Abstract
The loss of telomeric DNA with each cell division contributes to the limited replicative lifespan of human T lymphocytes. Although telomerase is transiently expressed in T lymphocytes upon activation, it is insufficient to confer immortality. We have previously shown that immortalization of human CD8+ T lymphocytes can be achieved by ectopic expression of the human telomerase reverse transcriptase (hTERT) gene, which encodes for the catalytic component of the telomerase complex. To study the role of endogenous hTERT in the lifespan of human T cells, we blocked endogenous hTERT expression by ectopic expression of dominant-negative (DN) hTERT. Cells expressing DN-hTERT had a decreased lifespan and showed cytogenetic abnormalities, including chromosome ends without detectable telomeric DNA as well as chromosome fusions. These results indicate that while endogenous hTERT cannot prevent overall telomere shortening, it has a major influence on the longevity of human T cells. Furthermore, we show that up-regulation of hTERT in T cells upon activation decreases over time in culture. Long-term–cultured T cells also show a decreased expression of c-myc upon activation, resulting in less c-myc–induced transcription of hTERT. Moreover, memory T cells, which have expanded in vivo upon antigen encounter, expressed a lower level of hTERT upon activation than naive cells from the same donor. The observed inverse correlation between telomerase levels and replicative history suggests that telomerase levels in T cells are limiting and increasingly insufficient to sustain their proliferation.
Introduction
Human T lymphocytes have a limited lifespan. During long-term culture, human T cells proliferate for a restricted number of cell divisions, after which the cells cease to proliferate and become senescent.1 Therefore, in vitro–established antigen-specific CD8+ or CD4+ T clones can usually not be expanded beyond 20 to 30 population doublings,2,3 and subcloning of established human T-cell clones usually fails or yields subclones with very little replicative potential. The proliferative capacity of T cells may therefore be linked to the replicative history of the cells. Replicative senescence of T cells has also been demonstrated in vivo: memory T cells have a decreased proliferative capacity and shorter telomeres compared with naive T cells,4,5 and the telomere length in lymphocytes shows a progressive decrease with donor age.4-7
It has been suggested that the limited lifespan of most human somatic cells is due to progressive telomere shortening.8,9 Telomeres shorten during each cell division until they reach a critical length, at which point the cells undergo cell-cycle arrest and enter the nondividing state known as replicative senescence. Replicative senescence can be overcome by expression of the enzyme complex telomerase, which consists of a catalytic subunit with reverse transcriptase activity, human telomerase reverse transcriptase (hTERT), a RNA template, human telomerase RNA component (hTERC), and associated proteins.10 Ectopic expression of the hTERT gene in human cells has been shown to result in telomere elongation and immortalization of various cell types, including fibroblasts, endothelial cells, and retinal pigment epithelial cells.11,12 We have previously reported that ectopic hTERT expression in CD8+ T cells leads to an extended lifespan, which demonstrates that loss of telomere ends is a limiting factor for the lifespan of T cells.13,14
In contrast to most other somatic cell types, T cells express endogenous telomerase activity, which is highly regulated during both T-cell development and activation.15 Telomerase RNA expression and enzymatic activities are transiently up-regulated by T cells upon activation through T-cell–receptor (TCR) ligation in the presence of appropriate costimulatory signals.16 Despite the fact that forced expression of hTERT results in immortality, the endogenous telomerase activity is insufficient to prevent telomere erosion in vitro and in vivo5,6,17-20 and does not confer immortality to T cells in vitro. This raises the question whether endogenous telomerase plays a role in the lifespan of human T cells. To investigate this, we used a dominant-negative (DN)–hTERT mutant, which was shown to inhibit endogenous telomerase activity in tumor cells, most likely by competing with hTERT for limiting amounts of telomerase RNA. In tumor cells, expression of DN-hTERT led to an enhanced degree of telomere erosion upon proliferation.21,22 We examined the consequences of inhibition of endogenous hTERT activity in human T cells by forced expression of 2 different DN versions of the hTERT gene (DN-hTERT). We show here that T cells transduced with DN-hTERT have a shorter lifespan than untransduced cells and accumulate cytogenetic abnormalities, which indicates that endogenous telomerase activity is involved in regulating the lifespan of primary human T cells. Strikingly, DN-hTERT expression led to loss of detectable telomere repeats on multiple chromosomes as well as chromosome fusions. However, no generalized telomere shortening was observed, suggesting a role of endogenous telomerase activity in the repair of critically short telomeres. In T cells after extensive in vitro expansion and in freshly isolated memory and naive T cells, we observed that hTERT transcription upon activation decreased with the replicative age of the cells. This decrease correlated with decreased levels in c-myc expression. These results demonstrate the importance of endogenous telomerase activity for the lifespan of telomerase-positive somatic cells and provide a plausible explanation for the limited lifespan of T cells.
Materials and methods
Retroviral constructs
The hTERT retroviral construct hTERT–enhanced green fluorescence protein (GFP) was generated by insertion of hTERT cDNA into the polylinker of LZRS-linker internal ribosomal entry site (IRES)–GFP, as described.13 Amphotropic retrovirus was made by transfection of the retroviral construct into ΦNX-A cells. DN-hTERT (Asp712Ala, Val713Ile), kindly provided by Dr Robert Weinberg (Massachusetts Institute of Technology [MIT], Boston), was generated by substitution of the aspartic acid and valine residues at positions 712 and 713 in the third reverse transcriptase (RT) motif of hTERT with alanine and isoleucine, respectively, as described.21 The DN-hTERT (Asp712Ala, Val713Ile) cDNA was inserted into the polylinker of retroviral vector LZRS-IRES-GFP to enable retrovirus production.
DN-hTERT (Asp868Ala, Asp869Ala) complementary DNA, kindly provided by Dr Lea Harrington (University of Toronto, ON, Canada) was generated by substitution of the aspartic acid residues at position 868 and 869 with alanine residues.22 DN-hTERT (Asp868Ala, Asp869Ala) or the full-length hTERT (cDNA kindly provided by Dr Robert Weinberg) were inserted into a murine stem cell virus (MSCV)–based retroviral vector containing the gene for GFP (Clontech, Palo Alto, CA) under the control of phosphoglycerate kinase 1 (PGK) promoter.23 Helper-free retrovirus, pseudotyped with the gibbon ape leukemia virus (GALV) envelope for efficient infection of human cells, was generated using PG13 packaging cells.24
Isolation of human CD4+ and CD8+ T cells
The Netherlands Cancer Institute's institutional review board approved these studies. Informed consent was provided according to the Declaration of Helsinki. Peripheral blood mononuclear cells (PBMCs) were isolated from healthy individuals by density centrifugation using Ficoll-Hypaque (Pharmacia, Uppsala, Sweden). PBMCs were stained with CD4-PE (phycoerythrin; Becton Dickinson, San Jose, CA) and CD8-FITC (fluorescein isothiocyanate; Becton Dickinson). CD4+ or CD8+ cells were sorted by fluorescence-activated cell sorting (FACStar Plus, Vantage SE; Becton Dickinson) and cultured in RPMI 1640 medium (Gibco, Grand Island, NY) containing 10% human serum (HS) supplemented with phytohemagglutinin (PHA; Gibco), 100 U/mL recombinant human interleukin 2 (rhIL-2; Roche, Nutley, NJ) and 0.5 × 106/mL irradiated (30 Gy) allogeneic mononuclear feeder cells. The cultures were stimulated weekly with the feeder cells and PHA until sufficient cells were obtained for transduction. The number of population doublings (PDs) was calculated from the average cell count using the following equation: PD = 10log (number of cells counted after expansion) - 10log (number of cells seeded)/10log 2.
Human T-cell clones
The human CD4+ T-cell clones MoT-72 and MoT-81, specific for the tetanus toxoid peptides (941-960) and (16-35), respectively, were generated from the PBMCs of a healthy individual vaccinated with tetanus toxoid. The human CD4+ T-cell clone BOY JF161 was obtained from CD4+ T cells isolated from a skin biopsy taken from a healthy donor following sensitization and challenge with dinitrochlorobenzene.25
Retrovirus-mediated hTERT transduction in T cells and monitoring of cell growth
CD4+ or CD8+ T cells were stimulated with PHA (Gibco) and irradiated allogeneic mononuclear feeder cells prior to transduction. T cells were transduced with retroviral supernatant using 24-well plates coated with fibronectin fragments as described.26,27 CD4+ T-cell clone MoT-72 was transduced on day 2 after stimulation followed by a second transduction overnight with retrovirus encoding DN-hTERT (Asp712Ala, Val713Ile)–GFP, full-length hTERT-GFP, or control GFP only. Transduction efficiency ranged from 15% to 35%. The bulk of transduced and untransduced cells was cultured by weekly stimulation with a feeder cell mixture consisting of irradiated allogeneic PBMCs (40 Gy), the Epstein-Barr virus (EBV) B-cell line JY (80 Gy), 100 ng/mL PHA and 40 U/mL rhIL-2 for 4 months.28 All cultures were maintained under equal conditions and the number of population doublings was calculated weekly. The percentage GFP-positive cells in the bulk cultures of DN-hTERT (Asp712Ala, Val713Ile)–GFP, full-length hTERT-GFP, or control GFP-transduced cells and untransduced cells was measured weekly by FACS analysis. Polyclonal T cells, as well as CD8+ T-cell clones obtained by fluorescence-activated single cell sorting (FACStar Plus; Becton Dickinson) were transduced with the DN-hTERT (Asp868Ala, Asp869Ala), full-length hTERT, or control GFP-encoding retrovirus. These retroviruses were all produced as GALV envelope pseudotyped retrovirus. The transduction procedure was repeated on 2 consecutive days. The efficiency of transduction ranged from 1% to 40%. After transduction, cells were expanded for 5 to 6 days and then sorted for GFP expression. DN-hTERT (Asp868Ala, Asp869Ala)–GFP, full-length hTERT-GFP, or control GFP-positive cells were mixed with untransduced T cells in a 50%:50% ratio and cultured for 55 to 65 days. The percentage of GFP-positive cells was measured during the culture to estimate the expansion of the DN-hTERT (Asp868Ala, Asp869Ala)–expressing cells compared with untransduced T cells.
Limiting dilution of GFP-positive cells of clone MoT-72 transduced with DN-hTERT (Asp712Ala, Val713Ile)–GFP, full-length hTERT-GFP, or control GFP was performed by FACStar Plus (Becton Dickinson) in 100-, 30-, 10-, 3-, or 1-cell-per-well densities in round-bottom 96-well plates. Sorted cells were cultured by biweekly stimulation with the feeder cell mixture and rhIL-2 until growing cultures were visible for cloning efficiency determination. The cloning efficiency was calculated as follows: in a negative linear regression curve, the number of cells seeded per well (x-axis) was plotted as a function of the fraction of negative wells (y-axis). The x-value corresponding to a y-value of 0.37 was determined from the graph and indicates the number of cells of which one cell grew out as a clone. This value was recalculated as a percentage indicating the cloning efficiency. Subclones were analyzed for GFP expression and cultured by weekly stimulation with the feeder cell mixture and rhIL-2.
TRAP assay
Telomerase activity was measured by the telomeric repeat amplification protocol (TRAP) assay using an end-labeled telomerase substrate (TS) primer as described.29 DN-hTERT (Asp868Ala, Asp869Ala)–GFP, hTERT-GFP, or control GFP-transduced T cells were sorted 5 days after transduction and cell extracts were obtained from 1 × 105 T cells stimulated with 1 μg/mL PHA, 100 U/mL rhIL-2, and irradiated feeder cells for 2, 4, or 6 days. Cells of the erythroid-myeloid cell line K562 were tested as a positive control cell line for telomerase expression. Control cultures containing feeder cells only were tested to exclude the presence of telomerase activity in the irradiated feeder cells. Elongation of the TS primer by telomerase was performed at room temperature for 30 minutes, and the products were amplified by 30 cycles of polymerase chain reactions (PCR) at 95°C for 60 seconds, 50°C for 45 seconds, and 72°C for 60 seconds using the anchored return primer. The amplified products were resolved on a 12% polyacrylamide gel and visualized by a phosphoimaging system (Storm 820; Molecular Dynamics, Sunnyvale, CA).
RT-PCR
CD4+ T-cell clones were stimulated with anti-CD3 mAb SPV-T3b30 and anti-CD28 mAb B-T3 (a kind gift from Dr John Wijdenes, Diaclone, Besançon, France) in the presence of 20 U/mL rhIL-2. Cell pellets of 105 cells were snap frozen at day 0, 1, 3, 5, 7, and 11. RNA was isolated from the cell pellets using the RNAeasy kit (Qiagen, Crawley, West Sussex, United Kingdom) and eluted in a volume of 50 μL. RNA (12 μL) was transcribed into cDNA using Superscript II reverse transcriptase (GIBCO-BRL, Life Sciences; Breda, The Netherlands) according to the manufacturer's protocol. PCR reactions were performing using primers 5′-GACACAAACATGATTCAAATCCCTGA-3′ and 5′-TATGGACAGGACTGAACGTCTTGC-3′ to detect hypoxanthine-guanine phosphoribosyltransferase (HPRT) expression for cDNA quantification. Endogenous hTERT expression was detected by primer hTERT 3, 5′-CCTCAGACTCCCAGCGGTGC-3′, which anneals in the nontranslated region of hTERT mRNA that is absent in the ectopic hTERT construct in combination with primer hTERT 5, 5′-CTGCAGGCGTACAGGTTTCACG-3′. PCR reactions of primer hTERT 5 and primer IRES 3, 5′-GAGAGGGGCGGAATTTACGTAG-3′, which anneals to the IRES sequence present in the hTERT retroviral construct, were used to detect ectopic hTERT expression. PCR reactions were run at 94°C for 4 minutes followed by 30 cycles of 94°C for 30 seconds, 68°C for 30 seconds, and 72°C for 1 minute. For endogenous hTERT detection 35 cycles were run to detect low levels of hTERT expression. PCR reactions specific for c-myc were performed using primers, 5′-TCGGATTCTCTGCTCTCCTC-3′ and 5′-TTCCGCAACAAGTCCTCTTC-3′, and run in parallel with HPRT PCR reactions at 94°C for 4 minutes followed by 30 cycles of 94°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds. PCR products were visualized on a 1.5% agarose gel by ethidium bromide staining. PCR product bands were quantified relative to HPRT expression using the Phoretix 1D Advanced imager software (Nonlinear Dynamics, Newcastle upon Tyne, United Kingdom). PCR reactions were performed on serial 2-fold dilutions of the cDNA samples. Relative levels of gene expression were calculated from the linear part of the dilution curve, normalized to the HPRT expression level.
Real-time PCR
PBMCs from 3 donors were incubated with PE-conjugated anti-CD27 mAbs (Becton Dickinson) and Cy-5–conjugated anti-CD45RA mAbs (Becton Dickinson), washed, and CD27+CD45RA+ (naive) and CD27+CD45RA- (memory) T cells were sorted by fluorescence-activated cell sorting. Sorted cells, which were of more than 95% purity and consisted of equal numbers of CD28+ T cells, were stimulated with anti-CD3 and anti-CD28 mAb–coated beads (Dynabeads CD3/CD28 T-cell expander; Dynal, Oslo, Norway) and 20 U/mL rhIL-2 or cultured in rhIL-2 alone for 2 days, and snap frozen at -80°C as pellets of 2 × 105 cells. RNA was extracted from the cells using the RNAeasy kit and eluted in a volume of 50 μL. RNA (11 μL) was transcribed into cDNA by 5 U/μL reverse transcriptase (Superscript II; Invitrogen, Breda, The Netherlands) using 12.5 μM random hexamers (Applied Biosystems, Foster City, CA), in the presence of 1 mM deoxynucleoside triphosphates (dNTPs), 0.01 M dithiothreitol (DTT), and 1 U/μL RNAse inhibitor (Applied Biosystems), in a volume of 40 μL. Per reaction, 5 μL cDNA was amplified in a Sequence Detector (Perkin Elmer, Norwalk, CT) using 5′ primer 5′-TTTTCTACCGGAAGAGTGTCTGG-3′, 3′ primer 5′-GCTTCCCGATGCTGCCT-3′, and carboxyfluorescein (FAM; Applied Biosystems)–labeled probe 5′-TTGCAAAGCATTGGAATCAGACAGC ACT-3′ for hTERT detection. hTERT PCR reactions were run in triplicate. Expression of the housekeeping genes, human TATA box–binding protein (TBP), glyceraldehyde phosphate dehydrogenase (GAPDH), and β-actin (Applied Biosystems) were tested in duplicate on the cDNA samples. Expression of the housekeeping gene porphobilinogen deaminase (PBGD) was tested using 5′ primer 5′-ACGATCCCGAGACTCTGC-3′, 3′ primer 5′-GCACGGCTACTGGCACACT-3′, and VIC-labeled probe 5′-CCTGAGGCACCTGGAAGGAGGCTG-3′ (kindly provided by C. Bosch and Dr E. Robanus-Maandag, The Netherlands Cancer Institute, Amsterdam). A 10-fold dilution range of cDNA obtained from the telomerase-expressing B-cell line JY was tested in duplicate to define a standard curve for the expression level. Standard curves were linear with a correlation coefficient higher than 0.998 and a negative slope of more than 3.5 to ensure reliable quantitative amplification detection. Gene expression of the samples was analyzed in the linear range of the amplification plot using the Sequence Detector System software (Perkin Elmer) and expressed as a value relative to the standard curve.
Telomere analysis by quantitative FISH on metaphase chromosomes
Cloned CD4+ T lymphocytes that had undergone approximately 24 PDs were transduced with DN-hTERT (Asp868Ala, Asp869Ala)–GFP, full-length hTERT-GFP, or control GFP. Cells of an approximate age of 45 PDs were stimulated with 1 μg/ml PHA, 100 U/ml rhIL-2, and 0.5 × 106/mL irradiated allogeneic feeder cells for 4 to 5 days before addition of colcemid for 1 hour. Cells were then treated with hypotonic KCl for 50 minutes at 37°C and fixed in methanol–acetic acid. Quantitative fluorescence in situ hybridization (Q-FISH) on metaphase chromosomes was performed with Cy-3–labeled (CCCTAA)3 PNA probe and subsequent quantitative analysis of digital images as previously described.31,32 Briefly, slides were observed with an Axioplan microscope (Zeiss, Thornwood, NY) equipped with a charged coupled device (CCD) camera. Separate images were captured for DAPI (4,6 diamidino-2-phenylindole) and Cy-3 and subjected to telomere fluorescence measurements using TFL-Telo software.33 Metaphases were analyzed for the presence of chromosomal abnormalities, such as dicentric chromosomes, and for the number of detectable telomeres per chromosome. Individual telomere length was quantified by the level fluorescence intensity of each telomere spot, expressed in telomere fluorescence units (TFUs).
Results
Dominant-negative hTERT expression decreases the lifespan of human CD4+ T cells
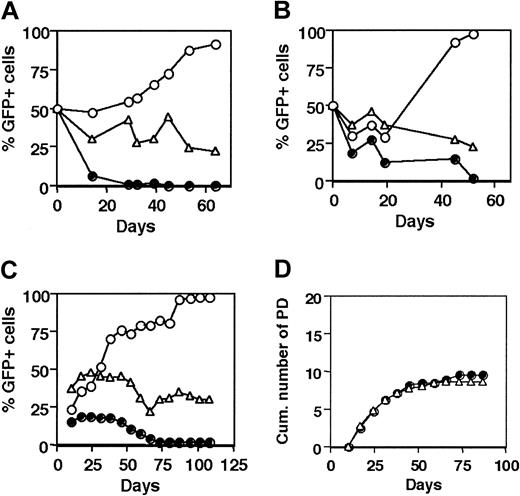
We tested the contribution of endogenous hTERT to the lifespan of human T cells by expression of 2 different DN mutants of the hTERT gene, DN-hTERT (Asp868Ala, Asp869Ala) and DN-hTERT (Asp712Ala, Val713Ile), which both inhibit telomerase activity in tumor cells.21,22 In human polyclonal T cells isolated from PBMCs and transduced with DN-hTERT (Asp868Ala, Asp869Ala)–GFP, hTERT-GFP, or the empty vector–encoding GFP only, the telomerase activity upon stimulation was reduced in DN-hTERT–transduced cells compared with control GFP-transduced cells (Figure 1). Polyclonal CD4+ T cells that had been cultured for a short period of time before transduction with DN-hTERT (Asp868Ala, Asp869Ala)–GFP were cultured in a 50%:50% ratio with untransduced cells and the percentage of GFP-expressing cells was measured during the culture (Figure 2A). The percentage of DN-hTERT–GFP–positive cells decreased over time during the culture and disappeared from the culture within 30 to 50 days, demonstrating that these cells did not expand and died earlier than the untransduced cells in the same culture. Similar results were obtained in experiments of cloned CD8+ cells transduced with DN-hTERT–GFP, indicating that DN-hTERT expression also led to early death of cells of an at least 20-PD higher replicative age acquired during T-cell cloning (Figure 2B). In control cultures of cells transduced with the empty GFP-containing vector, the percentage of GFP-positive cells did not change over time. In contrast, hTERT-GFP–transduced cultures showed an accumulation of cells expressing the hTERT-GFP gene. The limiting effect of DN-hTERT expression on the lifespan of human T cells was confirmed with a second DN mutant of the hTERT gene, DN-hTERT (Asp712Ala, Val713Ile), expressed in the CD4+ T-cell clone MoT-72. MoT-72 cells transduced with DN-hTERT (Asp712Ala, Val713Ile)–GFP, were cleared from mixed cultures of transduced and untransduced cells within 75 days (Figure 2C). These results indicate that, compared with untransduced or control GFP-transduced T cells, expression of a dominant-negative mutant of the hTERT gene confers a growth disadvantage in both CD4+ and CD8+ T cells.
Inhibition of telomerase activity by ectopic DN-hTERT expression. Telomerase activity in a culture of polyclonal T cells isolated from donor PBMCs transduced with DN-hTERT (Asp868Ala, Asp869Ala)–GFP, hTERT-GFP, or control GFP after 2, 4, and 6 days of stimulation. Telomerase activity was detected in control GFP-transduced cells 2 to 4 days after stimulation and was absent in the DN-hTERT–transduced cells. In hTERT-transduced cells telomerase activity was present at all time points analyzed. Each lane shows the telomerase activity present in 2000 cells analyzed by the TRAP assay. K562 cells were tested as a positive control for telomerase activity. Results are representative of 3 experiments.
Inhibition of telomerase activity by ectopic DN-hTERT expression. Telomerase activity in a culture of polyclonal T cells isolated from donor PBMCs transduced with DN-hTERT (Asp868Ala, Asp869Ala)–GFP, hTERT-GFP, or control GFP after 2, 4, and 6 days of stimulation. Telomerase activity was detected in control GFP-transduced cells 2 to 4 days after stimulation and was absent in the DN-hTERT–transduced cells. In hTERT-transduced cells telomerase activity was present at all time points analyzed. Each lane shows the telomerase activity present in 2000 cells analyzed by the TRAP assay. K562 cells were tested as a positive control for telomerase activity. Results are representative of 3 experiments.
Dominant-negative hTERT-transduced T cells disappear early from bulk cultures. (A) Human polyclonal CD4+ T cells transduced with either DN-hTERT (Asp868Ala, Asp869Ala)–GFP (•), hTERT-GFP (○), or control GFP (▵) were mixed with untransduced cells in a 50%:50% ratio and cultured in parallel for 53 days to determine the lifespan compared with the untransduced T cells. The graph shows the percentage of GFP-positive cells during the culture. Results are representative of 3 experiments. (B) CD8+ T-cell clone transduced and cultured as described in panel A. Results are representative of 2 experiments. (C) CD4+ T-cell clone MoT-72 was transduced with either DN-hTERT (Asp712Ala, Val713Ile)–GFP (•), hTERT-GFP (○), or control GFP (▵), and the percentage of GFP-positive cells was measured weekly during the culture. The graph shows the percentage GFP-positive cells during the culture. Transduction efficiencies at the onset of the cultures were as follows: hTERT-GFP, 20%; GFP control, 35%; and DN-hTERT–GFP, 15%. Results are representative of experiments with 6 different CD4+ T-cell clones. Panels A-C show that in both CD4+ and CD8+ T-cell cultures, ectopic expression of DN-hTERT decreased the lifespan compared with untransduced T cells. (D) Cumulative number of population doublings of the untransduced GFP-negative cells in the mixed culture containing DN-hTERT (•) or GFP-transduced T cells (▵). The graph shows that the untransduced T cells were growing equally well in both mixed cultures, indicating that the decrease in percentage DN-hTERT–GFP–expressing T cells in DN-hTERT-GFP–transduced cultures was not due to an enhanced proliferation of the untransduced T cells compared with untransduced T cells in the control GFP-transduced cultures.
Dominant-negative hTERT-transduced T cells disappear early from bulk cultures. (A) Human polyclonal CD4+ T cells transduced with either DN-hTERT (Asp868Ala, Asp869Ala)–GFP (•), hTERT-GFP (○), or control GFP (▵) were mixed with untransduced cells in a 50%:50% ratio and cultured in parallel for 53 days to determine the lifespan compared with the untransduced T cells. The graph shows the percentage of GFP-positive cells during the culture. Results are representative of 3 experiments. (B) CD8+ T-cell clone transduced and cultured as described in panel A. Results are representative of 2 experiments. (C) CD4+ T-cell clone MoT-72 was transduced with either DN-hTERT (Asp712Ala, Val713Ile)–GFP (•), hTERT-GFP (○), or control GFP (▵), and the percentage of GFP-positive cells was measured weekly during the culture. The graph shows the percentage GFP-positive cells during the culture. Transduction efficiencies at the onset of the cultures were as follows: hTERT-GFP, 20%; GFP control, 35%; and DN-hTERT–GFP, 15%. Results are representative of experiments with 6 different CD4+ T-cell clones. Panels A-C show that in both CD4+ and CD8+ T-cell cultures, ectopic expression of DN-hTERT decreased the lifespan compared with untransduced T cells. (D) Cumulative number of population doublings of the untransduced GFP-negative cells in the mixed culture containing DN-hTERT (•) or GFP-transduced T cells (▵). The graph shows that the untransduced T cells were growing equally well in both mixed cultures, indicating that the decrease in percentage DN-hTERT–GFP–expressing T cells in DN-hTERT-GFP–transduced cultures was not due to an enhanced proliferation of the untransduced T cells compared with untransduced T cells in the control GFP-transduced cultures.
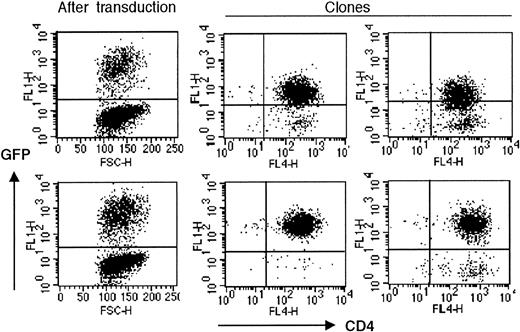
The observed growth disadvantage following DN-hTERT expression was further investigated by limiting dilution experiments of sorted DN-hTERT (Asp712Ala, Val713Ile)–GFP expressing cells. In 2 independent experiments, the efficiency of subcloning DN-hTERT (Asp712Ala, Val713Ile)–GFP–transduced cells of clone MoT-72 was lower than the subcloning efficiency of untransduced cells (Table 1), or cells transduced with hTERT-GFP, indicating that many DN-hTERT–transduced T cells could not expand another 20 PDs in culture to establish a subclone. The few subclones that were obtained from the cloning of DN-hTERT (Asp712Ala, Val713Ile)–GFP–expressing cells all died soon after cloning at lower population doublings than control GFP-transduced subclones. No GFP-negative clones were obtained in any experiments. Since the DN-hTERT–expressing subclones expressed low levels of GFP, these subclones had probably arisen by selective outgrowth of cells with a relatively low transgene expression, in which inhibition of endogenous telomerase activity is less efficient (Figure 3). Thus, cells with higher levels of GFP and DN-hTERT expression had selectively disappeared from the cultures at earlier time points. In contrast, all subclones obtained from hTERT-GFP or control GFP-transduced cells expressed GFP at levels comparable to those right after transduction, indicating that, in these cultures, cells with a high level of GFP expression had no selective disadvantage during cloning (Figure 3).
Subclones of DN-hTERT–transduced T cells display lower DN-hTERT gene expression. The level of GFP expression is decreased in DN-hTERT–GFP–transduced clones. Two clones isolated from limiting dilution of MoT-72 transduced with DN-hTERT (Asp712Ala, Val713Ile)–GFP (top middle and top right), displayed a lower GFP expression than the average GFP level expressed by the bulk culture right after transduction (top left). Graphs are representative of the GFP expression by all DN-hTERT clones obtained. All DN-hTERT (Asp712Ala, Val713Ile)–GFP–expressing clones died after cloning. The GFP expression remained high in clones isolated from cloning of MoT-72 transduced with hTERT-GFP (bottom middle and bottom right). This level of GFP expression was equal to the GFP expression level after transduction (bottom left). Graphs are representative of 16 hTERT-GFP–expressing clones analyzed.
Subclones of DN-hTERT–transduced T cells display lower DN-hTERT gene expression. The level of GFP expression is decreased in DN-hTERT–GFP–transduced clones. Two clones isolated from limiting dilution of MoT-72 transduced with DN-hTERT (Asp712Ala, Val713Ile)–GFP (top middle and top right), displayed a lower GFP expression than the average GFP level expressed by the bulk culture right after transduction (top left). Graphs are representative of the GFP expression by all DN-hTERT clones obtained. All DN-hTERT (Asp712Ala, Val713Ile)–GFP–expressing clones died after cloning. The GFP expression remained high in clones isolated from cloning of MoT-72 transduced with hTERT-GFP (bottom middle and bottom right). This level of GFP expression was equal to the GFP expression level after transduction (bottom left). Graphs are representative of 16 hTERT-GFP–expressing clones analyzed.
Cytogenetic abnormalities in DN-hTERT–transduced cells
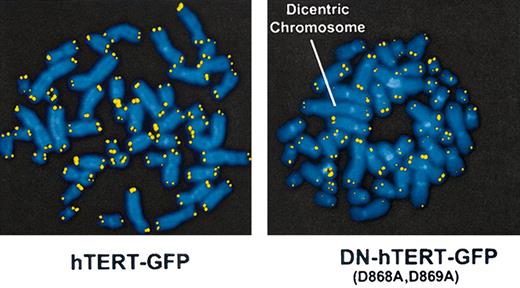
Next, we studied the effect of endogenous telomerase inhibition on chromosomal stability and telomere length by Q-FISH (Tables 2 and 3; Figure 4). For this purpose, we transduced the progeny of a single CD4+ T cell with retroviral vectors containing GFP, GFP-hTERT, or GFP-DN (Asp868Ala, Asp869Ala)–hTERT. Figure 4 shows preparations of metaphase chromosomes of sorted GFP-positive cells hybridized with Cy-3–labeled (CCCTAA)3 peptide nucleic acid probe specific for telomeric DNA. Metaphase chromosomes of cells that were transduced with the GFP-DN (Asp868Ala, Asp869Ala)–hTERT display a higher frequency of chromosomes without detectable telomeric DNA than those in control GFP-transduced cells. On average, only 3.5 telomeres were detectable per chromosome in DN (Asp868Ala, Asp869Ala)–hTERT–transduced cells, whereas all 4 telomeres per chromosome were detected on most chromosomes in control GFP-transduced cells or hTERT-transduced cells (Tables 2, 3). Expression of DN-hTERT significantly decreased the number of detectable telomeres per chromosome compared with control GFP-transduced cells (P < .001) or with hTERT-GFP–transduced cells (P < .001; Table 3).
DN-hTERT–transduced cells show chromosomal abnormalities. Q-FISH analysis of metaphase chromosomes isolated from human CD4+ T cells that have undergone approximately 45 PD and were transduced with DN-hTERT (Asp868Ala, Asp869Ala)–GFP or hTERT-GFP at approximately 24 PD. Telomeres are visualized in yellow by hybridization with the Cy-3–labeled (CCCTAA)3 PNA probe. The arrow indicates a dicentric chromosome found in DN-hTERT (Asp868Ala, Asp869Ala)–GFP–transduced cells. Results are representative of 16 metaphases of DN-hTERT–transduced cells and 20 metaphases of hTERT-transduced cells analyzed.
DN-hTERT–transduced cells show chromosomal abnormalities. Q-FISH analysis of metaphase chromosomes isolated from human CD4+ T cells that have undergone approximately 45 PD and were transduced with DN-hTERT (Asp868Ala, Asp869Ala)–GFP or hTERT-GFP at approximately 24 PD. Telomeres are visualized in yellow by hybridization with the Cy-3–labeled (CCCTAA)3 PNA probe. The arrow indicates a dicentric chromosome found in DN-hTERT (Asp868Ala, Asp869Ala)–GFP–transduced cells. Results are representative of 16 metaphases of DN-hTERT–transduced cells and 20 metaphases of hTERT-transduced cells analyzed.
In addition, several dicentric chromosomes were observed in the GFP-DN (Asp868Ala, Asp869Ala)–hTERT–transduced cells, which were absent in the GFP- or hTERT-GFP–transduced cells. No telomeric DNA was detected at the junction site in the dicentric chromosomes. The chromosome fusions observed in GFP-DN (Asp868Ala, Asp869Ala)–hTERT–transduced cells involved fusions of chromosomes 1, 7, and 6 with chromosome 20, 22, and 10, respectively (Figure 4; Table 2). Q-FISH analysis revealed that the average length of the telomeres in DN (Asp868Ala, Asp869Ala)–hTERT-GFP–transduced cells was comparable to control GFP- or hTERT-GFP–transduced cells. Strikingly, Q-FISH analysis also revealed that DN-hTERT expression increased the frequency of very short telomeres that are undetectable by in situ hybridization (Figure 5). Most likely some of the chromosomes with undetectable telomere repeats in DN-hTERT expressing cells underwent chromosome end-to-end fusions. The observation that inhibition of endogenous telomerase activity did not lead to a decrease in the average telomere length, but instead to an increase in the number of undetectable telomeres and to the occurrence of chromosome fusions, suggests a role of endogenous telomerase activity in the repair of critically short telomeres rather than in mediating telomere elongation in all chromosomes.
Telomere length analysis of human CD4+ T cells transduced with DN-hTERT. Graphs show the quantification of individual telomere length expressed as the distribution of TFUs of Q-FISH telomere analysis of metaphase chromosomes from human CD4+ T cells that have undergone approximately 45 PD and were transduced with DN-hTERT (Asp868Ala, Asp869Ala)–GFP, hTERT-GFP, or control GFP. Analyses were performed on 16 metaphases of DN-hTERT (Asp868Ala, Asp869Ala)–GFP–transduced CD4+ T cells, 20 metaphases of hTERT-GFP–transduced cells, and 28 metaphases of GFP-transduced cells. The average telomere fluorescence of individual chromosome ends was calculated to be 1.35 TFU for DN-hTERT (Asp868Ala, Asp869Ala)–GFP–transduced cells, 1.92 TFU for hTERT-GFP–transduced cells, and 1.60 TFU for control GFP-transduced cells.
Telomere length analysis of human CD4+ T cells transduced with DN-hTERT. Graphs show the quantification of individual telomere length expressed as the distribution of TFUs of Q-FISH telomere analysis of metaphase chromosomes from human CD4+ T cells that have undergone approximately 45 PD and were transduced with DN-hTERT (Asp868Ala, Asp869Ala)–GFP, hTERT-GFP, or control GFP. Analyses were performed on 16 metaphases of DN-hTERT (Asp868Ala, Asp869Ala)–GFP–transduced CD4+ T cells, 20 metaphases of hTERT-GFP–transduced cells, and 28 metaphases of GFP-transduced cells. The average telomere fluorescence of individual chromosome ends was calculated to be 1.35 TFU for DN-hTERT (Asp868Ala, Asp869Ala)–GFP–transduced cells, 1.92 TFU for hTERT-GFP–transduced cells, and 1.60 TFU for control GFP-transduced cells.
Activation-induced endogenous hTERT expression in human T cells decreases with age
The results described here indicate that endogenous hTERT expression is important for the survival of human T cells, raising the question as to why this is not sufficient to confer immortality to the T cells. One possibility is that the limited lifespan of T cells might result from a decrease in the expression of the telomerase complex. We therefore determined the endogenous hTERT expression levels in T cells of different replicative age by comparing the up-regulation of endogenous hTERT expression in early cultures with that in long-term–cultured T cells.
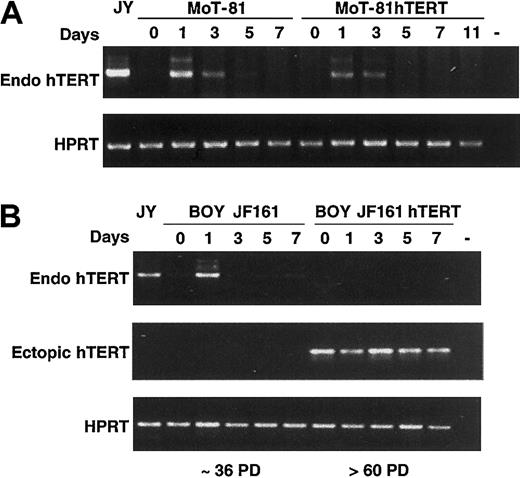
To determine variations in the expression of endogenous hTERT in long-term cultures of hTERT-transduced cells, we developed an RT-PCR analysis using sets of primers that discriminate between endogenous hTERT and transduced hTERT expression. In Figure 6A the expression of endogenous hTERT, 1 to 7 days after antibody-mediated activation in an early culture of the CD4+ T-cell clone MoT-81 (20 PD), is shown. Both wild-type and hTERT-transduced MoT-81 cells with identical replicative age show hTERT expression one day after stimulation, which is no longer detectable after 5 days. Furthermore, these results show that ectopic hTERT expression did not affect the up-regulation of endogenous hTERT expression following TCR/CD3-mediated activation.
Expression of endogenous hTERT upon stimulation during culture. (A) Wild-type MoT-81 cells and hTERT-transduced cells of identical age were stimulated with anti-CD3 and anti-CD28 mAbs, and the expression of endogenous hTERT at day 0, 1, 3, 5, 7, and 11 was determined by RT-PCR. All samples were tested for HPRT expression to check for equal amounts of RNA. (B) RT-PCR analysis of endogenous and ectopic hTERT expression in wild-type BOY JF161 cells after approximately 36 PD and in hTERT-GFP–transduced cells after more than 60 PD.
Expression of endogenous hTERT upon stimulation during culture. (A) Wild-type MoT-81 cells and hTERT-transduced cells of identical age were stimulated with anti-CD3 and anti-CD28 mAbs, and the expression of endogenous hTERT at day 0, 1, 3, 5, 7, and 11 was determined by RT-PCR. All samples were tested for HPRT expression to check for equal amounts of RNA. (B) RT-PCR analysis of endogenous and ectopic hTERT expression in wild-type BOY JF161 cells after approximately 36 PD and in hTERT-GFP–transduced cells after more than 60 PD.
In contrast, in a long-term culture of an hTERT-transduced clone that had undergone more than 60 PDs after its generation, no endogenous hTERT could be detected (Figure 6B). Wild-type cells at an earlier time point (36 PDs) of culture were still able to induce endogenous hTERT expression one day after stimulation with anti-CD3 and anti-CD28 monoclonal antibodies (Figure 6B). Both wild-type and hTERT-transduced cells proliferated extensively following stimulation (data not shown), excluding the possibility that the absence of endogenous hTERT expression in the latter cells was due to the lack of T-cell activation. Taken together, these results show that upon long-term culture, T cells lose the ability to increase endogenous hTERT expression upon activation.
Next, we investigated what caused the decreased hTERT expression in long-term–cultured T cells. The c-myc transcription factor directly activates hTERT transcription by dimerizing with the Max protein and binding to E-box binding sites that are present in the hTERT promoter.34 We analyzed c-myc expression upon activation by RT-PCR analysis with primers that are specific for c-myc (Figure 7). In resting T cells from both long-term and short-term cultures, low levels of c-myc expression were detected (Figure 7 day 0). One day after stimulation with anti-CD3 and anti-CD28 mAbs, the c-myc expression in the relatively young T cells increased more than 70-fold compared with levels of c-myc expression in unstimulated cells (Figure 7A day 0), with a somewhat lower increase (27-fold) after 3 days (Figure 7A). In the long-term–cultured T cells of the same clone, one day after stimulation the activated cells expressed only 7 times more c-myc RNA than unstimulated cells (Figure 7B). Taken together, these results show that during long-term culture T cells have a strongly reduced capacity to up-regulate hTERT expression upon activation, which is correlated with a strongly reduced up-regulation of c-myc.
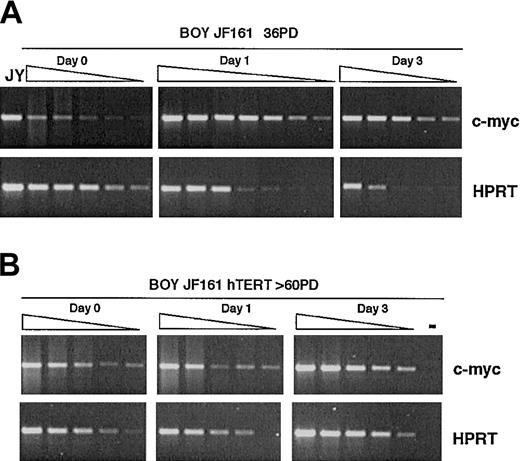
Expression of c-myc upon activation. BOY JF161 T cells (after approximately 36 PD, A) and long-term–cultured BOY JF161-TERT T cells (after more than 60 PD, B) were stimulated with anti-CD3 and anti-CD28 mAbs, and c-myc expression was determined at day 1 and day 3 after stimulation. Day 0 indicates unstimulated cells. c-myc and HPRT PCR reactions were run in parallel on a 1:2 dilution range of the cDNA samples as indicated by the triangles above the lanes. The increase in c-myc expression after activation relative to unstimulated cells (day 0) was quantified by comparing the relative peak intensity of c-myc PCR products at equal intensity of HPRT amplification.
Expression of c-myc upon activation. BOY JF161 T cells (after approximately 36 PD, A) and long-term–cultured BOY JF161-TERT T cells (after more than 60 PD, B) were stimulated with anti-CD3 and anti-CD28 mAbs, and c-myc expression was determined at day 1 and day 3 after stimulation. Day 0 indicates unstimulated cells. c-myc and HPRT PCR reactions were run in parallel on a 1:2 dilution range of the cDNA samples as indicated by the triangles above the lanes. The increase in c-myc expression after activation relative to unstimulated cells (day 0) was quantified by comparing the relative peak intensity of c-myc PCR products at equal intensity of HPRT amplification.
The loss of hTERT expression upon activation may also occur upon expansion in vivo. Memory and effector T cells have expanded upon antigen encounter in vivo and on average have divided considerably more frequently than naive T cells of the same donor. We compared the level of hTERT expression upon stimulation in naive and memory cells isolated from donor PBMCs by quantitative real-time PCR. T cells were sorted by their expression of CD27 and CD45RA to isolate the CD27+CD45RA+ naive and CD27+CD45RA- memory T-cell population.35 The percentage of CD28-expressing cells in the sorted naive and memory T-cell population was identical in each donor, and comprised 80% to 90% (data not shown). Following stimulation with anti-CD3 and anti-CD28 antibodies, we observed an up-regulation of hTERT expression in both activated naive and memory T cells compared with the nonactivated cells. However, hTERT expression levels were lower in activated memory cells than in activated naive cells of the same donor (Table 4). These results suggest that expansion of T cells upon antigen encounter in vivo may eventually result in a lower hTERT expression upon activation.
Discussion
We report here that endogenous telomerase activity plays a role in regulating the lifespan of human T cells. Blocking endogenous telomerase activity by ectopic expression of either the DN-hTERT (Asp712Aala, Val713Ile) or the DN-hTERT (Asp868Ala, Asp869Ala) mutant of the hTERT gene was shown to shorten the lifespan of both short-term– and long-term–cultured CD4+ T cells and CD8+ T cells. The observation that endogenous telomerase does not confer immortality to human T cells might imply that endogenous telomerase is not involved in lifespan regulation. However, our results indicate that this assumption is incorrect, since inactivation of endogenous telomerase resulted in a shortened lifespan. Our observation that upon in vitro culture T cells progressively lose their capacity to up-regulate hTERT expression upon in vitro culture suggests that, upon aging, levels of endogenous telomerase are increasingly insufficient to maintain telomere length of more than a few short telomeres. Importantly, memory T cells were found to express less hTERT upon activation than naive cells from the same donor, indicating that a loss of hTERT up-regulating ability may also occur in vivo.
T cells that had been transduced with DN-hTERT showed a loss of detectable telomeres at many chromosome ends and a high frequency of dicentric chromosomes. The observed chromosome fusions and chromosome ends without detectable telomeric DNA are similar to the type of abnormalities seen in various cells from late generations of the telomerase mTERC knock-out mice.36 The dramatic effect of telomerase inhibition on some, but not all, telomeres is compatible with a role of endogenous telomerase in the repair of sporadic telomere attrition. One possibility is that telomerase is essential for the repair of telomeric DNA following damage by oxygen radicals.37,38 It has been shown that G-rich telomeric DNA is particularly vulnerable to oxidative stress,39,40 and it has been proposed that such damage is the primary cause of telomere attrition in human cells.41 Our results suggest that while endogenous telomerase may be essential to repair sporadic telomere shortening, overall telomere shortening and replicative senescence is not prevented.
One explanation is that endogenous telomerase levels are simply insufficient to deal with more than a few critically short telomeres, resulting in a net telomere loss that increases gradually over time. The observation that ectopic expression of hTERT immortalizes human T lymphocytes13,14 indicates that increased levels of hTERT expression contribute to longevity in T cells. Moreover, the telomerase activity upon hTERT expression may depend on phosphorylation and nuclear translocation of the hTERT protein.42 Another possibility is that broken ends (perhaps following some processing event) are better substrates for telomerase than natural chromosome ends that may be folded into a T loop,43 which is underscored by the recently described possible differences in the processing of telomeres.44 Furthermore, the ability of T cells to up-regulate hTERT expression upon activation may decrease over time, resulting in accelerated telomere erosion in aged T cells.45
Our results strengthen the observation of Weng et al45 of a progressive decrease in telomerase activity at each sequential stimulation in a polyclonal culture of human CD4+ T cells. The loss of telomerase expression upon activation may also occur upon extensive expansion of T cells in vivo. We have shown that memory T cells express lower levels of hTERT when activated ex vivo than naive T cells of the same donor. This decrease in hTERT up-regulation correlates with the greater replicative history of memory T cells compared with naive T cells. However, Weng and coworkers20 reported that activation-induced telomerase activity in polyclonal CD4+ or CD8+ T cells did not decrease with the donor age, whereas the average telomere length was shorter in older individuals. This suggests that donor age may not reveal the differences in replication of the entire CD4+ or CD8+ T-cell population and that variations in telomerase expression may be more evident between T-cell populations within the same donor. The same authors also reported comparable levels of telomerase activity in activated CD45RA+ naive and CD45RO+ memory T cells.46 It cannot be excluded, however, that CD45RA+ effector cells were present in the naive T-cell fraction, which may have a lower level of telomerase induction. We have observed that the total CD45RA+ cell fraction in donor PBMCs contained up to 50% CD45RA+CD27- effector cells. Therefore, contaminating effector cells may have lowered the average telomerase level that was detected in the CD45RA+ T-cell population and thereby may have concealed possible differences in telomerase activity between activated naive and memory T cells. Moreover, the effect of aging on the telomerase expression was also observed in T cells of patients with X-linked lymphoproliferative disorder, characterized by a primary immunodeficiency disease that leads to an inability to regulate the immune response to EBV. In these patients, the EBV-reactive T cells were found to have greatly reduced telomere lengths, as well as a reduced telomerase activity and proliferative capacity upon stimulation in vitro compared with EBV-reactive T cells of chronically infected control individuals (Dr A. Akbar, personal communication).
The impaired hTERT expression may be caused by the reduced c-myc expression upon activation that we have observed in long-term–cultured T cells, since hTERT is a direct target for c-myc.47-49 Strikingly, c-myc expression levels of activated T cells of humans of old age is decreased compared with those in young donors and coincided with a reduced proliferative capacity.50 Transcription of hTERT is tightly regulated by c-myc, by other activators of transcription such as Sp1 and estrogen, as well as by repressors of hTERT transcription, such as Mad1, WT1, p53, and MZF-2.51 Aging of T cells may have an effect on more regulators of hTERT transcription, resulting in a decreased telomerase activity in older cells. The remarkable effect of inhibition and overexpression of telomerase on T-cell growth indicates that telomerase levels in vivo are very tightly controlled. This conclusion is supported by the observation that individuals with a heterozygous defect in telomerase RNA suffer from dyskeratosis congenita, a rare genetic disorder typically resulting in early death form aplastic anemia, cancer, or immune deficiency.52
Prepublished online as Blood First Edition Paper, April 10, 2003; DOI 10.1182/blood-2002-07-2015.
Supported by grant AI29524 from the National Institutes of Health and by a grant from the National Cancer Institute of Canada with funds from the Terry Fox Run.A.R. is funded by a grant from the Deutsche Forschungsgemeinschaft. R.M.L. is supported by a grant from the Dutch Cancer Society (NKI 99-2048).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Robert Weinberg (MIT, Boston, MA) for the DN-hTERT (Asp712Ala, Val713Ile) cDNA, Dr Lea Harrington (University of Toronto) for the DN-hTERT (Asp868Ala, Asp869Ala) cDNA, Dr Marianne Naspetti for designing hTERT and c-myc PCR primers, and Cathy Bosch and Astrid Bosma for providing real-time PCR primers and probes and their excellent help with the Taqman analyses.