It is believed that polyploidy induces an orchestrated increase in gene expression. To know whether all alleles remain functional during megakaryocyte polyploidization, we used a well-established fluorescence in situ hybridization technique which allows one to simultaneously detect pre-mRNAs and assess ploidy level in a single cell. All alleles of GPIIb, GPIIIa,VWF, β-actin, hsp70,c-mpl, Fli-1, and FOG-1 genes are transcriptionally active in megakaryocytes from 4N to 32N. All X chromosomes in male cells are transcriptionally active but only half of them are transcriptionally active in female megakaryocytes, as revealed by the transcriptional activity of the GATA-1gene. Nuclear untranslated XIST RNA accumulates on the inactivated X chromosomes, indicating that they are subjected to a normal inactivation process. Altogether, our results demonstrate that megakaryocyte polyploidization results in a functional gene amplification whose likely function is an increase in protein synthesis parallel with cell enlargement.
Introduction
Megakaryocytes are one of the rare mammalian cells in which polyploidization is a normal process controlled by the differentiation program.1 Polyploidization is associated with an increase in cellular size and is an efficient manner in which to increase platelet production, since this production depends essentially on the megakaryocyte mass.2 In addition to its capacity to increase cell size, megakaryocyte polyploidization may participate in the regulation of genes involved in platelet function.3,4 To date, there have been very few reports on the effects of polyploidization on gene expression in megakaryocytes. In plants and fish, polyploidization is associated with a rapid silencing of some genes, due to chromosome rearrangements or epigenetic regulations.5 6 It is presently unknown if such a mechanism also operates in human megakaryocytes. We sought to determine by fluorescence in situ hybridization (FISH) if all alleles of several genes important for megakaryocyte differentiation and platelet function are transcriptionally active in polyploid megakaryocytes.
Materials and methods
In vitro growth of megakaryocytes from CD34+cells
CD34+ cells were obtained with informed consent from peripheral blood of healthy individuals undergoing hip surgery and grown as previously reported in the presence of thrombopoietin (10 ng/mL) (a generous gift from Kirin Brewery, Tokyo, Japan).7
RNA-FISH analysis
Probes.
Different genomic DNA probes were used to detect nuclear RNAs: plasmid clones for the β-actin locus (14 kb), for theXIST gene (10 kb), for the GPIIb gene (9 kb) and for human hsp70 gene (2.3 kb)8; 2 cosmid clones for the Fli-1 gene (36 kb and 23.2 kb); λ phage clones for the mpl gene (3 9-kb to 10-kb clones), for theVWF gene (20 kb), for the GPIIIa gene (2 9-kb clones); and a BAC clone containing the FOG-1 human genomic locus. A 3-kb genomic DNA fragment containing the second intron of theGATA-1 gene was amplified by polymerase chain reaction (PCR) and cloned into the pGEM-T plasmid.
Sample preparation.
Megakaryocytes were harvested at day 6 or 8 of culture, and cytocentrifuged onto slides at 500 rpm for 5 minutes (cytospin; Shandon, Pittsburgh, PA).
Hybridization.
The FISH procedure on premessenger RNAs was performed as previously described by Jolly et al9 without denaturation of cellular DNA. Briefly, probes were labeled by nick-translation in the presence of digoxigenin-16-dUTP (Roche Diagnostics, Meylan, France). Cells were fixed in 4% formaldehyde/phosphate-buffered saline (PBS) and permeabilized by a treatment with 0.1 M HCl for 10 minutes followed by three 5-minute incubations in 0.5% saponin/0.5% Triton X-100/PBS. As a second permeabilization step, cells were incubated in 20% glycerol/PBS and freeze-thawed 3 times by dipping the slides into liquid nitrogen. Cells were then dehydrated and incubated overnight with the denatured probe. After hybridization, the slides were washed and incubated for 30 minutes at 37°C in blocking buffer (3% bovine serum albumin [BSA]/0.3% Triton X-100/4× standard saline citrate [SSC]). The hybridized probe was detected using an anti–digoxigenin fluorescein isothiocyanate (FITC) antibody (Roche Diagnostics). After washing, the slides were mounted using Vectashield with DAPI (Vector Laboratories, Burlingame, CA).
Image acquisition and analysis
Images were acquired using a confocal laser scanning microscope (Leica, Heidelberg, Germany) equipped with a Planapochromat 63X/NA1.4 immersion objective or using an epifluorescence microscope (Nikon Eclipse 600, Tokyo, Japan) equipped with a 60× objective (Nikon) and processed using the Adobe Photoshop 6.0 software.
Results and discussion
RNA-FISH allows the detection of nuclear RNA at the site of transcription in individual nuclei and has been developed for allelic analysis of gene expression.10 11 In this work, we have optimized this procedure to study the proportion of functional alleles of several genes in megakaryocytes at different ploidy levels. Nearly pure populations of human megakaryocytes were obtained by growing CD34+ blood precursors in liquid cultures in the presence of thrombopoietin alone. We used confocal microscopy to detect the hybridization spots because it allows the precise enumeration of all the spots in thick cells such as megakaryocytes, especially when the spots are close to each other. After the RNA-FISH step, cell coordinates were recorded and cell images were stored. The same cell preparation was then subjected to a denaturation step and a second hybridization step was carried out to assess the ploidy level of the cells.
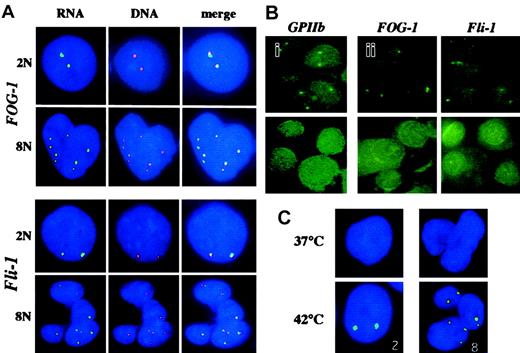
For RNA-FISH experiments, we used specific genomic probes of different sizes (between 3 kb for GATA-1 to about 100 kb forFOG-1). Because the size of the probe might be limiting in DNA-FISH experiments, we first checked if we could get a signal with the different probes in DNA-FISH experiments. A DNA signal was obtained only with the 2 largest probes (more than 50 kb in length), ie, FOG-1 and Fli-1, as illustrated in Figure 1A. We decided for further experiments to use a centromeric probe yielding a strong signal in order to precisely determine the ploidy level.
RNA- and DNA-FISH analysis of polyploid megakaryocytes.
Cells were cultured for 6 days and hybridized to different specific genomic probes. (A) Simultaneous RNA- and DNA-FISH analysis of polyploid megakaryocytes using FOG-1 and Fli-1 probes. RNA-FISH was performed using digoxigenin-labeled FOG-1 or Fli-1 probes revealed by an anti–digoxigenin FITC antibody (green) as described in “Materials and methods.” After image analysis, the cover slides were removed, the cells were washed in 4× SSC/0.1%Tween-20, and incubated in the presence of 0.1 mg/mL RNAse A for 30 minutes at 37°C. DNA-FISH was then performed after denaturation of cells in 70% formamide/2× SSC (3 minutes at 75°C) using the same digoxigenin-labeled FOG-1 or Fli-1 probes revealed by anti–digoxigenin TRITC antibody (red). Nuclear DNA was counterstained with DAPI (blue). The ploidy level determined from the number of red spots is indicated as 2N or 8N. Merging of the pictures obtained by RNA-FISH and DNA-FISH (merge) produced yellow spots, demonstrating the accumulation of nuclear FOG-1 or Fli-1 RNAs at the location of the corresponding gene. Note that most of the RNA and DNA spots superimpose in the case of the FOG-1, whereas the majority of RNA and DNA spots for Fli-1 were disposed side by side. (B) Effect of RNAse A or actinomycin D treatment on the signals obtained in RNA-FISH experiments. RNA-FISH experiments were performed with probes corresponding to the indicated genes. (i) The slides were treated (lower panel) or not (upper panel) with 0.1 mg/mL RNAse A for 30 minutes before hybridization. Absence of signals after RNAse treatment confirmed that the signals observed after hybridization without RNAse treatment correspond to RNA hybridization. (ii) The cells were incubated for 90 minutes in the absence (upper panels) or in the presence (lower panels) of 5 μg/mL actinomycin D (ActD) before being processed for RNA-FISH analysis. (C) Hsp70 RNAs detection. Megakaryocytes were heat-shock–treated by immersion for 30 minutes in a waterbath at 42°C or not treated (37°C). The number of transcription sites detected is indicated. Original magnifications × 60.
RNA- and DNA-FISH analysis of polyploid megakaryocytes.
Cells were cultured for 6 days and hybridized to different specific genomic probes. (A) Simultaneous RNA- and DNA-FISH analysis of polyploid megakaryocytes using FOG-1 and Fli-1 probes. RNA-FISH was performed using digoxigenin-labeled FOG-1 or Fli-1 probes revealed by an anti–digoxigenin FITC antibody (green) as described in “Materials and methods.” After image analysis, the cover slides were removed, the cells were washed in 4× SSC/0.1%Tween-20, and incubated in the presence of 0.1 mg/mL RNAse A for 30 minutes at 37°C. DNA-FISH was then performed after denaturation of cells in 70% formamide/2× SSC (3 minutes at 75°C) using the same digoxigenin-labeled FOG-1 or Fli-1 probes revealed by anti–digoxigenin TRITC antibody (red). Nuclear DNA was counterstained with DAPI (blue). The ploidy level determined from the number of red spots is indicated as 2N or 8N. Merging of the pictures obtained by RNA-FISH and DNA-FISH (merge) produced yellow spots, demonstrating the accumulation of nuclear FOG-1 or Fli-1 RNAs at the location of the corresponding gene. Note that most of the RNA and DNA spots superimpose in the case of the FOG-1, whereas the majority of RNA and DNA spots for Fli-1 were disposed side by side. (B) Effect of RNAse A or actinomycin D treatment on the signals obtained in RNA-FISH experiments. RNA-FISH experiments were performed with probes corresponding to the indicated genes. (i) The slides were treated (lower panel) or not (upper panel) with 0.1 mg/mL RNAse A for 30 minutes before hybridization. Absence of signals after RNAse treatment confirmed that the signals observed after hybridization without RNAse treatment correspond to RNA hybridization. (ii) The cells were incubated for 90 minutes in the absence (upper panels) or in the presence (lower panels) of 5 μg/mL actinomycin D (ActD) before being processed for RNA-FISH analysis. (C) Hsp70 RNAs detection. Megakaryocytes were heat-shock–treated by immersion for 30 minutes in a waterbath at 42°C or not treated (37°C). The number of transcription sites detected is indicated. Original magnifications × 60.
In order to specifically detect nuclear transcripts, we used the protocol developed by Jolly et al9 (see “Materials and methods”). It allowed us to obtain bright signals with all the different genomic probes that we used. In order to demonstrate that the signals we detected resulted from hybridization with RNA and not with DNA, we carried out 2 control experiments. First, cells were treated with RNAse before hybridization with the different genomic probes. This treatment totally abrogated the signal as illustrated in Figure 1Bi for the GPIIb gene. Second, the cells were treated with actinomycin D, an inhibitor of transcription.12 As illustrated in Figure 1Bii, the FOG-1 and Fli-1 transcripts completely disappeared from their sites of transcription after a 90-minute treatment.
Then, we tested the specificity of hybridization in 2 ways. First, we investigated the transcription of the hsp70 gene, an intronless heat shock gene whose expression can be induced by incubation of the cells at 42°C. As previously shown in fibroblasts,13 we found that in unstressed (37°C) cultured megakaryocytes with different ploidy levels, the hsp70 transcripts were not detected (Figure 1C). In contrast, after incubation at 42°C, the hsp70 nuclear transcripts appeared as discrete foci. Moreover, the number of foci was in most cases a power of 2 (2N) (Figure 1C), suggesting that all alleles of thehsp70 gene are transcriptionally active. As a second approach to confirm the specificity of the technique, RNA-FISH was followed by DNA-FISH on the same cells, using the same FOG-1 or Fli-1 probes. RNA-FISH revelation was carried out with an FITC-conjugated antibody and the cells were subsequently treated with RNAse A. After a denaturation step, hybridization was carried out with the same probes revealed with a tetramethyl rhodamine isothiocyanate (TRITC)–conjugated antibody. Nuclear transcripts accumulate at the sites of transcription and, thus, a colocalization of the RNA and DNA signals must be found if the hybridization is specific. The results are shown in Figure 1A. The spots corresponding to FOG-1 and Fli-1 transcripts are slightly larger than the corresponding DNA signals but the RNA and DNA spots colocalize in all cases.
Next, we investigated the expression of the GPIIb andGPIIIa genes encoding the 2 chains of αIIbβ3 integrin, the platelet receptor for fibrinogen. In all megakaryocytes examined, GPIIb RNA accumulation appeared as punctuated signals distributed in the nucleus which was counterstained with DAPI (4,6 diamidino-2-phenylindole) (Figure 2A). Confocal microscopy allowed us to quantify the number of nuclear spots corresponding to the sites of GPIIb transcription. After RNA-FISH, cell preparations were incubated with a centromeric probe from chromosome 12 after a denaturation step. The number of GPIIb transcription sites was equivalent to the level of ploidy from 8N to 32N, demonstrating that all alleles are transcriptionally active in polyploid megakaryocytes and that no transcriptional silencing occurs during ploidization (Figure 2B). Similar results were obtained for the GPIIIagene, for the von Willebrand factor (VWF) gene, and for a housekeeping β-actin gene (data not shown).
Analysis of
GPIIb or Fli-1 gene nuclear RNA accumulation and of ploidy level in polyploid megakaryocytes using confocal microscopy. Megakaryocytes were cultured for 8 days and double RNA- and DNA-FISH experiments were performed as in Figure 1A using a GPIIb or a Fli-1 probe (green) and a centromeric probe (red) staining chromosome 12 (Oncor, Gaithersburg, MD). Nuclear DNA was counterstained with DAPI (blue). The ploidy level determined from the number of red spots is indicated as 8N, 16N, or 32N. (A) Representative cells for each ploidy class. Original magnification × 63. (B) Spots corresponding to GPIIb or Fli-1 transcription sites were counted in 15 cells for each ploidy class using a confocal microscope. There were 3 laser excitation wavelengths used: 360 nm, 488 nm, and 543 nm, for DAPI, FITC, and TRITC, respectively. Serial optical sections of 2.5 μm (images collected at 0.25-μm intervals) in the z-axis of the cell were collected sequentially for each marker and overlaid to obtain a 2-dimensional reconstruction. Note that in megakaryocytes with a high ploidy level the spots were counted separately in serial optical sections through the cell. After superposition, 2 or 3 spots could yield a single strong signal. This approach allows a precise counting of the spots.
Analysis of
GPIIb or Fli-1 gene nuclear RNA accumulation and of ploidy level in polyploid megakaryocytes using confocal microscopy. Megakaryocytes were cultured for 8 days and double RNA- and DNA-FISH experiments were performed as in Figure 1A using a GPIIb or a Fli-1 probe (green) and a centromeric probe (red) staining chromosome 12 (Oncor, Gaithersburg, MD). Nuclear DNA was counterstained with DAPI (blue). The ploidy level determined from the number of red spots is indicated as 8N, 16N, or 32N. (A) Representative cells for each ploidy class. Original magnification × 63. (B) Spots corresponding to GPIIb or Fli-1 transcription sites were counted in 15 cells for each ploidy class using a confocal microscope. There were 3 laser excitation wavelengths used: 360 nm, 488 nm, and 543 nm, for DAPI, FITC, and TRITC, respectively. Serial optical sections of 2.5 μm (images collected at 0.25-μm intervals) in the z-axis of the cell were collected sequentially for each marker and overlaid to obtain a 2-dimensional reconstruction. Note that in megakaryocytes with a high ploidy level the spots were counted separately in serial optical sections through the cell. After superposition, 2 or 3 spots could yield a single strong signal. This approach allows a precise counting of the spots.
Finally, we investigated the allelic expression of the Fli-1and FOG-1 genes encoding 2 transcription factors playing a crucial role in megakaryocyte differentiation14-16 and that of the receptor for thrombopoietin, c-mpl.17 18 Spots corresponding to Fli-1 transcription sites were counted in 15 cells for each ploidy class (Figure 2A-B). The results obtained confirmed that all alleles of the Fli-1 gene are functional, whatever the ploidy level. Combined RNA-FISH and DNA-FISH revealed that this was also the case for the FOG-1 and c-mpl genes in polyploid megakaryocytes (data not shown).
Taken together, our results indicate that at least some genes important for megakaryocyte differentiation and platelet function as well as some housekeeping genes are not subjected to an allele-specific regulation during polyploidization. In addition, preliminary experiments suggest that expression of GPIIIa, Fli-1, GATA-1, VWF, c-mpl, GAPDH, and β-actin mRNAs increases in parallel with the ploidy level (H.R., unpublished, January 2002). However, we cannot exclude that other genes whose high expression is associated with cell toxicity may be submitted to such an allelic regulation leading to their down-regulation.
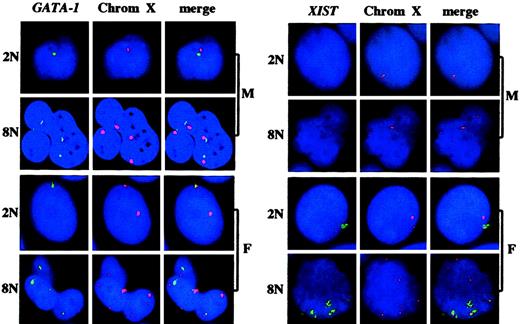
The GATA-1 gene is located on the X chromosome and encodes a transcription factor which is essential for the expression of several platelet-specific genes.19,20 As illustrated in Figure3, several GATA-1 genes per cell were active, their number corresponding to half of the ploidy level. Thus, several X chromosomes are active in the same cell in polyploid megakaryocytes. During embryogenesis, one X chromosome is inactivated in cells of female mammals. In diploid cells, with the exception of undifferentiated embryonic female cells where both X chromosomes are active, the inactive X chromosome produces a nuclear untranslated RNA called XIST,21-24 that spreads along the inactive chromosome to coat it.25 Thus, this XIST RNA identifies the inactive X chromosome. As shown in Figure 3, the XIST RNA signal colocalized with half of the X chromosomes in megakaryocyte cells from female individuals. In contrast, XIST RNA was not detected in male cells, demonstrating that X inactivation takes place normally in polyploid megakaryocytes from female donors.
Analysis of the accumulation of
GATA-1 gene nuclear RNA and XIST RNA and of the ploidy level in polyploid megakaryocytes from male and female individuals.The experiments were performed and the results are presented as in Figure 1A except that a GATA-1 or XIST probe and a centromeric probe for chromosome X (Oncor) were used. M indicates male; F, female. 2N, 8N indicate the ploidy level. Original magnification × 60.
Analysis of the accumulation of
GATA-1 gene nuclear RNA and XIST RNA and of the ploidy level in polyploid megakaryocytes from male and female individuals.The experiments were performed and the results are presented as in Figure 1A except that a GATA-1 or XIST probe and a centromeric probe for chromosome X (Oncor) were used. M indicates male; F, female. 2N, 8N indicate the ploidy level. Original magnification × 60.
Altogether these results indicate that polyploidization in megakaryocytes leads to a functional amplification of the genome, a phenomenon which was previously hypothesized but never demonstrated. However, the polyploidization process may also be a mechanism that regulates gene expression as recently demonstrated in yeast.26 Hancock et al3 have quantified the mRNA expression level of GPIIb andβ-actin genes as a function of the ploidy level and they have shown that the expression level of the GPIIb andβ-actin genes is differentially regulated during polyploidization of megakaryocytes, although, as shown here, all alleles of these 2 genes remain functional in polyploid megakaryocytes. As reviewed by Ravid et al,27 polyploidy may modify the profile of gene expression and hence, the repertoire of proteins in the cell. This mechanism of gene regulation may be independent of the number of functional alleles. Thus, to know if polyploidization regulates gene expression in megakaryocytes, as demonstrated in yeast, it should be interesting to investigate the accumulation levels of a large panel of mRNAs and proteins involved in differentiation in megakaryocytes of different ploidy levels.
We wish to thank J. B. Lawrence (University of Massachusetts Medical School, Worcester), O. Delattre (Institut Curie, INSERM U509, Paris, France), V. Mignotte (INSERM U484, Institut Cochin, Paris, France), D. Kerbiriou-Nabias (Hôpital de Bicêtre, INSERM U143, Kremlin-Bicêtre, France), P. F. Bray (Baylor College of Medicine, Houston, TX), J.-P. Rosa (Hôpital Lariboisière, INSERM U348, Paris, France), and P. Vyas (John Radcliffe Hospital, Oxford, United Kingdom) for providing us with the β-actin, Fli-1, c-mpl, VWF, GPIIb, GPIIIa, and FOG-1 human genomic probes, respectively. We thank F. Wendling for critical reading of the manuscript and helpful suggestions.
Prepublished online as Blood First Edition Paper, September 5, 2002; DOI 10.1182/blood-2002-05-1553.
Supported by La Ligue Nationale contre le Cancer, France (équipe labellisée). H.R. was funded by the Institut Gustave Roussy (comité de recherche clinique).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
W. Vainchenker, INSERM U362, Institut Gustave Roussy, 39 rue Camille Desmoulins, Villejuif, 94805, France; e-mail: verpre@igr.fr.