The CCAAT/enhancer binding protein alpha (C/EBPα) belongs to a family of transcription factors that are involved in the differentiation process of numerous tissues, including the liver and hematopoietic cells. C/EBPα−/− mice show a block in hematopoietic differentiation, with an accumulation of myeloblasts and an absence of mature granulocytes, whereas expression of C/EBPα in leukemia cell lines leads to granulocytic differentiation. Recently, dominant-negative mutations in the C/EBPα gene and down-regulation of C/EBPα by AML1-ETO, an AML associated fusion protein, have been identified in acute myelogenous leukemia (AML). To better understand the role of C/EBPα in the lineage commitment and differentiation of hematopoietic progenitors, we transduced primary human CD34+ cells with a retroviral construct that expresses the C/EBPα cDNA fused in-frame with the estrogen receptor ligand-binding domain. Induction of C/EBPα function in primary human CD34+ cells, by the addition of β-estradiol, leads to granulocytic differentiation and inhibits erythrocyte differentiation. Using Affymetrix (Santa Clara, CA) oligonucleotide arrays we have identified C/EBPα target genes in primary human hematopoietic cells, including granulocyte-specific genes that are involved in hematopoietic differentiation and inhibitor ofdifferentiation 1 (Id1), a transcriptional repressor known to interfere with erythrocyte differentiation. Given the known differences in murine and human promoter regulatory sequences, this inducible system allows the identification of transcription factor target genes in a physiologic, human hematopoietic progenitor cell background.
Introduction
Pluripotent hematopoietic stem cells have the potential for self-renewal and for differentiating into mature blood cells of all lineages.1,2 This is a highly regulated process and data from genetically modified mice suggest that transcription factors play an important role in the differentiation toward specific lineages, by inducing the expression of lineage-determining genes.3 The transcription factors C/EBPα and PU.1 play a crucial role in hematopoietic differentiation; C/EBPα−/− mice lack mature granulocytes and show an accumulation of myeloblasts in the blood and bone marrow.4Mice lacking PU.1 show defects in the generation of lymphocytes, granulocytes, and monocytes, while differentiation toward the erythrocytic or megakaryocytic lineages is not affected.5-7
The CCAAT/enhancer binding protein alpha (C/EBPα) belongs to the leucine zipper family of transcription factors, which bind DNA as homodimers. Expression of C/EBPα in a myeloid cell line and in multipotential avian progenitor cells leads to granulocytic differentiation.8,9 Expression of C/EBPα follows the expression of C/EBPβ and C/EBPγ in adipocytes, and plays an important role in adipocyte differentiation. Similarly, the specific role of C/EBPα in myeloid lineage commitment and in myeloid differentiation likely reflects the lack of redundancy in some of the functions of the C/EBP proteins. C/EBPα appears to play an important role in the pathogenesis of human acute myelogenous leukemia (AML): mutations in the C/EBPα gene have been identified in AML of the M2 French-American-British classification (FAB) subtype that lack the t(8;21),10,11 whereas expression of the t(8;21)-associated AML1-ETO fusion protein leads to the down-regulation of C/EBPα.12,13 Several of the identified mutations in the C/EBPα gene lead to the expression of a 30-kDa protein that lacks the amino-terminal transactivation domain. These mutations are generally monoallelic and the fact that the remaining copy ofC/EBPα is unaffected argues for a dominant-negative function of these mutations.10,11 C/EBPα can affect transcription in a pleiomorphic way. For instance, wild-type C/EBPα can down-regulate c-myc by binding to E2F,14,15 however a mutant C/EBPα that retains its transactivation properties but cannot bind to E2F does not promote granulocytic differentiation.16 Thus, the role of C/EBPα in differentiation and growth arrest may be linked yet mechanistically distinct (for review see McKnight17).
To evaluate the effects of C/EBPα on hematopoietic differentiation and lineage commitment, we transduced primary human hematopoietic cells with a bicistronic retroviral vector that expresses C/EBPα fused to the ligand-binding domain of the estrogen receptor (C/EBPα-ER) and also expresses green fluorescence protein (GFP). Induction of C/EBPα activity in human CD34+ cells by β-estradiol (β-ES) blocks erythrocyte differentiation (as shown by decreased burst-forming unit erythroid [BFU-E] formation) and promotes granulocytic differentiation. We have used this novel system to identify C/EBPα target genes using Affymetrix (Santa Clara, CA) oligonucleotide arrays, comparing the RNA expression profile of primary human hematopoietic cells transduced with the empty MIGR1 retroviral vector, with cells transduced with the MIGR1 C/EBPα-ER expression vector. There were 2 different approaches used to identify “direct” target genes of C/EBPα: a 2-hour time point for RNA collection, to minimize the possibility of detecting secondarily affected target genes, and an analysis of the transcriptional profile in the absence of protein synthesis, which was accomplished by adding cycloheximide (CHX; since activation of the C/EBPα-ER fusion protein is not dependent on protein synthesis). This approach takes advantage of several powerful techniques to identify target genes of lineage-specifying transcription factors, such as C/EBPα, and link the expression profile to effects on the behavior of primary human hematopoietic progenitor cells.
Materials and methods
Cell culture
RD18 cells (kindly provided by Dr M. Collins, Imperial College London, United Kingdom) and 293T cells were grown in Dulbecco modified essential medium with 10% heat-inactivated fetal calf serum (FCS), 100 U/mL penicillin/streptomycin, and 2 mMl-glutamine. Charcoal-stripped fetal calf serum (Gemini Bioproducts, Woodland, CA) and medium without phenol red were used to eliminate estrogen derivates that would lead to activation of the C/EBPα-ER fusion protein.
Retroviral production by transient transfection or using the producer cell line RD18
The rat C/EBPα-ER cDNA (kindly provided by Dr A. Friedman, Johns Hopkins University) was cloned into the EcoR1 restriction site of the MIGR1 vector (kindly provided by Dr W. Pear, University of Pennsylvania). The MIGR1 retrovirus contains the murine stem cell virus (MSCV) promoter and an internal ribosomal entry site (IRES) element followed by the GFP gene (Figure1). Retrovirus was made by transient cotransfection of the MIGR1, pEQ-PAM3(-E) (kindly provided by Dr E. Vanin, St Jude's Children's Hospital) and pSVampho plasmids into 293T cells. Retroviral supernatant was collected after 36 and 48 hours. The supernatant was filtered through a 0.45-μm filter to avoid contamination of the hematopoietic cells with 293T cells.
Expression of C/EBPα-ER in primary human CD34+ cells.
(A) The retroviral vector MIGR1 is shown schematically. The rat C/EBPα cDNA fused in-frame with the ligand-binding domain of the estrogen receptor (C/EBPα-ER) was cloned into the EcoRI restriction site of MIGR1. (B) Western blot analysis of human CD34+ cells transduced with MIGR1 or MIGR1 C/EBPα-ER using an α-C/EBPα antibody (lanes 1-2). Expression of C/EBPα-ER in the producer cell line RD18 was used as a positive control (lane 3). (Note that the 45-kDa band shown in lanes 2 and 3 is not present in lane 1 and likely represents a degradation product of the ER fusion protein.) (C) The experimental design to evaluate the effects of C/EBPα on human hematopoietic progenitor cell behavior and to identify C/EBPα target genes is shown schematically (IFA indicates immunofluorescence assay).
Expression of C/EBPα-ER in primary human CD34+ cells.
(A) The retroviral vector MIGR1 is shown schematically. The rat C/EBPα cDNA fused in-frame with the ligand-binding domain of the estrogen receptor (C/EBPα-ER) was cloned into the EcoRI restriction site of MIGR1. (B) Western blot analysis of human CD34+ cells transduced with MIGR1 or MIGR1 C/EBPα-ER using an α-C/EBPα antibody (lanes 1-2). Expression of C/EBPα-ER in the producer cell line RD18 was used as a positive control (lane 3). (Note that the 45-kDa band shown in lanes 2 and 3 is not present in lane 1 and likely represents a degradation product of the ER fusion protein.) (C) The experimental design to evaluate the effects of C/EBPα on human hematopoietic progenitor cell behavior and to identify C/EBPα target genes is shown schematically (IFA indicates immunofluorescence assay).
Retroviral producer cell lines were generated for the MIGR1 and MIGR1 C/EBPα-ER vectors by transducing the RD18 cells with transiently made retrovirus and selecting clonal green RD18 cells. The multiplicity of infection (MOI) of the obtained clones was tested by transduction of U937 cells. Retroviral supernatant from selected RD18 producer cell lines was centrifuged at 27 000 rpm for 90 minutes to concentrate the retrovirus. The retroviral pellet was resuspended in 0.5 mL Iscoves modified Dulbecco medium (IMDM) with 20% charcoal-stripped FCS, 100 U/mL penicillin/streptomycin, and 2 mML-glutamine and either used immediately or frozen in aliquots at −70°C.
Transduction of human hematopoietic progenitors
Human CD34+ hematopoietic cells were selected using a magnetic CD34 selection kit system (Miltenyi Biotec, Auburn, CA) from small aliquots of leukapheresis products collected from one healthy donor and one patient undergoing stem/progenitor cell collection after granulocyte–colony stimulating factor (G-CSF) treatment for the treatment of a nonhematologic malignancy at Memorial Hospital, following their informed consent. After magnetic selection, more than 95% of the cells expressed the CD34 antigen. An aliquot containing 5 × 106 CD34+ cells was cultured for 72 hours in IMDM with 20% heat-inactivated FCS, 100 ng/mL Flt3-ligand (Flt3-L) and 20 ng/mL granulocytic-macrophage colony stimulating factor (GM-CSF) (both kindly provided by Immunex, Seattle, WA); 100 ng/mL of stem cell factor (SCF; kindly provided by Amgen, Thousand Oaks, CA); 100 ng/mL thrombopoietin (TPO); 100 ng/mL of interleukin-6 (IL-6) and 50 ng/mL of IL-3 (generous gifts from Kirin Brewery, Gumma, Japan); 100 U/mL penicillin/streptomycin; and 2 mMl-glutamine. The stimulated cells were transduced with retroviral supernatant using the same concentrations of cytokines and 4 μg/mL polybrene. Spinoculation (4 rounds) was performed at 1800 rpm for 45 minutes at room temperature on retronectin-coated plates (TaKaRa Shuzo, Shiga, Japan). The CD34+ cells were removed from the retronectin-coated plates using 0.02% EDTA (ethylenediaminetetraacetic acid) in phosphate-buffered saline (PBS) and expanded in the cytokine mix described above for 2 additional days. In general, beginning with approximately 5 × 106 cells, we would have approximately 4 × 107 cells (before sorting) for further analyses (including Western blot, microarray analysis, and in vitro culture studies). After 48 hours the cells were sorted for GFP expression and expanded for 2 or 8 hours in IMDM with the addition of the described human cytokines and 1 μM β-estradiol or an equal volume of ethanol as a control.
Clonogenic assay
The indicated number of hematopoietic cells were resuspended in 0.9% methylcellulose containing 1% bovine serum albumin, 10 μg/mL bovine pancreatic insulin, 200 μg/mL human transferrin (iron saturated), 0.1 mM 2-mercaptoethanol, 2 mM l-glutamine, 100 U/mL penicillin/streptomycin (MethoCultSFBIT; Stem Cell Technologies, Vancouver, Canada) and 20 ng/mL human IL-6, 20 ng/mL human IL-3, 20 ng/mL human SCF, 10 ng/mL G-CSF (Amgen) and 6 U/mL human erythropoietin (EPO) (Amgen). Colonies were evaluated and counted after 12 to 14 days in culture. Cells were collected from the clonogenic assays by washing with PBS. Cytospins were performed using 8 × 104 cells for immunofluorescence staining or morphologic analysis after Wright-Giemsa staining.
Flow cytometry
Human hematopoietic progenitor cells transduced with MIGR1 or MIGR1 C/EBPα-ER were washed with PBS, resuspended in 2% FCS/PBS and stained with CD71–phycoerythrin (PE) antibody (BD Biosciences PharMingen, San Diego, CA) for 45 minutes at 4°C. Stained cells were sorted for expression of GFP and CD71 (< 102 mean fluorescence intensity) using a MoFlo (Cytomation, Fort Collins, CO) or Vantage cell sorter (Becton Dickinson, Clearwater, FL). Cells collected from the clonogenic assays were washed with PBS, resuspended in 2% FBS in PBS, and stained with anti-CD11b, CD14, or glycophorin A PE-conjugated antibodies (BD Biosciences PharMingen). Idiotype controls were used accordingly. Data were analyzed using the software CellQuest 3.1 (Clearwater, FL).
Immunofluorescence staining and histochemical analysis
Cells were fixed with 2% paraformaldehyde in PBS for 10 minutes and washed twice with PBS. Fixed cells were incubated with 0.1% saponin (Sigma Chemical, St Louis, MO) in PBS for 1 hour at room temperature and washed twice with PBS. Incubation with a 1:150 dilution of the anti–inhibitor of differentiation 1 (Id1) antibody (Santa Cruz Biotechnology, Santa Cruz, CA) in PBS with 10% FCS was performed for one hour in the dark at room temperature. The cells were washed 4 times with 0.02% Tween20 (Sigma Chemical) in PBS. Cells were incubated with an antirabbit antibody conjugated to Cy3 (Jackson ImmunoResearch Laboratories) for 1 hour at room temperature and counterstained with DAPI (4,6 diamidino-2-phenylindole), washed once with water, and mounted in SlowFade (Molecular Probes, Eugene, Oregon). Cells were analyzed using a fluorescence microscope (Olympus, Melville, NY).
Preparation of labeled cRNA and oligonucleotide array
Total RNA was extracted from cells, which were approximately 50% CD34+ at the end of the retroviral transduction period, using the RNeasy Mini Kit (Qiagen, Valencia, CA) following the manufacturer's instruction. Total RNA (5-10 μg) was reverse transcribed with a cDNA synthesis kit (Gibco BRL/Life Technologies, Rockville, MD) in the presence of an oligo dT-T7 primer. After phenol/chloroform extraction and ethanol precipitation, the cDNA pellet was air dried and resuspended in 12 μL of Rnase-free H2O. Ten microliters was used for the in vitro transcription amplification reaction, in the presence of biotinylated nucleotides (Enzo Diagnostics, Farmingdale, NY). Labeled cRNA (15 μg) was fragmented by incubation at 95°C for 35 minutes in fragmentation buffer (40 mM Tris-acetate, pH 8.1, 100 mM KOAc, and 30 mM MgOAc) and the fragmented cRNA was then hybridized against the Affymetrix HG_U95Av2 oligonucleotide arrays. The arrays were scanned using a Hewlett Packard confocal laser scanner and analyzed using MicroArray Suite 5.0 (Affymetrix), GeneSpring 4.0 (Silicon Genetics, Redwood City, CA), and in-house analytical tools.
The default parameters were used for the statistical algorithm and for probe set scaling using Microarray Suite 5.0 (with a target intensity of 500). The data were then filtered so that the absolute value of the fold change was more than 1.5 and the “fold change” Pvalue was less than .005. Additionally, we removed genes that were scored as absent in experimental and baseline files (both numerator and denominator of the fold change), as well as genes scored as increasing but absent in the experimental file (numerator of the fold change). We chose a conservative procedure for combining the duplicate data (intersecting the filtered list for each replicate based on a list of genes that passed the filtering criteria for both replicates).
Real time reverse transcriptase–polymerase chain reaction (RT-PCR) analysis
To quantify the expression of the Id1, c-myc, and calgranulin mRNAs, PCR amplification was carried out using the 7700 Sequence detector (PE Applied Biosystems, Norwalk, CT) and the PCR products detected using SYBR Green I chemistry. Each cDNA was quantified relative to glyceraldehyde phosphate dehydrogenase (GAPDH), which served as the reference gene transcript. The following primer sequences were used: Id1, forward primer 5′-GGACGAGCAGCAGGTAAACG-3′ and reverse primer 5′-TGCTCACCTTGCGGTTCTG-3′; c-myc, forward primer 5′-GCTGCTTAGACGCTGGATTTTT-3′ and reverse primer 5′-TCGAGGTCATAGTTCCTGTTGGT-3′; calgranulin, forward primer 5′-TTCCATGCCGTCTACAGGGA-3′ and reverse primer 5′-TCCAACTCTTTGAACCAGACGTC-3′; GAPDH, forward primer 5′-ATTGGGCGCCTGGTCAC-3′ and reverse primer 5′-AAGATGTAAACCATGTAGTTGAGGTCA-3′. All primers were designed using PrimerExpress 1.0 software (PE Applied Biosystems), and the default TaqMan parameters. Primer sets spanning intron-exon junctions were chosen to prevent amplification of any possible residual, contaminating genomic DNA in the cDNA sample. The reaction mixture consisted of 1X SYBR Green PCR master mix (PE Applied Biosystems), 0.1 μL of each primer (100 ng/μL) and oligo-dT–generated cDNA in a final volume of 25 μL. Amplification conditions were as follows: 50°C for 2 minutes, 95°C for 10 minutes, then 40 cycles at 95°C for 15 seconds and 60°C for 1 minute. A negative control (lacking the cDNA template) was included in every assay and the size of the PCR products was confirmed by agarose gel electrophoresis. The comparative threshold cycle (CT) method was used for analysis after a validation experiment was performed for all primer sets.
Western blot analysis
Cells were washed with PBS and either frozen as a pellet or lysed directly with protein lysis buffer (1% Triton X-100, 5 mM EDTA, 50 mM HEPES (4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid), pH 7.5, 50 mM NaCl, 10 mM Na Pyrophosphate, and 50 mM NaF) and protease inhibitors (Roche, Indianapolis, IN). Cell lysates were sonicated and centrifugated for 10 minutes. Protein concentrations were measured using the Bradford method (Bio-Rad Laboratories, Hercules, CA). The indicated amounts of protein were mixed with an equal volume of 2 × sodium dodecyl sulfate (SDS) loading buffer and boiled for 10 minutes. Proteins were electrophoretically separated on a 8% to 16% SDS polyacrylamide gel electrophoresis (PAGE) gel (Bio-Rad Laboratories) and blotted on a polyvinylidene fluoride (PVDF) membrane (Amersham Pharmacia Biotech, Piscataway, NJ). The membranes were blocked with 5% nonfat dry milk in Tris-buffered saline (TBS) with 0.1% Tween20, and incubated first with the anti-C/EBPα primary antibody (Santa Cruz Biotechnology) at a 1:200 dilution overnight at 4°C, washed 4 times for 15 minutes with 0.1% Tween20 in TBS followed by the secondary antibody (conjugated to horseradish peroxidase) for one hour at room temperature. After being washed, the blot was developed using the enhanced chemiluminescence (ECL) Western blotting detection reagent (Amersham Pharmacia Biotech) and exposed to film (Amersham Pharmacia Biotech).
Results
C/EBPα blocks erythrocyte differentiation and enhances neutrophil maturation of human hematopoietic progenitors
To define the effects of C/EBPα on the differentiation and lineage commitment of primary hematopoietic stem/progenitor cells, we expressed the rat C/EBPα-ER fusion cDNA (which has cross-species activity and is capable of differentiating a human leukemia cell line13) in human CD34+ cells by retroviral gene transfer (Figure 1A).
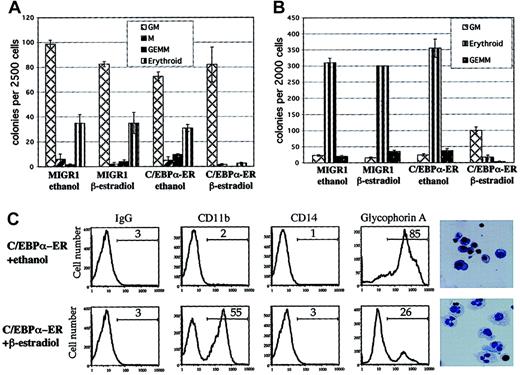
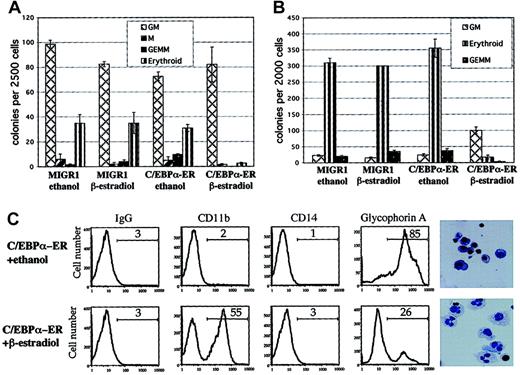
Transduced cells were sorted for GFP expression and the levels of C/EBPα-ER and the endogenous C/EBPα protein were evaluated by Western blot analysis, using an anti-C/EBPα antibody. Expression of the 72-kDa C/EBPα-ER protein was readily detectable, whereas the 42-kDa wild-type C/EBPα protein was not detectable in these immature hematopoietic cells (Figure 1B). The C/EBPα-ER fusion protein translocates from the cytoplasm to the nucleus in the presence of β-estradiol, which leads to its activation (Pabst et al13 and data not shown). Hematopoietic progenitors transduced with MIGR1 C/EBPα-ER (or MIGR1 as a control) were sorted for GFP expression and plated into clonogenic assays with or without β-estradiol (shown schematically in Figure 1C). The addition of β-estradiol to the C/EBPα-ER–transduced cells led to significantly reduced numbers of colony-forming unit erythrocyte (CFU-E) and BFU-E (Figure 2A), demonstrating a strong inhibitory effect of C/EBPα on erythrocyte differentiation. The decrease in colony-forming unit granulocyte, erythrocyte, megakaryocyte and monocyte (CFU-GEMMs), coupled with a minimal increase in colony-forming unit granulocyte macrophage (CFU-GMs), may reflect inhibition of the erythroid cells in the CFU-GEMM leading to the misidentification of CFU-GEMM as CFU-GM (or it could represent a stimulatory effect of C/EBPα on the granulocytic lineage).
C/EBPα inhibits erythrocyte differentiation and leads to granulocytic differentiation.
(A) Hematopoietic cells transduced with MIGR1 or MIGR1 C/EBPα-ER were sorted for expression of GFP and plated in clonogenic assays in triplicate with or without β-estradiol. Colonies were scored and counted 12 to 14 days later. One of 2 representative experiments is shown. (B) Transduced hematopoietic cells were sorted for GFP and CD71 expression higher than 102 mean fluorescence intensity (CD71bright) and plated in clonogenic assays in duplicate. Shown is 1 of 3 independent experiments. (C) Cells were collected from the clonogenic assays (see Figure 2B) and analyzed for surface marker expression (CD11b, CD14, and glycophorin A) by flow cytometry. Cytospins were performed and stained with Wright-Giemsa (last panels on the right; original magnification, × 600). Cells transduced with MIGR1 and treated with ethanol or β-estradiol showed the same phenotype as cells transduced with C/EBPα-ER and treated with ethanol (data not shown).
C/EBPα inhibits erythrocyte differentiation and leads to granulocytic differentiation.
(A) Hematopoietic cells transduced with MIGR1 or MIGR1 C/EBPα-ER were sorted for expression of GFP and plated in clonogenic assays in triplicate with or without β-estradiol. Colonies were scored and counted 12 to 14 days later. One of 2 representative experiments is shown. (B) Transduced hematopoietic cells were sorted for GFP and CD71 expression higher than 102 mean fluorescence intensity (CD71bright) and plated in clonogenic assays in duplicate. Shown is 1 of 3 independent experiments. (C) Cells were collected from the clonogenic assays (see Figure 2B) and analyzed for surface marker expression (CD11b, CD14, and glycophorin A) by flow cytometry. Cytospins were performed and stained with Wright-Giemsa (last panels on the right; original magnification, × 600). Cells transduced with MIGR1 and treated with ethanol or β-estradiol showed the same phenotype as cells transduced with C/EBPα-ER and treated with ethanol (data not shown).
To evaluate whether C/EBPα expression could alter the phenotype of cells already displaying erythroid characteristics we sorted the transduced hematopoietic cells for expression of GFP and for high expression of the CD71 antigen (CD71bright) and plated these cells into clonogenic assays with or without β-estradiol. The CD71 antigen (transferrin receptor) is an activation marker and is also highly expressed on BFU-E cells.18 Most of the CD71bright cells transduced with control vector (with or without β-estradiol) formed erythrocyte colonies (BFU-E/CFU-E) or mixed colonies (CFU-GEMM), while cells expressing active C/EBPα protein showed a dramatic reduction of erythrocyte colonies, as well as a significant reduction of CFU-GEMM and increased numbers of CFU-GM (Figure 2B). No such effect was seen in the absence of β-estradiol. The CFU-GM colonies expressing the active C/EBPα protein were smaller, yet less compact, which implies an accelerated differentiation and earlier acquisition of mobility (data not shown).
The GFP+/CD71bright cells from the clonogenic assays were collected, stained for lineage-specific cell surface markers, and analyzed by flow cytometry. The progenitor cells transduced with the control vector or those that contained the inactive form of C/EBPα showed high-level expression of the erythrocyte surface marker glycophorin A, while cells that contained the active C/EBPα protein predominantly expressed the differentiation marker CD11b, but not CD14 (Figure 2C). Cytospins revealed that the cells expressing C/EBPα-ER protein in the presence of β-estradiol were predominantly granulocytes, while the cells plated in ethanol showed predominantly an erythroblastic morphology. Cells expressing only GFP, in the presence or absence of β-estradiol, had a phenotype indistinguishable from cells expressing C/EBPα-ER in the absence of β-estradiol (data not shown).
Identification of C/EBPα target genes in primary human hematopoietic cells
To identify C/EBPα target genes involved in hematopoietic differentiation and commitment we transduced hematopoietic progenitor cells with control or C/EBPα-ER–expressing retrovirus, sorted the cells for GFP expression, and grew them in the presence of β-estradiol for 8 hours. RNA was prepared and the transcriptional profiles of the GFP+ cells were analyzed using Affymetrix HU95A oligonucleotide arrays (which contain probe sets that can analyze the level of expression of 12 000 transcripts; see thewww.affymetrix.com website for more information). Genes whose level of expression changed at least 1.5-fold 8 hours after the addition of β-estradiol, in each of 3 independent experiments, were considered up- or down-regulated by C/EBPα. Although the probes on the oligonucleotide arrays generally hybridize to the highly specific 3′ untranslated region of each gene, the probe for estrogen receptor expression hybridizes to sequences in the coding region. Because we used a C/EBPα-ER fusion protein, estrogen receptor expression was up-regulated in all experiments, serving as an (unexpected) internal control, but not as a target gene of C/EBPα. A list of C/EBPα-regulated genes is shown in Table1.
Identification of direct C/EBPα target genes
An advantage of using the C/EBPα-ER construct is that activation of C/EBPα function occurs independently of protein translation because the fusion protein is present but is inactive in the absence of ligand, because of its cytoplasmic location. The C/EBPα-ER protein translocates to the nucleus and becomes functional upon the addition of β-estradiol.13 Therefore, we used cycloheximide (CHX), which inhibits protein synthesis, to confirm our identification of “direct” C/EBPα target genes. Transduced, GFP-sorted hematopoietic cells were treated with 5 μg/mL CHX 30 minutes before exposure to β-estradiol and the transcriptional profile was analyzed as described in the preceding paragraph. It is well known that CHX can alter RNA levels for some genes by affecting proteins that control RNA stability (Kannan et al19). Therefore, as an alternative approach to confirming the identity of direct C/EBPα target genes, we prepared RNA after a brief (2 hour) exposure to β-estradiol alone; these potential direct target genes are shown in Table 2. Genes identified as “direct” targets of C/EBPα based on the triplicate experiments (following an 8-hour treatment with β-estradiol) that were also found after either a short exposure to β-estradiol or to CHX are shown in the seventh and eighth columns of Table 1.
Identification of C/EBPα target genes in erythroid progenitor cells
The most dramatic effect of C/EBPα expression on hematopoietic colony growth was the inhibition of BFU-E and CFU-E formation (Figure 2). Therefore, to identify genes involved in the inhibition of erythrocyte differentiation by C/EBPα we transduced human CD34+ cells with MIGR1 or MIGR1 C/EBPα-ER, sorted for GFP+/CD71bright–expressing cells, and incubated the cells with β-estradiol for 8 hours before preparing RNA for transcriptional profiling. A list of genes up- or down-regulated is shown in Table 3. C/EBPα target genes identified in these CD71bright cells, which were also identified as “direct” target genes in Table 2, are listed in the sixth and seventh columns of Table 3.
Target genes commonly regulated by C/EBPα
The transcriptional repressor Id1 and adipophilin were up-regulated after the expression of C/EBPα in all experiments, although a higher fold induction was seen in the erythroid precursors. Both genes were also identified as “direct” C/EBPα target genes according to the experiments using either CHX (to inhibit translation) or the early time point (Tables 1 and 3). No genes were significantly and consistently down-regulated by C/EBPα in both the multipotent and the erythroid progenitors. Given the described impairment in IL-6R and G-CSFR expression in C/EBP−/− mice, we checked whether changes in the expression of these genes were also found in our experiments: G-CSFR expression was low and remained low despite induction of C/EBPα activity. Unfortunately, probe sets for detecting IL-6R mRNA are not present on the U95A chip.
To validate the C/EBPα target genes identified by the oligonucleotide arrays, we also analyzed changes in gene expression using real-time RT-PCR to quantify the levels of Id1, calgranulin B, and c-myc mRNAs. Using RNA from 2 of the 3 triplicate experiments described in Table1, we performed the real time PCR assays in triplicate. Similar levels of Id1 and calgranulin up-regulation were seen comparing real-time PCR with the microarray data (6.7- and 4.6-fold for each gene); down-regulation of c-myc was also seen in both of these experiments.
Id1 protein is up-regulated in primary hematopoietic cells after C/EBPα activation
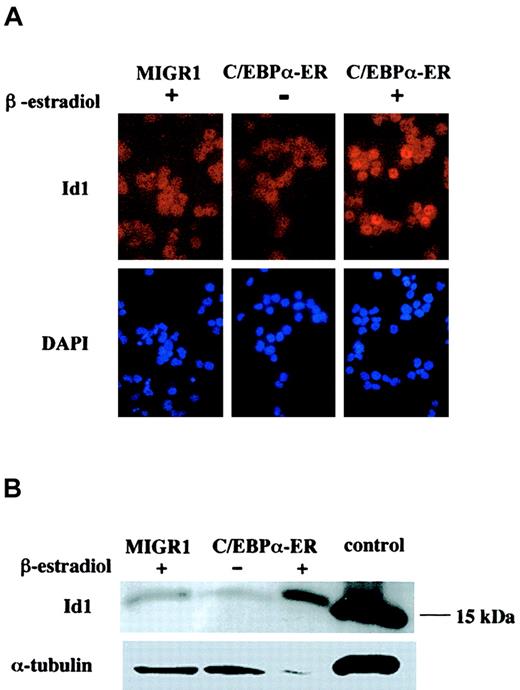
Id1 mRNA was up-regulated after the expression of C/EBPα in all experiments, and especially in the erythroid precursors. To evaluate whether the level of Id1 protein correlated with Id1 mRNA, we performed immunofluorescence staining on cytospins obtained from GFP-sorted cells after 16 hours of β-estradiol treatment. Cells expressing the active form of C/EBPα showed an increase in Id1 protein, with a nuclear localization pattern (Figure 3A). Western blot analysis showed high-level expression of Id1 protein in cells expressing the active C/EBPα protein, compared with the low Id1 expression seen in cells transduced with the empty MIGR1 vector or in cells expressing the inactive form of C/EBPα (Figure 3B).
Id1 protein levels are increased after expression of active C/EBPα.
(A) Human CD34+ cells transduced with MIGR1 or MIGR1 C/EBPα-ER, were sorted for the expression of GFP and treated with β-estradiol (+) or ethanol (−) for 16 hours. Immunofluorescence was performed using an α-Id1 antibody and counter stained with DAPI to identify the nucleus (original magnification, × 600). (B) Cells sorted for GFP expression and treated for 16 hours with β-estradiol or ethanol were analyzed for expression of Id1 protein by Western blot analysis using an α-Id1 antibody. The control lane contains cell lysate from 3T3 cells. The level of α-tubulin expression is indicated, to control for equal protein loading.
Id1 protein levels are increased after expression of active C/EBPα.
(A) Human CD34+ cells transduced with MIGR1 or MIGR1 C/EBPα-ER, were sorted for the expression of GFP and treated with β-estradiol (+) or ethanol (−) for 16 hours. Immunofluorescence was performed using an α-Id1 antibody and counter stained with DAPI to identify the nucleus (original magnification, × 600). (B) Cells sorted for GFP expression and treated for 16 hours with β-estradiol or ethanol were analyzed for expression of Id1 protein by Western blot analysis using an α-Id1 antibody. The control lane contains cell lysate from 3T3 cells. The level of α-tubulin expression is indicated, to control for equal protein loading.
Discussion
C/EBPα−/− mice lack mature granulocytes and show an accumulation of myeloblasts, providing strong evidence that this transcription factor is involved in hematopoietic differentiation.4 C/EBPα is specifically up-regulated during granulocytic differentiation,20 and overexpression of C/EBPα leads to differentiation of myeloid leukemia cell lines4,8,13 and to eosinophil/neutrophil differentiation of multipotential avian hematopoietic progenitor cells.9
We used retroviral transduction of human CD34+hematopoietic cells to evaluate the role of C/EBPα in hematopoietic differentiation and commitment of primary human progenitor cells, and to identify target genes of C/EBPα involved in these processes. Following the expression of functional C/EBPα protein, we observed a dramatic inhibition of erythrocyte differentiation, with a decrease in BFU-E, CFU-E, and CFU-GEMM formation. We also observed increased CFU-GM colony formation using purified erythrocytic progenitor cells (CD71bright) following induction of C/EBPα. To identify the genes involved in this process we analyzed the transcriptional profiles of GFP+ or GFP+/CD71bright-sorted cells following transduction with either the MIGR1 or MIGR1 C/EBPα-ER retrovirus. Using this approach, we have identified a number of C/EBPα target genes, many of which are linked to C/EBPα for the first time. Previous studies, which compared myeloblasts from C/EBPα−∖− mice with granulocytes from wild-type littermates, may have identified many differentiation-specific genes that are not necessarily regulated by C/EBPα4,21 Other genes, involved in the lineage commitment of primitive progenitor cells, may have been missed because C/EBPα was inducibly expressed in lineage-restricted leukemia cell lines.8 15
C/EBPα target genes that are involved in granulocytic differentiation include calgranulin A and calgranulin B. Calgranulin A (MRP-8) is induced during myeloid differentiation, and it was recently identified as a C/EBPα target gene.15,22 We have confirmed its up-regulation using both Affymetrix microarrays and quantitative (real-time) RT-PCR. The MRP-8 promoter has 5 potential C/EBP binding sites and it has been successfully used to express oncogenic fusion proteins in transgenic mouse models of myeloid leukemia.23-25 Having identified it as a “direct” C/EBPα target gene (using cycloheximide), the potential regulation of its promoter by C/EBPα could make its use problematic for expressing fusion proteins, such as AML1-ETO, or dominant-negative C/EBPα proteins in transgenic mice.13 23
We observed C/EBPα-induced down-regulation of c-myc mRNA in primary CD34+ hematopoietic cells, arguing for its key role in granulocytic differentiation and confirming previous observations that C/EBPα expression in a leukemia cell line leads to c-myc down-regulation.15 Down-regulation of c-myc appears to be mediated through repression of E2F-dependent transcription15 (depicted in Figure4B) and was required for granulocytic differentiation in that system.15 Similarly, knock-in mice that lack wild-type C/EBPα and express only C/EBPα mutants that do not suppress E2F-dependent transcription, lack granulocytic differentiation.16 This demonstrates that down-regulation of c-myc by C/EBPα is required for the differentiation of c-myc–expressing leukemia cells and also that its down-regulation plays a pivotal role in the differentiation of multipotential progenitor cells. Inhibitory effects of C/EBPα on the cell cycle (mediated via suppressing E2F, c-myc, and other proteins) may explain our observation that C/EBPα expression leads to smaller colonies and seems to accelerate myeloid differentiation in clonogenic assays.
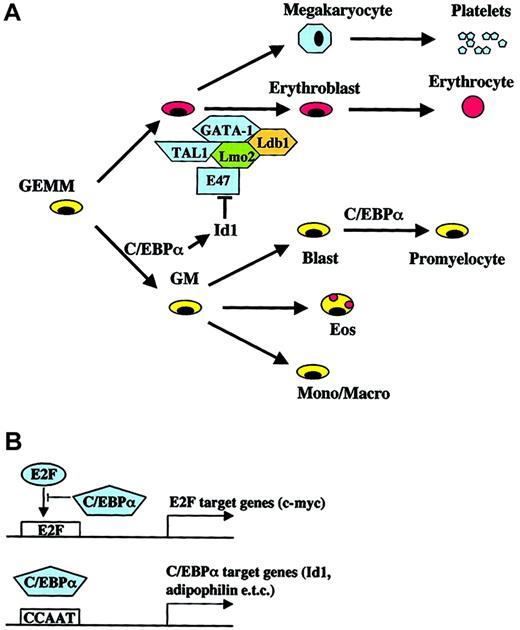
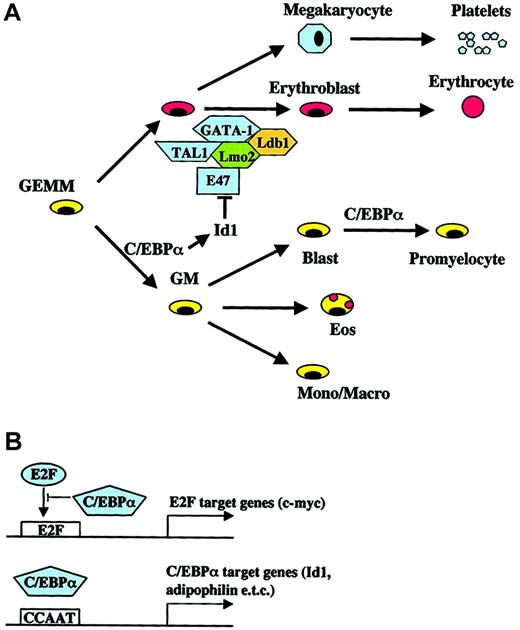
Models for the role of C/EBPα in hematopoietic differentiation and lineage commitment.
(A) Known effects of Id1 expression on erythroid differentiation and the complex of GATA-1, TAL1 (SCL), Lmo2, and Ldb1 are shown. (B) Different mechanisms of “direct” target gene activation/repression by C/EBPα (for review see Loken et al18).
Models for the role of C/EBPα in hematopoietic differentiation and lineage commitment.
(A) Known effects of Id1 expression on erythroid differentiation and the complex of GATA-1, TAL1 (SCL), Lmo2, and Ldb1 are shown. (B) Different mechanisms of “direct” target gene activation/repression by C/EBPα (for review see Loken et al18).
C/EBPα expression in primary human hematopoietic cells also induced the expression of genes involved in the acute-phase response to bacterial infection or inflammation, such as those regulated by tumor necrosis factor α (TNFα; TSG-6, TSG-14, IL-6, COX-2), and exodus, those induced by IL-1 (IEX-1, IL-6, and IL-1RA), and several cytokine genes directly (pro–IL-1β and IL-6) (shown in Table 4). These cytokines also promote the proliferation and/or survival of early myeloid cells. Several of the C/EBPα target genes are also regulated by nuclear factor (NF)–κB (eg, IL-6, IEX-1, and COX-2), and C/EBPα and NF-κB may cooperatively regulate the transcription of these genes. This is of interest because C/EBPα−/− mice lack the acute-phase response to inflammation and other members of the C/EBP family cannot substitute for this function.26 Mature granulocytes are required for the nonspecific host defense against bacterial infections, and bacterial products induce a rapid and persistent increase in granulocyte numbers in the peripheral blood. Besides regulating acute-phase response genes, C/EBPα expression leads to a faster maturation of granulocytes, which may also contribute to this response.
We observed up-regulation of cyclin A1 and cyclin D3 expression following C/EBPα activation (in both of the CD71brightexperiments and 2 of the 3 GFP experiments), but their role in the observed phenotypic changes is unclear. The identification of cyclin A1 as a C/EBPα target gene is perhaps surprising because cyclin A1 is highly expressed in leukemic cell lines and expression of cyclin A1 leads to leukemia in mice.27,28 Furthermore, cyclin A1 is induced after expression of the oncogenic fusion proteins PML/RARα and promyelocytic leukemia zinc finger/retinoic acid receptor gene (PLZF/RARα) and the transcription factor c-myb.29 30
We identified 2 genes that could help explain the phenotype of the C/EBPα−/− mice, namely adipophilin and glycogen phosphorylase. Glycogen phosphorylase is important for glycogenolysis, while adipophilin is involved in lipid storage. These proteins are relevant because C/EBPα−/− mice have no white fat, and they die shortly after birth from severe hypoglycemia due to defects in energy homeostasis.31,32 Adipophilin is expressed in monocytes/macrophages but its role in hematopoietic cell commitment and differentiation needs further investigation.33 34
C/EBPα is a key transcription factor involved in hematopoiesis that can enhance commitment toward a specific hematopoietic lineage but also block differentiation toward the “competing” lineage. Such effects can occur either via protein-protein interactions with other lineage-directing transcription factors or through direct binding to the promoter of target genes.35-38 A major phenotypic effect of C/EBPα expression in human CD34+ cells was the block in erythrocytic differentiation. A direct C/EBPα target gene, which was strongly up-regulated in erythroid precursors, is the transcriptional repressor Id1. Id1 is a helix-loop-helix (HLH) protein, which lacks the basic region required for DNA binding but heterodimerizes with and represses other HLH transcription factors.39 Erythroid differentiation is associated with down-regulation of the Id1 protein, whereas constitutive expression of Id1 leads to a block in erythrocyte differentiation even in the presence of transcription factors that promote erythrocyte-specific gene expression.40 The HLH proteins E47 and Tal-1 (SCL) associate with the zinc finger transcription factor GATA-1 and the LIM proteins Lmo2 and Ldb1 in erythroid cells,41 and this complex is disrupted by Id1.42 The Id1 promoter contains 7 potential C/EBP binding sites,43 and we have shown that Id1 mRNA and protein levels are increased in cells expressing active C/EBPα. We propose a molecular mechanism for C/EBPα in lineage commitment that involves up-regulating Id1 expression (and other proteins) that interfere with the erythrocyte differentiation transcriptional program, and at the same time activating genes important in neutrophilic differentiation (Figure 4A). In preliminary experiments, overexpressing murine Id1 in human hematopoietic progenitor cells did not lead to a significant block in erythrocyte differentiation in clonogenic assays (data not shown). It is likely that other genes must cooperate with Id1 to shift the differentiation program away from the erythroid lineage. It is also possible that the level of Id1 expression must be within a certain range to generate a detectable block in differentiation. Similar to the effects of C/EBPα on granulocytic versus erythroid differentiation, constitutive expression of PU.1 in murine hematopoietic progenitor cells blocks T-cell development without impairing macrophage development. The block in T-cell differentiation is accompanied by, among other things, an increase in Id2, which has been shown to inhibit T-cell differentiation.44 45
Unlike prior studies, we have used primary human hematopoietic cells to identify C/EBPα target genes, representing the first attempt to identify target genes of a transcription factor in the physiologic cellular background. The use of multipotential human hematopoietic progenitor cells and the brief period of exposure to active C/EBPα (achieved using the C/EBPα-ER fusion protein) allow us to detect changes in gene expression that are not due to changes in differentiation but rather reflect a change in hematopoietic lineage commitment.
We would like to thank Dr Alan Friedman for providing the C/EBPα-ER cDNA; KIRIN Brewery (Gumma, Japan), Amgen (Thousand Oaks, CA), and Immunex (Seattle, WA) for generously providing us with human cytokines; Dr Malcolm Moore, Dr Robert Benezra, and Paola De Candia (Sloan-Kettering Institute, New York, NY) for providing technical and scientific advice; and Diane Domingo for flow cytometry. The RD18 cell line was a generous gift from Dr Mary Collins. The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infections Diseases (NIAID), NIH from Dr Nathaniel Landau and Dr Dan Littman: SV-A-MLV-env.
Prepublished online as Blood First Edition Paper, October 24, 2002; DOI 10.1182/blood-2002-05-1546.
Supported by National Institutes of Health (NIH) grant DK52208 (S.D.N.), the Leukemia and Lymphoma Society SCOR grant on Myeloid Malignancies, and the Renny Saltzman Leukemia Research Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Stephen D. Nimer, MSKCC, 1275 York Ave, Box 575, New York, NY 10021; e-mail: s-nimer@mskcc.org.