Flow cytometry confirmed the myeloid nature of the ND13 BM cells (62% ± 25% and 67% ± 27% of GFP+ cells for GR-1 and Mac-1, respectively). Morphologic examination of the marrow of ND13 mice revealed no significant increase in the proportion of blasts cells (7% ± 4.5% vs 6.2% ± 0.5% for control BM) and increased levels of myeloid cells in their spleens (50%-100%). Histopathologic analysis of 4 diseased ND13 mice failed to reveal any significant perivascular infiltration in the kidney, lung, and liver (with the exception of 1 for the latter). These data underline that ND13 as a single factor induces significant perturbations of the proliferation and differentiation program of hematopoietic progenitor cells in vivo. Over time, the fusion gene can cause a myeloproliferative syndrome.
Survival curves. Survival curves are given for mice that received transplants of BM cells transduced with ND13 (n = 23), GFP control (n = 8), Meis1 (n = 6), or ND13 plus Meis1 (n = 21) viruses. Mice that received transplants of ND13-N51S (n = 6) and ND13-N51S-Meis1–transduced BM cells (n = 3) remained healthy (data not shown). Shaded box indicates the range of survival time for secondary transplant recipients of cells recovered from leukemic ND13-Meis1 primary mice. Two diseased ND13 mice that died 398 and 410 days after transplantation are not represented on this survival curve.
Survival curves. Survival curves are given for mice that received transplants of BM cells transduced with ND13 (n = 23), GFP control (n = 8), Meis1 (n = 6), or ND13 plus Meis1 (n = 21) viruses. Mice that received transplants of ND13-N51S (n = 6) and ND13-N51S-Meis1–transduced BM cells (n = 3) remained healthy (data not shown). Shaded box indicates the range of survival time for secondary transplant recipients of cells recovered from leukemic ND13-Meis1 primary mice. Two diseased ND13 mice that died 398 and 410 days after transplantation are not represented on this survival curve.
ND13 in collaboration with Meis1 induces fatal acute myeloid leukemia in vivo
Because the expression of ND13 alone was not associated with AML over the observation time extending up to 13 months, we were interested in assessing whether ND13 would be leukemogenic on the coengineered overexpression of Meis1, a nonclustered homeobox gene previously implicated in HOX-associated leukemogenesis.14,40,41 Lethally irradiated mice received transplanted BM cells infected singly with a Meis1-YFP virus (40%-60% YFP+ [n = 7]) or with enriched (80% GFP/YFP+ [n = 10]) or nonpurified (0.3%-10% GFP/YFP+(n = 9) BM cells coinfected with the ND13 and the Meis1 viruses (Table 2). In striking contrast to the ND13 and Meis1 mice, all mice that received transplanted BM cells cotransduced with ND13 and Meis1 rapidly fell ill. Their conditions quickly deteriorated, and they became moribund with a median latency of 75 days, as evidenced by significant weight loss, shortness of breath, and lethargy. They were killed for further analysis (Figure 4; Table 2).
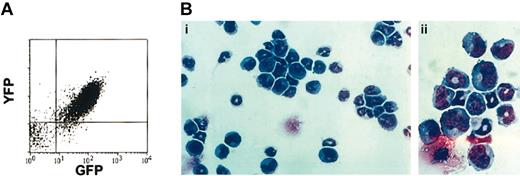
Diseased ND13-Meis1 mice were characterized by extremely elevated peripheral nucleated cell counts (range, 100-575 × 106/mL), severe anemia, and splenomegaly (Table 3). Proviral integration of ND13 and Meis1 was confirmed by Southern blot analysis of leukemic marrow (data not shown) and coexpression of the IRES-linked reporter genes GFP and YFP, respectively, as shown by flow cytometry (Figure 5A). Detailed hematologic analyses of the leukemic mice revealed a high percentage of blasts in the BM (65.0% ± 11.4%), peripheral blood (45.7% ± 13.7%), and spleen (47.2% ± 8.0%), confirming the diagnosis of AML (Table 3; Figure 5B). Peripheral blood and BM blast cells were negative for N-acetyl-chloroacetate esterase staining, consistent with a primitive myeloblastic rather than a myelomonocytic phenotype.
Hematologic characteristics of diseased mice given transplants of ND13-Meis1-cotransduced BM cells
. | . | Peripheral blood . | . | Blast, % . | . | Spleen . | ||
|---|---|---|---|---|---|---|---|---|
| Primary mouse . | Disease onset, d . | WBC, × 106/mL . | RBC, × 109/mL . | PB . | BM . | Weight, g . | ||
| Experiment 3 | ||||||||
| ND13-Meis1 no. 1* | 112 | 385 | 2.05 | ND | ND | 0.39 | ||
| Experiment 4 | ||||||||
| ND13-Meis1 no. 2 | 98 | 210 | 1.20 | ND | ND | 0.58 | ||
| ND13-Meis1 no. 3 | 110 | 100 | 0.85 | 36.0 | 49.3 | 0.31 | ||
| ND13-Meis1 no. 4† | 88 | 107 | 3.05 | 63.0 | 80.3 | 0.41 | ||
| ND13-Meis1 no. 5† | 88 | 151 | 10.0 | ND | ND | 0.37 | ||
| Experiment 5 | ||||||||
| ND13-Meis1 no. 6 | 65 | 573 | 3.65 | 39.0 | 62.5 | 0.51 | ||
| ND13-Meis1 no. 7 | 61 | 100 | 1.3 | ND | ND | 0.34 | ||
| Experiment 6 | ||||||||
| ND13-Meis1 no. 8† | 64 | 352 | NA | NA | NA | NA | ||
| ND13-Meis1 no. 9† | 74 | 245 | 0.9 | 52.1 | 66.1 | 0.43 | ||
| ND13-Meis1 no. 10 | 76 | 150 | 1.7 | 27.4 | 57.0 | 0.35 | ||
. | . | Peripheral blood . | . | Blast, % . | . | Spleen . | ||
|---|---|---|---|---|---|---|---|---|
| Primary mouse . | Disease onset, d . | WBC, × 106/mL . | RBC, × 109/mL . | PB . | BM . | Weight, g . | ||
| Experiment 3 | ||||||||
| ND13-Meis1 no. 1* | 112 | 385 | 2.05 | ND | ND | 0.39 | ||
| Experiment 4 | ||||||||
| ND13-Meis1 no. 2 | 98 | 210 | 1.20 | ND | ND | 0.58 | ||
| ND13-Meis1 no. 3 | 110 | 100 | 0.85 | 36.0 | 49.3 | 0.31 | ||
| ND13-Meis1 no. 4† | 88 | 107 | 3.05 | 63.0 | 80.3 | 0.41 | ||
| ND13-Meis1 no. 5† | 88 | 151 | 10.0 | ND | ND | 0.37 | ||
| Experiment 5 | ||||||||
| ND13-Meis1 no. 6 | 65 | 573 | 3.65 | 39.0 | 62.5 | 0.51 | ||
| ND13-Meis1 no. 7 | 61 | 100 | 1.3 | ND | ND | 0.34 | ||
| Experiment 6 | ||||||||
| ND13-Meis1 no. 8† | 64 | 352 | NA | NA | NA | NA | ||
| ND13-Meis1 no. 9† | 74 | 245 | 0.9 | 52.1 | 66.1 | 0.43 | ||
| ND13-Meis1 no. 10 | 76 | 150 | 1.7 | 27.4 | 57.0 | 0.35 | ||
ND indicates not determined.
Unsorted BM cells injected.
Received a mix of ND13, Meis1, and ND13-Meis1 BM cells.
Diseased mice that received transplanted BM cotransduced with ND13 and Meis1 died of AML. (A) Analysis of YFP and GFP expression in leukemic marrow cells. (B) Wright-Giemsa staining of ND13-Meis1 BM cells. Original magnification, × 32 (i); × 80 (ii).
Diseased mice that received transplanted BM cotransduced with ND13 and Meis1 died of AML. (A) Analysis of YFP and GFP expression in leukemic marrow cells. (B) Wright-Giemsa staining of ND13-Meis1 BM cells. Original magnification, × 32 (i); × 80 (ii).
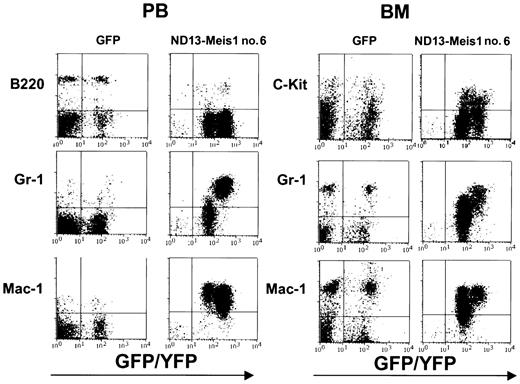
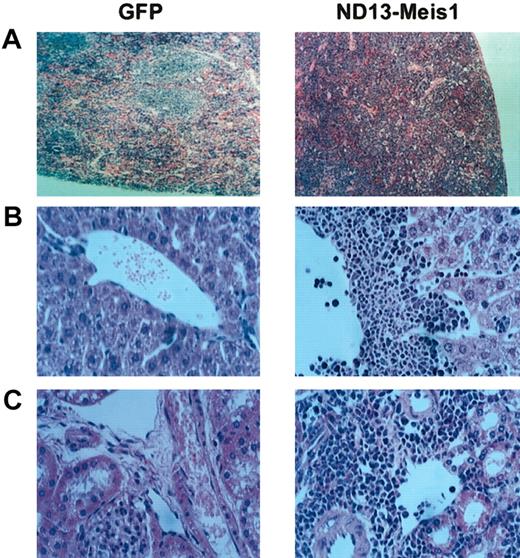
The myeloblastic characteristic of the disease was further revealed by immunophenotyping (Figure 6), which showed the expression of Mac-1 and Gr-1 on most of the transduced cells (86.6 ± 8.9% and 74.0 ± 7.6%, respectively (± SEM; 10 mice analyzed from 4 experiments) in the BM. The same phenotype was also observed for circulating WBCs (Figure 6) and those isolated from the spleen (not shown). Histopathologic analysis of the diseased mice showed massive cell infiltration in the spleen with an effacement of its normal germinal center architecture (Figure 7A). This cell infiltration extended to other organs with prominent perivascular infiltrations in the liver, kidneys (Figure 7B-C), and, to a lesser extent, in the lungs.
Immunophenotyping of leukemic cells derived from an ND13-Meis1 mouse. Shown are flow cytometric profiles for peripheral blood and BM of a GFP control and a representative ND13-Meis1 leukemic mouse.
Immunophenotyping of leukemic cells derived from an ND13-Meis1 mouse. Shown are flow cytometric profiles for peripheral blood and BM of a GFP control and a representative ND13-Meis1 leukemic mouse.
Diseased ND13-Meis1 mice are characterized by massive perivascular infiltration of leukemic myeloblastic cells. (A) Histologic analysis of spleen (original magnification, × 10). (B) Histologic analysis of liver (original magnification, × 32). (C) Histologic analysis of kidneys (original magnification, × 32).
Diseased ND13-Meis1 mice are characterized by massive perivascular infiltration of leukemic myeloblastic cells. (A) Histologic analysis of spleen (original magnification, × 10). (B) Histologic analysis of liver (original magnification, × 32). (C) Histologic analysis of kidneys (original magnification, × 32).
The strong oncogenic collaboration between these 2 genes was corroborated by the fact that all recipients (n = 9) of transplants with a nonpurified transduced BM cell mix, initially containing a low (0.3%-10.0%) proportion of doubly transduced cells, also developed leukemia. As few as 3000 doubly infected BM cells (out of an inoculum of 1 × 106 cells) were found sufficient to induce leukemia in all mice that received transplants. Finally, all 4 mice that received transplants with a mixture of ND13, Meis1, and ND13-Meis1 transduced cells (GFP+, YFP+, or both sorted cells) (experiment 4) also rapidly succumbed to leukemia originating exclusively in the GFP+/YFP+ doubly transduced cells, thus demonstrating the leukemogenic potential of only ND13-Meis1 cotransduced cells.
ND13-Meis1–induced acute leukemia was transplantable, and all secondary irradiated recipients that received 1.5 × 106 BM cells (one third normal and two thirds leukemic cells) developed aggressive fatal acute myeloblastic leukemia, with a disease-onset median of 29 days (Figure 4; 6 primary animals tested [3 secondary transplants per primary mouse]). As previously observed, the diseased transplants were characterized by elevated levels of WBCs (70-469 × 106 WBC/mL), anemia (1.5-4.6 × 109 RBC/mL), and splenomegaly (0.2-0.5 g).
Expression of Hoxa7 and Hoxa9 but not Meis1 is up-regulated in NUP98-HOXD13 BM cells
Given the strong synergy between Meis1 and ND13 to induce AML, we asked whether BM cells from mice that became sick after the transplantation of ND13-transduced cells had increased expression of the endogenous Meis1 gene. Semiquantitative RT-PCR showed no increase in the level of endogenous Meis1 transcripts in diseased ND13 mice compared with healthy mice or in ND13 versus GFP control transduced cells placed in extended liquid culture (LC) and analyzed after 28 days (Figure 8A-B). In contrast, BM from diseased ND13-Meis1 mice or doubly transduced cells recovered after extended liquid culture had highly elevated total Meis1 transcripts (proviral derived plus endogenous; Figure 8A) compared with levels in normal BM (more than 60-fold higher) or GFP control-transduced cells in liquid culture (more than 5-fold), respectively. This increase was clearly the result of retrovirally driven Meis1 expression because expression of the endogenous Meis1 gene (Figure 8B) was reduced in doubly transduced cells compared with controls, suggesting a possible negative autoregulatory mechanism of regulation for Meis1 expression.
Expression ofMeis1,Hoxa7,andHoxa9in BM cells from sick ND13 and ND13-Meis1 mice. Expression of Meis1 transcripts (A), endogenous Meis1 (B), and Hoxa7 and Hoxa9 (C) in BM cells harvested from sick mice or transduced cells grown in LC. Mean expression shown for ND13 (n = 5), ND13-Meis1 (n = 5), sick mice, and control mouse (n = 1). LC samples consisted of transduced BM cells isolated by FACS and cultured for more than 28 days in 15% FBS, IL-3, hIL-6, and mSF (n = 1 for GFP+ control and ND13-Meis1 [ND/M] and n = 2 for ND13). For semiquantitative RT-PCR, all expression was normalized to GAPDH and then relative to control marrow, which was set at an arbitrary value of 1, except for the expression of endogenous Meis1 in the LC samples, which is given relative to GFP+ control. See “Materials and methods” for details. Endogenous Meis1-specific primers were designed not to amplify retrovirally driven Meis1 transcript. Mean ± SD shown. Similar analysis carried out for Hoxd13 was negative for expression (data not shown).
Expression ofMeis1,Hoxa7,andHoxa9in BM cells from sick ND13 and ND13-Meis1 mice. Expression of Meis1 transcripts (A), endogenous Meis1 (B), and Hoxa7 and Hoxa9 (C) in BM cells harvested from sick mice or transduced cells grown in LC. Mean expression shown for ND13 (n = 5), ND13-Meis1 (n = 5), sick mice, and control mouse (n = 1). LC samples consisted of transduced BM cells isolated by FACS and cultured for more than 28 days in 15% FBS, IL-3, hIL-6, and mSF (n = 1 for GFP+ control and ND13-Meis1 [ND/M] and n = 2 for ND13). For semiquantitative RT-PCR, all expression was normalized to GAPDH and then relative to control marrow, which was set at an arbitrary value of 1, except for the expression of endogenous Meis1 in the LC samples, which is given relative to GFP+ control. See “Materials and methods” for details. Endogenous Meis1-specific primers were designed not to amplify retrovirally driven Meis1 transcript. Mean ± SD shown. Similar analysis carried out for Hoxd13 was negative for expression (data not shown).
Given that Hoxa7 and Hoxa9 have been implicated previously in leukemia with or without Meis114,25,42 and that expression of the normal HOXD13 allele has been reported in a patient with ND13 leukemia,21 we also investigated whether the expression of these Hox genes would be increased in sick mice or ND13 and ND13-Meis1 cell lines. Expression of Hoxa7 and Hoxa9 was elevated in sick ND13 and ND13-Meis1 mice and in ND13 and ND13-Meis–transduced cell lines compared with low levels observed for control whole marrow or GFP control–transduced cell lines (Figure 8C). Endogenous Hoxd13 expression was not detected in any samples (more than 40 amplification cycles; data not shown). Whether the observed elevations in Hoxa7 and Hoxa9 expression are causally linked to the transformation process or simply reflect the differentiation block induced by the fusion gene remains unclear.43 Consistent with the latter interpretation, ND13-transduced BM cells in extended liquid culture were poorly differentiated (small cytoplasm) and formed large colonies in methylcellulose with a high plating efficiency (approximately 1 of 15), whereas control GFP cultures had a differentiated phenotype (larger cytoplasm, increased granules) and only formed small colonies in methylcellulose with a low plating efficiency (approximately 1 of 50).
Discussion
Chromosomal rearrangements involving the nucleoporin gene NUP98 on chromosome 11p15 have defined a novel group of t-MDS/AML and de novo AML.7, 8, 9,12,44 ND13 t(2;11), however, describes a novel class of HOX gene-associated leukemias, linking de novo expression of a nonhematopoietic clustered HOX gene, normally involved in embryogenesis, to human leukemogenesis. We now have directly demonstrated that ND13 itself induces severe perturbations of the normal proliferation and differentiation program of early hematopoietic progenitor cells. We also demonstrated that ND13 can rapidly induce acute myeloblastic leukemia when coexpressed with the TALE homeobox gene Meis1. Furthermore, our data emphasize the key role of the HOXD13 portion of the fusion gene for its hematopoietic activity.
The direct involvement of HOXD13 in human leukemia is provocative given that HOXD cluster genes are not normally expressed in hematopoietic cells and that little is known about the involvement of the 13-paralog group in the regulation of hematopoiesis. Remarkably then, many of the in vitro and in vivo hematopoietic effects induced by ND13 expression are highly reminiscent of the effects of retrovirally overexpressed hematopoietic HOX genes.3 Thus, for example, the dramatic growth stimulation on primitive CFU-S myeloid progenitors in liquid culture induced by ND13 mimics that seen for HOXA10.15 Similarly, the inhibitory effect of ND13 on lymphoid growth in vivo is reminiscent of that seen following the overexpression of HOXB3 or HOXA10, each of which can induce AML after a long latency period.15,36 The striking similarities induced by these 2 HOX proteins from 2 different clusters and that of the chimeric HOXD13 proteins suggest a high level of functional redundancy among HOX proteins in hematopoiesis, as previously observed in embryogenesis.45,46
Our results, coupled with observations of other groups, underline that the NUP98 portion plays a major role in the transforming potential of the NUP98-HOX fusion genes. Although juxtaposition of the HOXD13 second exon next to NUP98 following t(2;11) leads to the aberrant expression of the HOXD13 homeodomain, this is clearly not the sole function of the NUP98 portion because expression of this short truncated HOXD13 portion alone did not cause any significant hematopoietic perturbations. The activity of ND13 is also strictly dependent on the HOXD13 portion and, more precisely, on its DNA-binding homeodomain. Our findings are consistent with the structure–function analysis of NUP98-HOXA9 done by Kasper et al,47 who showed that the ability of NUP98-HOXA9 to transform fibroblasts is dependent on the NUP98 portion, especially of its FG repeats. The latter, found conserved in all NUP98-HOX fusion genes, may promote transcription by mediating the recruitment of CBP and p300,47 2 transcriptional coactivators of a variety of transcription factors including HOX proteins.24,48 Together, these data support a model in which the ND13 chimeric protein forms an aberrant HOX DNA-binding transcription factor molecule, in which the HOXD13 and NUP98 portions provide the DNA binding motif and transcriptional activation domain, respectively.
The long latency time observed before the onset of hematologic disease in the ND13 recipient suggests that additional mutation(s) are required for the development of frank leukemia. Our data reveal that the TALE homeobox gene Meis1 is a strong potential collaborative gene in ND13-associated leukemia, converting ND13-induced nonleukemic alterations in growth and differentiation of early progenitor cells to frank aggressive myeloblastic leukemia. Most mice (more than 66%) receiving ND13-Meis1–cotransduced BM cells developed AML within 75 days, suggesting that the synergy between both genes, without further mutation, may be sufficient for full oncogenicity.
HOXD13, as an AbdB-like HOX gene, can directly interact with Meis1, forming heterodimeric DNA-binding complexes.38,49,50 However, the mode of synergy between Meis1 and the ND13 fusion gene is unclear. Two models are conceivable: (1) both proteins may physically interact together and deregulate HOX-dependent target gene expression, or (2) they 2 act on independent pathways that combine to induce AML. Several observations argue in favor of the second model. First, the only Meis1-interacting site mapped on a HOX protein (HOXA9) was located to the first N-terminal 61 aa.50 Consequently, ND13 would also lack the putative Meis1-interacting motif, and, in agreement with this, we have been unable to successfully coimmunoprecipitate ND13 and Meis1 proteins from nuclear extracts (N.P., unpublished results, 2002). The second model is further supported by the collaboration of HOXB3 and Meis1 in inducing leukemia given that HOXB3, as a member of the 3′-located Antp-like HOX genes, lacks a Meis1-interacting site.27
The genetic collaboration between NUP98-HOX and several HOX genes with Meis1 in various murine models has identified Meis1 rather than Pbx1 as a strong oncogenic collaborating gene.14,27,28,51 This conclusion is supported by several independent expression studies that showed perturbed Meis1 expression patterns in human AML BM cells,52 the coexpression of Meis1 with HOXA9 and HOXA7 in AML and ALL samples, or both.40,41,53 Calvo et al54 recently suggested that the contribution of Meis1 to transformation might be to block G-CSF–induced differentiation and to promote SF-induced proliferation of HOX-transformed BM cells. As observed with NUP98-HOXA9,42 ND13 alone was sufficient to promote the survival and limited growth of transduced BM in media supplied with SF only. However, under the same conditions, ND13-Meis1–cotransduced BM cells achieved higher cell numbers (more than a 2-fold increase after 6 days compared with ND13 cells). Intriguingly, the functional cooperation observed between ND13 and Meis1 is more dramatic than that reported for NUP98-HOXA9, with a marked acceleration in rate of disease onset and a change in the nature of the disease, findings that more clearly parallel the interactions reported between HOXA9 and Meis1. These findings further highlight HOX gene-specific effects that likely reflect differential target genes.
This study now provides a murine model of NUP98-HOXD13-associated leukemogenesis. This model underlines the importance of HOX-dependent downstream pathways for NUP98-associated AML and will facilitate further elucidation of the molecular pathogenesis of this rapidly growing family of human leukemias.
Prepublished online as Blood First Edition Paper, January 23, 2003; DOI 10.1182/blood-2002-08-2484.
Supported in part by the National Cancer Institute of Canada, with funds from the Terry Fox Foundation; by Genome BC; and by National Institutes of Health (NIH) grant DK48642. N.P. was the recipient of a scholarship from the National Science and Engineering Research Council of Canada. C.A. was a recipient of a Canadian Institute of Health Research Fellowship and a Leukemia Research Foundation of Canada Fellowship. C.B. was supported by a grant from the Deutsche Forschungsgesellschaft, and M.F.-B. was supported by a grant from the Deutsche Krebshifle.
N.P. and C.B. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


![Figure 2. Fusion gene ND13 perturbs the differentiation and proliferative potential of primitive myeloid progenitors, and its activity is dependent on the HOXD13 homeodomain and NUP98 portion. (A) Total number of d12 CFU-S derived from 5000 FACS-purified control GFP or ND13-transduced injected BM cells (mean ± SD, n = 2). (B) Lineage distribution of GFP+ d12 CFU-S–derived transduced cells. 2 × 104 cells recovered after infection were injected without selection into irradiated mice, and single spleen cell suspensions (2 spleens/construct) were analyzed by FACS 12 days later for GFP and lineage-specific marker expression (n = 2; representative experiment shown). □ indicates GFP; ▦, ND13. (C) Expression of FLAG-tagged ND13 wild-type and mutant constructs in nuclear extracts of transfected 293T cells detected by Western blot using an anti-FLAG monoclonal antibody. (D) Total number of d12 CFU-S colonies derived per culture initiated with 1 × 106 cells transduced with the GFP, ND13, and mutant viruses after 1 week in liquid culture. Yields for TR-HOXD13 and ND13-N51S were significantly different from those for ND13 (P < .03) (mean ± SD; n = 3 for all except TR-HOXD13 [n = 2]).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/11/10.1182_blood-2002-08-2484/5/m_h81134397002.jpeg?Expires=1769096708&Signature=TZ9tLqbLvg1A6BZmAJJ-Xa5ZLoSMW26VaILmWTAI9YKkz2~OrPqj-lKcn66xWeNcG6hDuJLDKoA3J7pxc-zeO7Dyr9suqumtyO-BMv2QtldJSHTFY71ewCpkzfZ7x7EUw1yP62e-0sxaX~q5MgZfqEblCz0CR7JNTBpAEzv652iKMyDoCs4ydjPfRXYhLV~s8D6QvGN5VQE-EIMCs~J-~fU2oWCX2jG4R8CW6T77upZc3bHXrVYFnBsGfMs9KAkgmleqlTLcOwcU1EZOQiQsjKwVV4248iXD8u8uLdpIkG7Fp24DXASbJOBHxFMTw2mYDaV5sidY7EyfGWiV2LglRA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 8. Expression of Meis1, Hoxa7, and Hoxa9 in BM cells from sick ND13 and ND13-Meis1 mice. Expression of Meis1 transcripts (A), endogenous Meis1 (B), and Hoxa7 and Hoxa9 (C) in BM cells harvested from sick mice or transduced cells grown in LC. Mean expression shown for ND13 (n = 5), ND13-Meis1 (n = 5), sick mice, and control mouse (n = 1). LC samples consisted of transduced BM cells isolated by FACS and cultured for more than 28 days in 15% FBS, IL-3, hIL-6, and mSF (n = 1 for GFP+ control and ND13-Meis1 [ND/M] and n = 2 for ND13). For semiquantitative RT-PCR, all expression was normalized to GAPDH and then relative to control marrow, which was set at an arbitrary value of 1, except for the expression of endogenous Meis1 in the LC samples, which is given relative to GFP+ control. See “Materials and methods” for details. Endogenous Meis1-specific primers were designed not to amplify retrovirally driven Meis1 transcript. Mean ± SD shown. Similar analysis carried out for Hoxd13 was negative for expression (data not shown).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/11/10.1182_blood-2002-08-2484/5/m_h81134397008.jpeg?Expires=1769096708&Signature=hVEwAE26g19dRxcd3avIdwJlhdMrOui-4l7KNBmZps~kKKROIaDDOrG6eS4CZXf2NWXXcVrLVzZSmtxsG66KKgGw-A8u0RhdBheKiXbTh3272H8l5yCjkdrXMdEtzgUNE-eZrSfvh6RV1re1SkyCMsfMAHSsEifvoFYVc1T9GQ8uk8av5heUmvODgMSdaGbk~Gv~faXlTlcmMCBaNQvO7v1ytbsbvlEqMn5cph27UZooTuYVDJ1JzuoTPldjlb6UwfZDxPn6bhg9WpgGJmRPw~pgE62yX-WA4bAJ8Nk7SuDnIYfKyeb-2NbmEitj8dagJekIIjDGv2bokDTWOI0OEQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal