Abstract
In the maternal decidua, natural killer (NK) cells, characterized by lack of CD16, are found in direct contact with the fetal extravillous trophoblasts (EVTs). It is yet unknown which factors contribute to the specific homing of this unique NK subset to the decidua. In this study we analyze the chemokine receptor repertoire on various NK populations derived from the peripheral blood and decidua. We show that CXCR4 and CXCR3 receptors are preferentially expressed on CD16– NK subsets derived either from the peripheral blood or the decidua and that these receptors are involved in migration of all NK subsets to their ligands. We further demonstrate in vivo that invading EVTs that eventually perform endovascular invasion express CXCL12, the ligand for CXCR4, but not ligands for CXCR3. Indeed, specific accumulation of the CD16– NK cells at the expense of CD16+ cells was observed only when in vitro migration was performed with ligands for CXCR4. Finally, incubation of the peripheral blood CD16– NK cells with cytokines present in the decidua, especially interleukin 15 (IL-15), resulted in the expression of chemokine receptor repertoire similar to that observed on decidual NK cells, suggesting an additional important regulatory effect of local decidual cytokines.
Introduction
The phenomenon of maternal immune tolerance toward the allogeneic fetus is still not yet fully understood. The fact decidual lymphocytes (including natural killer [NK] cells) tute about 40% of the total tissue cells suggests that they might important in controlling proper decidualization.1 Early in pregnancy the trophoblast cells invade the maternal decidual (including the entire uterine endometrium and the inner third of myometrium).2 Eventually, trophoblast cells destroy the and elastic tissues surrounding maternal spiral arteries, creating a high-capacity low-resistance blood flow to the fetus. Furthermore, invasive trophoblasts start expressing endothelial markers and eventually replace the original endothelial cell of the maternal blood vessels and place themselves in direct with the maternal blood flow.4 The extravillous (EVTs) interact with decidual lymphocytes via an unusual toire of class I major histocompatibility complex (MHC) Proteins. They do not express the HLA-A and HLA-B molecules,5 probably prevent efficient interactions with cytotoxic T cytes (CTLs). The expression of HLA-C protein along with HLA-E and HLA-G proteins,5,6 however, probably inhibits NK attack.7 Decidual lymphocytes were also documented to unique functions in local cytokine production,8 promoting cular invasion9 and placental development.10
More than 70% of decidual lymphocytes are NK cells characterized by CD56brightCD16– (FcRγIII) phenotype.2,3 This outstanding enrichment of the decidua with this specific NK subset is further emphasized when it is compared with the composition of NK cells in the peripheral blood. Only 10% of the peripheral blood lymphocytes are NK cells, and the CD56brightCD16– population composes only 10% to 15% of the total peripheral NK population. Compared with peripheral NK cells, decidual NK cells also display different functional behavior. Decidual NK cells have decreased killing activity against class I MHC-negative target cells11 and are also more susceptible to the inhibition mediated by the MHC class I molecules (classical and nonclassical) expressed by EVTs.12,13
The reasons and the mechanisms controlling the accumulation of the CD56+CD16– NK cells in the decidua are still largely unknown.14 It was recently reported that the chemokine CCL3 is expressed by decidual trophoblasts and plays a fundamental role in attracting various cell populations, including both peripheral CD16+ and CD16– NK subsets.15 These observations, however, cannot explain the specific preferential accumulation of the CD16– NK cells in the decidua, and in addition, normal recruitment of peripheral NK cells was observed to the decidua of mice in which the receptor for CCL3, CCR5, was knocked out.16 We, therefore, explored whether other chemokine receptors and their ligands are involved in the preferential accumulation of the CD16– NK cells in the decidua.
Chemokines are a group of small (8-14 kd) structurally related proteins that are subdivided into 4 subfamilies.17 The various chemokines interact with a subset of 7 transmembrane G protein–coupled receptors termed chemokine receptors.18 Chemokines and chemokine receptors have been implicated as pivotal players in many physiologic and pathologic situations. Some of the notable examples are their involvement in processes such as trafficking of immune cells to multiple target organs.19-22 In addition, chemokines play an important role in central nervous system development,23 in HIV pathogenesis,24 in controlling and targeting cancer cell metastasis,25 and in homeostasis of the hematopoietic system.26,27
By comparing various NK subsets derived either from peripheral blood lymphocytes (PBLs) or from the decidua, we observed that the main chemokine receptors expressed by decidual NK cells are CXCR3 and CXCR4. These 2 receptors are also widely expressed on peripheral CD16– NK cells and at significantly lower levels on peripheral CD16+ NK cells. Ligands for both receptors were demonstrated in decidual extracts both at mRNA and protein levels. We demonstrate that CXCR4/CXCL12, CXCR3/CXCL9, and CXCR3/CXCL10 interactions are involved in the migration of decidual and peripheral NK cells. Most importantly, analyzing CD16 phenotype on total migrated peripheral NK cells to these chemokines demonstrated a dramatic accumulation of CD16– NK cells at the expense of CD16+ cells only when CXCR4 ligand (CXCL12) was used. In vivo immunohistochemistry analysis demonstrated specific CXCL12 expression on invasive trophoblasts, but not of CXCL9 or CXCL10. Invading trophoblasts continue to express CXCL12 even after replacing endothelial cell lining of maternal decidual blood vessels. Finally, incubation of peripheral CD16– NK cells with various decidual cytokines resulted in acquisition of chemokine receptor repertoire resembling that of the decidual CD16– NK subset. Cross talk between trophoblasts expressing CXCL12 and CD16– subpopulation of NK cells expressing CXCR4 may play a fundamental role in the recruitment and motility of these NK cells to the decidua.
Materials and methods
Isolation of peripheral NK cells
Blood samples were taken from healthy donors and loaded on Ficoll density gradient to purify the lymphocyte population. NK cells were purified using the human NK cell isolation kit and the autoMACS instrument (Miltenyi Biotec, Bergisch-Gladbach, Germany), according to the manufacturer's instructions. Identity of NK cells was confirmed as they stained positive for CD56 and negative for CD3.
Isolation of decidual NK cells
The institutional board of Hadassah Medical School approved obtaining deciduae and placentas from elective pregnancy termination procedures, according to the principles of the Declaration of Helsinki. Decidual lymphocytes were isolated as previously described.8,28,29 Briefly, the tissue was trimmed into 1-mm pieces and enzymatically digested for 20 minutes, using vigorous shaking, with 1.5 mg type I DNAse and 24 mg type IV Colagenase present in 15 mL RPMI-1640 medium. This procedure was repeated 3 times. After an additional 5-minute incubation at room temperature without shaking, the supernatants were collected and loaded on Ficoll density gradient to purify the lymphocyte population. NK cells were purified using the human NK cell isolation kit and the autoMACS instrument (Miltenyi Biotec). NK cells were viable as confirmed by surface expression of various inhibitory, adhesion, and lysis receptors and by functional assays such as migration, proliferation, and cytokine secretion.
Semiquantitative PCR analysis
Total RNA was isolated from various NK populations and from decidual and placental tissues, using the RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. cDNA was prepared according to standard protocol. The existence of human chemokine receptors and chemokines was verified by reverse transcriptase–polymerase chain reaction (RT-PCR). The primers used for the detection of chemokine receptors and chemokines are indicated in Table 1. Sequence of β-actin primer used was as follows: sense, 5′-CCCTGGACTTCGAGCAAGAG-3′; antisense, 5′-TCTCCTTCTGCATCCTGTCG-3′.
Antibodies and quadruple staining
For quadruple staining, the following fluorochrome-conjugated monoclonal antibodies were used: fluorescein isothiocyanate (FITC)–conjugated antihuman CD56 monoclonal antibody (mAb; Southern Biotechnology Associates, Birmingham, AL) and CyChrome-conjugated antihuman CD3 mAb (BD Pharmingen, San Diego, CA). Biotinylated anti-CD16 mAb (Serotec, Oxford, United Kingdom) was used, followed by Streptavidin-Cy5 (Jackson Immunoresearch, West Grove, PA) as a second reagent. As the fourth color, one of the following antibodies obtained from R&D Systems (Minneapolis, MN) was used: phycoerythrin (PE)–conjugated antihuman CXCR1 mAb, PE-conjugated antihuman CXCR2 mAb, PE-conjugated antihuman CXCR3 mAb, PE-conjugated antihuman CXCR4 mAb, PE-conjugated antihuman CCR1 mAb, PE-conjugated antihuman CCR2 mAb, PE-conjugated antihuman CCR3 mAb, PE-conjugated antihuman CCR5 mAb, PE-conjugated antihuman CCR6 mAb, PE-conjugated antihuman CCR7 mAb. When quadruple staining was performed with FITC-conjugated antihuman CX3CR1 mAb (Medical & Biological Laboratories, Nagoya, Japan), the PE-conjugated antihuman CD56 mAb (BD Pharmingen) was used. To block nonspecific binding, cells were first incubated for 1 hour on ice with 30% human serum (Sigma-Aldrich, St Louis, MO) and then incubated with the various antibodies.
ELISA assays
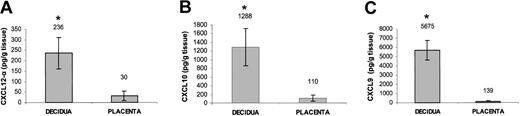
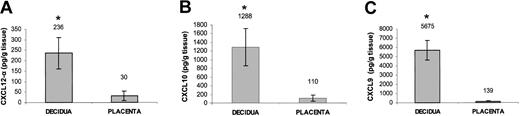
Freshly isolated first-trimester decidual and placental tissues were obtained from the same donor. The identity of the tissue specimens were confirmed by a pathologist. Protein extraction was performed by homogenization of preweighed (> 500 mg) sample 1:12 wt/vol in phosphate-buffered saline (PBS), pH 7.4. The homogenate was centrifuged and CXCL12-α, CXCL10, and CXCL9 concentration in the supernatant was determined by enzyme-linked immunosorbent assay (ELISA) using the quantikine kit (R&D Systems).
In vitro cell migration assay
Peripheral or decidual CD56+ NK cells (2 × 105) in 100 μL were loaded into each Transwell filter (5-μm pore filter Transwell, 24-well cells clusters; Corning, Corning, NY). Filters were then plated in each well containing 600 μL medium supplemented with previously described effective concentrations30-32 of various chemokines (obtained from R&D Systems), as indicated in Figures 4, 5. At least 3 wells were used for each chemokine concentration. To determine nonspecific or background migration, 6 wells that did not contain any chemokine were used as controls. After 3 hours of incubation at 37°C, 5% CO2, the upper chambers were removed, and cells in the bottom chamber were collected, counted, and analyzed by flow cytometry. Blocking for CXCR4 receptor was performed by pre-incubating the cells with T140 molecule33 for 30 minutes at 4°C. Percentage of migrated cells was calculated by subtracting the value of spontaneously migrating cells from the number of migrating cells at a given chemokine concentration, and afterward the result was divided by the total number of cells that were initially loaded on the transwell filter.
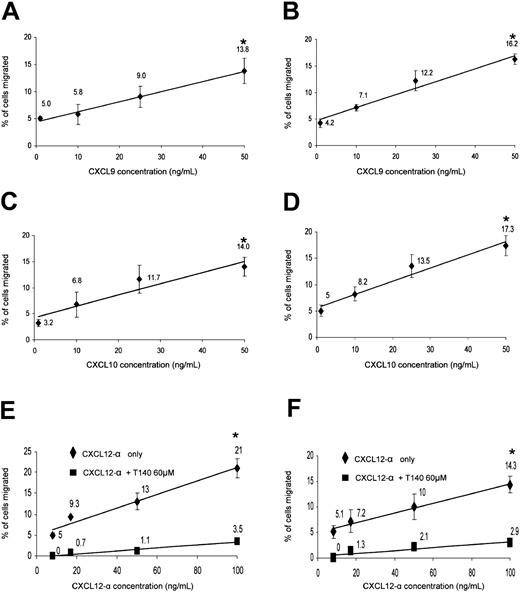
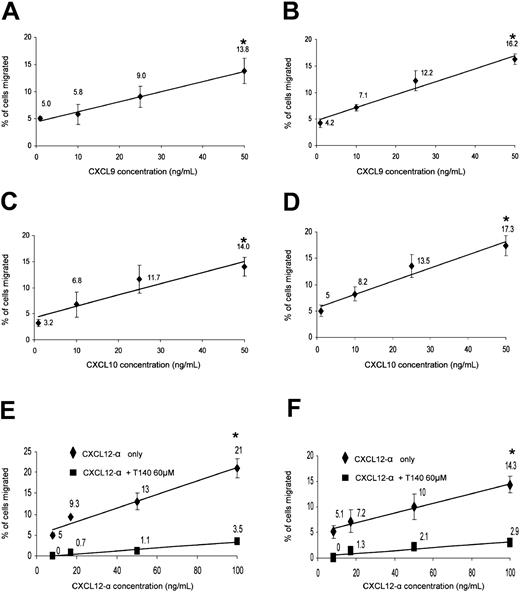
Migration of peripheral and decidual NK cells to recombinant human CXCL9, CXCL10, and CXCL12-α. Migration assays were performed as described in “Materials and methods.” The amount and identity of the migrated NK cells was analyzed by flow cytometry. Dose-dependent preferential migration of peripheral (left column) and decidual (right column) NK cells to CXCL9 (A-B), CXCL10 (C-D), and CXCL12-α with or without T140 blocking of CXCR4 (E-F). The percentages of migrated cells are calculated from the total cell population after the subtraction of the spontaneous migration. R-squared value for all linear trend lines displayed was above 0.95; *indicates P < .001 by Student t test. One representative experiment is shown of 4 performed. Error bars indicate SD.
Migration of peripheral and decidual NK cells to recombinant human CXCL9, CXCL10, and CXCL12-α. Migration assays were performed as described in “Materials and methods.” The amount and identity of the migrated NK cells was analyzed by flow cytometry. Dose-dependent preferential migration of peripheral (left column) and decidual (right column) NK cells to CXCL9 (A-B), CXCL10 (C-D), and CXCL12-α with or without T140 blocking of CXCR4 (E-F). The percentages of migrated cells are calculated from the total cell population after the subtraction of the spontaneous migration. R-squared value for all linear trend lines displayed was above 0.95; *indicates P < .001 by Student t test. One representative experiment is shown of 4 performed. Error bars indicate SD.
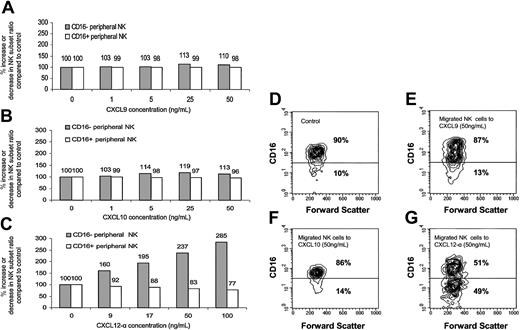
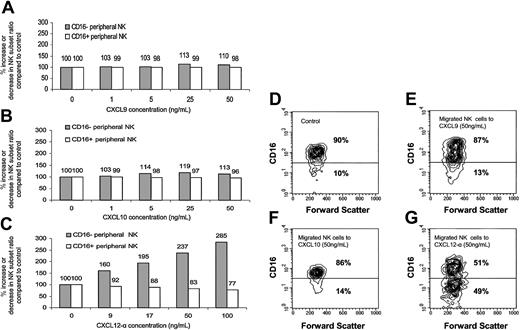
Enrichment of the CD16– subset in the migrated peripheral NK cells to CXCL12-α. The presence of the various NK populations in the migrated NK cells presented in Figure 4 was analyzed by quadruple staining. The percentage of increase or decrease of the CD16– or CD16+ subsets was calculated with regard to the starting point when no chemokine was present (100%). Migration was performed to CXCL9 (A), CXCL10 (B), and CXCL12-α (C). (D-G) Contour histogram of the CD16 phenotype of the migrated NK cells to CXCL9 (E), CXCL10 (F), and CXCL12-α (G). Control (D) represents the spontaneous migration when no chemokine was added. The percentages of CD16– and CD16+ subsets in the NK cells that spontaneously migrated when no chemokine was present were similar to those observed in the peripheral blood. Figure shows 1 representative experiment of 3 performed.
Enrichment of the CD16– subset in the migrated peripheral NK cells to CXCL12-α. The presence of the various NK populations in the migrated NK cells presented in Figure 4 was analyzed by quadruple staining. The percentage of increase or decrease of the CD16– or CD16+ subsets was calculated with regard to the starting point when no chemokine was present (100%). Migration was performed to CXCL9 (A), CXCL10 (B), and CXCL12-α (C). (D-G) Contour histogram of the CD16 phenotype of the migrated NK cells to CXCL9 (E), CXCL10 (F), and CXCL12-α (G). Control (D) represents the spontaneous migration when no chemokine was added. The percentages of CD16– and CD16+ subsets in the NK cells that spontaneously migrated when no chemokine was present were similar to those observed in the peripheral blood. Figure shows 1 representative experiment of 3 performed.
Immunohistochemistry
Placental tissue samples were routinely fixed with formalin and embedded in paraffin. Antigen retrieval (microwave treatment in citrate buffer) and immunohistochemistry were performed as previously described.34 Briefly, sections were incubated with either antihuman CXCL12 mAb, rabbit antihuman CXCL9 polyclonal antibody, goat antihuman CXCL10 polyclonal antibody (all obtained from R&D Systems), antihuman HLA-G mAb,34 or rabbit anti–von Willebrand factor polyclonal antibody (DAKO) overnight at 4°C followed by detection using the avidin-biotin Histostain Plus kit according to the manufacturer's instructions (Zymed Laboratories, San Francisco, CA). Color development was with 3-amino-9-ethylcarbazole (AEC), and sections were counterstained with hematoxylin. Immunohistochemistry results were confirmed by a pathologist.
Cytokine stimulation
PBLs were isolated from healthy donors and were incubated at a density of 4 × 106 cells in 4 mL medium containing cytokines that are known to be extensively expressed in human decidua.35-39 Cytokines used were recombinant human interleukin 8 (IL-8), IL-10, IL-12, IL-15, and transforming growth factor β 1 (TGFβ1) all obtained from R&D Systems and used at a concentration of 100 ng/mL each. After 24, 36, or 48 hours of incubation at 37°C 5% CO2, cells were collected and quadruple stained for chemokine receptor expression as mentioned before.
Results
mRNA for various chemokine receptors is expressed in decidual NK cells
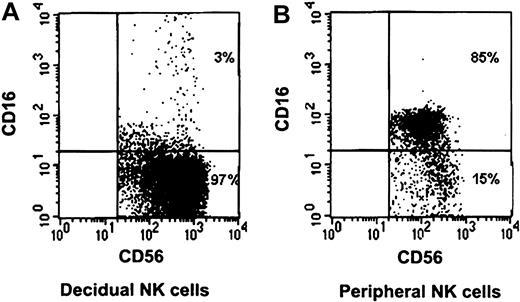
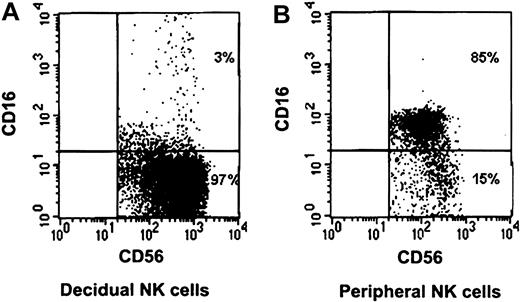
The NK cell population in the peripheral blood differs from that of the decidua.14 One of the most striking differences between the 2 populations is that the vast majority of the peripheral blood NK cells express the CD16 receptor, whereas the decidual NK cells do not (Figure 1).
CD16 characterization of decidual and peripheral NK cells. Freshly isolated peripheral and decidual lymphocytes were quadruple-stained as described in “Materials and methods.” NK cells were gated on the basis of the expression of CD56 and the lack of CD3 expression. Staining of decidual NK cells (A) and peripheral NK cells (B) is presented. The percentages of various populations, positive or negative, for CD16 expression are indicated. One representative staining is shown of 5 performed.
CD16 characterization of decidual and peripheral NK cells. Freshly isolated peripheral and decidual lymphocytes were quadruple-stained as described in “Materials and methods.” NK cells were gated on the basis of the expression of CD56 and the lack of CD3 expression. Staining of decidual NK cells (A) and peripheral NK cells (B) is presented. The percentages of various populations, positive or negative, for CD16 expression are indicated. One representative staining is shown of 5 performed.
The reasons why the CD16– NK cells are found in the decidua in such high numbers are still largely unknown. In this study we investigated whether the accumulation of the CD16– NK cells in the decidua might result from the expression of a different chemokine receptor repertoire on CD16– NK cells derived from the peripheral blood. We initially screened decidual NK cells for expression of mRNA for all known chemokine receptors. These cells were isolated from first-trimester elective pregnancy terminations obtained from 20 different donors. RT-PCR analysis for the expression of various chemokine receptors was performed by using the primers listed in Table 1. The PCR results show that decidual NK cells express mRNA for several chemokine receptors of both CXC (α) and CC (β) subclasses and CX3CR1 (Table 2).
Preferential expression of CXCR3 and CXCR4 on decidual and peripheral CD16– NK cells
Chemokine receptors are sensitive to a variety of posttranslational modifications and to endocytosis40,41 ; thus, the mRNA levels of the different chemokine receptors might not give an accurate measurement of the chemokine receptor expression on the NK cell surface.
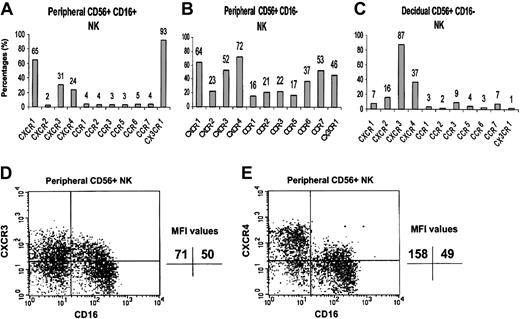
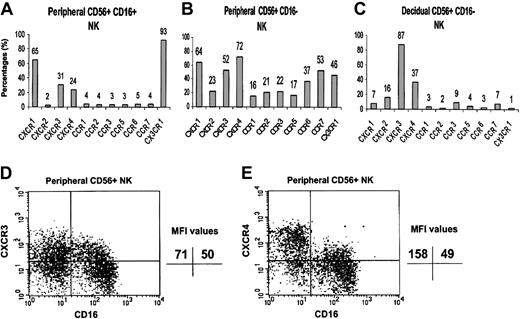
We, therefore, performed an analysis of the cell surface expression of the various chemokine receptors that were positively detected in the RT-PCR analysis, using commercially available specific mAbs. The expression of CCR8 was not tested because of the lack of a commercial anti-CCR8–conjugated antibody. Lymphocytes were isolated either from peripheral blood or from the deciduae, and quadruple staining was performed for the expression of CD3, CD16, CD56, and various chemokine receptors. Figure 2 shows that the chemokine receptor repertoire differs among the 3 NK cell populations tested (peripheral CD16+, peripheral CD16–, and decidual CD16– NK cells). In general, all of the chemokine receptors tested were found to be expressed to various degrees on the CD16– NK cells derived from peripheral blood. It is clear that the CD16– and CD16+ NK cells derived from peripheral blood are different populations with regard to the expression of chemokine receptors (Figure 2A-B). Significant percentages of peripheral CD16+ NK cells express CX3CR1, whereas all the rest of the chemokine receptors tested, except for CXCR1, were highly expressed only in the peripheral CD16– NK population.
Chemokine receptor distribution on the surface of peripheral and decidual NK cells. Peripheral and decidual lymphocytes were isolated and quadruple stained as described in “Materials and methods.” The figure shows the chemokine receptor expression on peripheral CD16+ NK cells (A), peripheral CD16– NK cells (B), and decidual CD16– NK cells (C). The indicated values represent the percentage of cells positive for a certain chemokine receptor of total cell population. FACS staining analysis of CXCR3 (D) and CXCR4 (E) on peripheral CD16– and CD16+ NK cells. The indicated MFI (median fluorescence intensity) values represent the expression level of a certain chemokine receptor on the NK cell population that stained positive for that specific chemokine receptor. One representative experiment is shown of 3 performed.
Chemokine receptor distribution on the surface of peripheral and decidual NK cells. Peripheral and decidual lymphocytes were isolated and quadruple stained as described in “Materials and methods.” The figure shows the chemokine receptor expression on peripheral CD16+ NK cells (A), peripheral CD16– NK cells (B), and decidual CD16– NK cells (C). The indicated values represent the percentage of cells positive for a certain chemokine receptor of total cell population. FACS staining analysis of CXCR3 (D) and CXCR4 (E) on peripheral CD16– and CD16+ NK cells. The indicated MFI (median fluorescence intensity) values represent the expression level of a certain chemokine receptor on the NK cell population that stained positive for that specific chemokine receptor. One representative experiment is shown of 3 performed.
Mice models suggested that the origin of decidual NK cells is from the PBLs.16 We therefore searched for chemokine receptors that are found to be expressed in high percentages in both decidual and peripheral CD16– NK cells and less in the peripheral CD16+ NK cells. Both the CXCR3 and CXCR4 receptors meet these criteria. Although 52% and 72% of peripheral CD16– NK cells and 87% and 37% of decidual NK cells expressed the CXCR3 and CXCR4 receptors, respectively, only 31% and 24% of CD16+ peripheral NK cells expressed the same receptors (Figure 2). The median fluorescence intensity (MFI), however, of CXCR3 expression was similar between the CD16+ and the CD16– NK population (Figure 2D, upper quadrants). In contrast, the MFI of CXCR4 on the CD16– NK population derived from peripheral blood was almost 3 times higher as compared with the CD16+ NK cells (Figure 2E). These results indicate that not only did many of the CD16– NK cells express the CXCR4 receptor, but also that the intensity of the CXCR4 receptor on these cells is much higher compared with the CD16+ NK population.
Ligands for CXCR3 and CXCR4 are expressed in decidual tissue
The results mentioned earlier suggest that the CXCR3 and CXCR4 receptors might be involved in the recruitment of the CD16– NK population from the peripheral blood to the decidua. We, therefore, tested whether the known chemokine ligands for these receptors are present in decidual tissues. mRNA was extracted from decidual and placental tissues obtained from the same donors. mRNA expression of HLA-G was consistently detected in both tissues (data not shown), as HLA-G is expressed only on EVTs.34 This result, together with microscopic and histologic analysis, confirmed the identity of the examined tissues to be deciduae and placentas. mRNA for CXCL10 and CXCL9 but not for CXCL11 (ligands for CXCR3) and mRNA for both isoforms of CXCL12 (ligand for CXCR4) were detected in the decidual and placental tissues (Table 3). We next directly tested whether chemokines can be observed in these tissues. ELISA assays were performed for the detection of CXCL12 (Figure 3A), CXCL10 (Figure 3B), and CXCL9 (Figure 3C) in both decidual and placental tissues. Strikingly, all 3 chemokines were found to be expressed in the decidua in higher levels (P < .01, Student t test) as compared with the placenta (Figure 3A-C), suggesting that the NK cells expressing either the CXCR3 or CXCR4 receptors might be attracted primarily to the decidua.
Expression of ligands for CXCR3 and CXCR4 in decidual and placental tissues. Placental and decidual tissues were obtained from the same donor. ELISA assays for the presence of CXCL12-α (A), CXCL10 (B), and CXCL9 (C) in decidual and placental tissue samples obtained from the same donor. Average values of 3 samples are shown. *Indicates P < .01 by Student t test. Error bars indicate SD.
Expression of ligands for CXCR3 and CXCR4 in decidual and placental tissues. Placental and decidual tissues were obtained from the same donor. ELISA assays for the presence of CXCL12-α (A), CXCL10 (B), and CXCL9 (C) in decidual and placental tissue samples obtained from the same donor. Average values of 3 samples are shown. *Indicates P < .01 by Student t test. Error bars indicate SD.
Preferential migration and enrichment of CD16– NK subset to CXCL12
Peripheral NK cells have been previously shown to migrate in vitro to various CXCR4 and CXCR3 ligands.32,42 We wanted to further test the functional significance of the CXCR3 and CXCR4 expression on decidual NK cells and whether the expression differences of CXCR3 and CXCR4 between peripheral CD16– and CD16+ subsets were able to result in preferential migration and enrichment of one of the subsets at the expense of the other. The significant expression levels of CXCR3 receptor on peripheral and decidual NK cells resulted in a dose-dependent migration to CXCL9 and CXCL10 (Figure 4A-D). Similarly, both peripheral and decidual NK cells consistently migrated to various concentrations of CXCL12-α in a dose-responsive manner (Figure 4E-F, respectively). The migration resulted from CXCR4 interaction with CXCL12-α because blocking of the migration was observed with the CXCR4 inhibitor T140 when either decidual NK (Figure 4F) or peripheral blood NK cells (Figure 4E) were used.
The migrating bulk NK cells were next analyzed for the expression of CD16. Migrated NK cells in the lower chambers of the migration transwells were collected and stained as described in “Materials and methods.” No significant preferential enrichment of peripheral CD16– NK cells of the total migrated NK cells was observed when migration was performed with either CXCL9 or CXCL10 (Figure 5A-B and D-F). In contrast, dose-dependent preferential enrichment of peripheral CD16– NK cells to CXCL12-α was evident (Figure 5C and G). This preferential enrichment is a result of the fact that CD16+ NK cells migrated in a much lower level than CD16– NK cells to CXCL12, and thus their relative ratio of total migrated NK cells was significantly reduced (Figure 5D and G). Similar results were obtained in 3 other independent experiments, and the enrichment ratio of the CD16– NK cells to 100 ng/mL CXCL12 was an average 3.28 with a P value of less than .02 (Student t test) (data not shown). When no chemokine was present in the assay, the relative ratios of both NK subsets of total NK population remained unchanged (Figure 5D and data not shown). No change in CD16 expression was observed when control incubations of peripheral blood NK cells with CXCL9, CXCL10, or CXCL12 were performed (data not shown).
The fact that CD16– NK enrichment was observed only in response to CXCR4 ligand is not surprising, because the differences observed in the intensity and number of cells expressing CXCR3 between the 2 peripheral NK subsets were much less prominent than those observed for CXCR4 (Figure 2D-E) and were probably not functionally significant in causing CD16– NK enrichment among total migrated NK cells.
CXCL12 is expressed in vivo on invasive trophoblasts and trophoblasts lining maternal decidual blood vessels
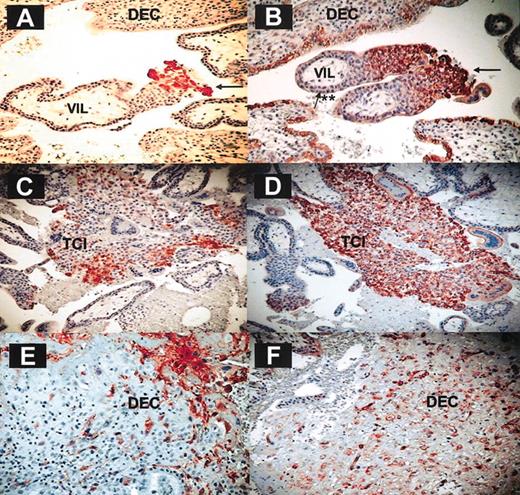
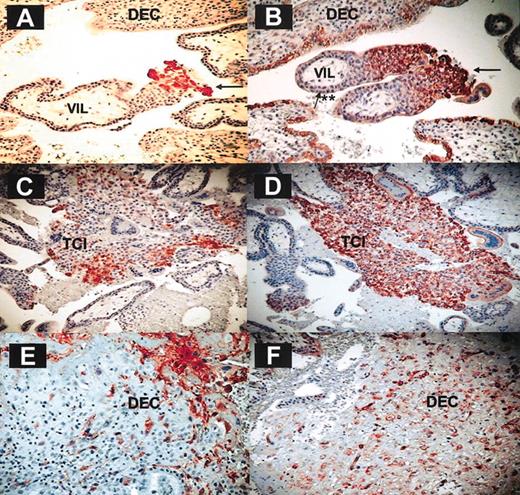
The result described earlier suggests a major role for CXCL12, that is present primarily in the decidua (Figure 3), in the specific enrichment of CD16– NK cells. Immunohistochemistry analysis of first-trimester decidual tissue sections was performed to identify the cells in the decidua that express CXCL12 (Figure 6). The identity of trophoblast cell columns (Figure 6A-B), trophoblast islands (Figure 6C-D), and invasive EVTs (Figure 6E-F) was confirmed by immunohistochemistry of these serial sections with anti–HLA-G antibody (Figure 6A,C,E). The presence of CXCL12 was detected in trophoblast cell columns especially in the distal end of the column (Figure 6B) and not in villous cytotrophoblasts or syncytiotrophoblasts. The trophoblast cell islands (Figure 6D) and the EVTs that have invaded the decidua also expressed CXCL12 (Figure 6F). The anti-CXCL12 antibody staining was consistently stronger than that observed with the anti–HLA-G antibody. Immunostaining with isotype-matched control immunoglobulin G (IgG) mAb yielded negative results (data not shown).
CXCL12 is expressed on invasive fetal trophoblasts. Immunohistochemistry on paraformaldehyde-fixed decidual tissue (9 weeks). Immunohistochemistry was performed on first-trimester serial sections derived from placental and decidual tissues. The figure shows HLA-G (left column) and CXCL12 (right column) staining on intermediate trophoblasts (A-B), trophoblast cell islands (C-D), and invasive EVTs (E-F) derived from first-trimester deciduae. One representative staining experiment is shown of 9 performed. Arrows indicate fetal trophoblasts; **, villous cytotrophoblasts and syncytiotrophoblasts; DEC, decidua; VIL, villous; and TCI, trophoblast cell island. Magnification is × 200 for all panels.
CXCL12 is expressed on invasive fetal trophoblasts. Immunohistochemistry on paraformaldehyde-fixed decidual tissue (9 weeks). Immunohistochemistry was performed on first-trimester serial sections derived from placental and decidual tissues. The figure shows HLA-G (left column) and CXCL12 (right column) staining on intermediate trophoblasts (A-B), trophoblast cell islands (C-D), and invasive EVTs (E-F) derived from first-trimester deciduae. One representative staining experiment is shown of 9 performed. Arrows indicate fetal trophoblasts; **, villous cytotrophoblasts and syncytiotrophoblasts; DEC, decidua; VIL, villous; and TCI, trophoblast cell island. Magnification is × 200 for all panels.
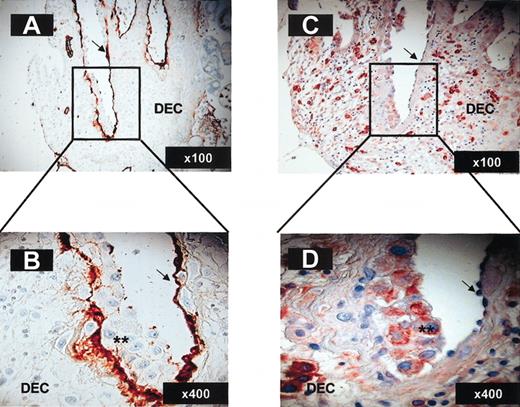
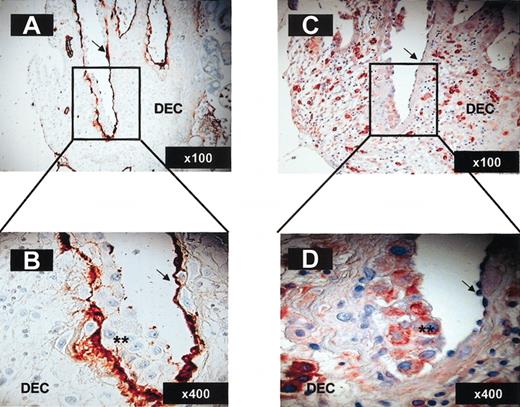
Importantly, trophoblasts invading maternal blood vessels that are in direct contact with maternal blood express CXCL12, whereas the original endothelial lining does not (Figure 7C-D). Endothelial cells were identified by staining with anti–von Willebrand factor polyclonal antibody (arrows in Figure 7A-B), whereas invading trophoblasts yielded negative staining with the same antibody (stars in Figure 7B). Immunostaining performed with anti-CXCL9 and anti-CXCL10 antibodies yielded negative expression on trophoblast cells at all stages of their development for both ligands (data not shown). These results are in agreement with previous work using in situ hybridization experiments demonstrating that decidual stroma, but not trophoblast cells, express CXCL9 and CXCL10.43 The decidual stroma is probably the source for the presence of these chemokines in decidual extracts (Figure 3).
Invasive trophoblasts lining maternal decidual blood vessels express CXCL12. Immunohistochemistry on paraformaldehyde-fixed serial decidua tissue sections (12 weeks; 6 μ). Staining with anti–von Willebrand factor polyclonal antibody (A-B) and CXCL12 (C-D) of decidual blood vessels undergoing endovascular invasion by trophoblasts. Arrows indicate endothelial cells; **, invasive trophoblasts. The magnification for each panel is indicated in the lower right corner.
Invasive trophoblasts lining maternal decidual blood vessels express CXCL12. Immunohistochemistry on paraformaldehyde-fixed serial decidua tissue sections (12 weeks; 6 μ). Staining with anti–von Willebrand factor polyclonal antibody (A-B) and CXCL12 (C-D) of decidual blood vessels undergoing endovascular invasion by trophoblasts. Arrows indicate endothelial cells; **, invasive trophoblasts. The magnification for each panel is indicated in the lower right corner.
Cytokine treatment of peripheral CD16– NK cells alters the cell surface chemokine receptor repertoire
Decidual CD16– NK cells expressed a different chemokine receptor repertoire when compared with peripheral CD16– NK cells (Figure 2B-C). Only a few chemokine receptors were expressed on freshly isolated decidual NK cells when compared with the peripheral CD16– subset (Figure 2B-C), indicating that the 2 cell populations are different. Decidual NK cells are constantly in direct contact with the semi-allogeneic invasive fetal trophoblasts, and different reports indicated that decidual NK cells are functionally different from those of the peripheral blood because of unique microenvironment conditions.11,12,28 Cytokines have been extensively studied and proven as major regulatory molecules in human decidua controlling adhesion molecules, vascularization, and NK cell activation.36 We, therefore, hypothesized that the unique decidual NK cells' chemokine receptor expression pattern might result from local cytokine stimulation. IL-8, IL-10, IL-12, IL-15, and TGFβ1 were reported to be the major regulatory cytokines found in human decidua.35-39 The expression of the various chemokine receptors on the peripheral CD16– NK cells was, therefore, tested following activation with these cytokines.
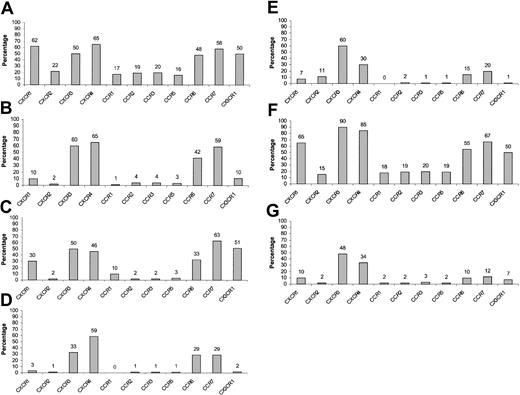
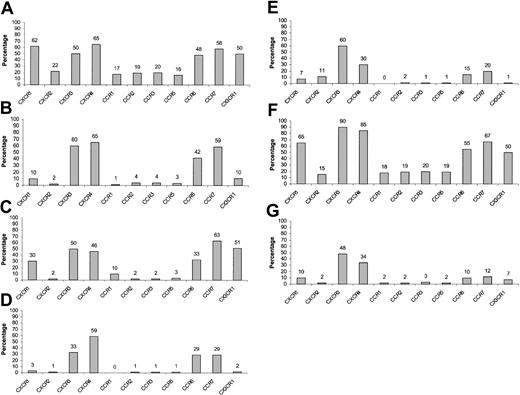
PBLs obtained from healthy donors were incubated with various cytokines for 48 hours as described in “Materials and methods” and quadruple stained for the expression of chemokine receptors. In general, CXCR1, CXCR2, CCR1, CCR2, CCR3, and CCR5 were significantly down-regulated by all cytokines used (Figure 8A-E) except for TGFβ1 (Figure 8F), whereas CCR6 and CCR7 were mainly down-regulated by IL-15 and to a lesser extent by IL-12 (Figure 8A and D-E). CX3CR1 expression was altered by all cytokines checked except for IL-10 and TGFβ1. Importantly, CXCR4 was subjected to up-regulation following TGFβ1 stimulation and to down-regulation mainly following IL-15 stimulation (by approximately 65%) (Figure 8A-E). Regarding CXCR3, treatment with IL-12 caused a small but consistent reduction in its expression levels, whereas TGFβ1 induced an opposite effect. A more prominent reduction in the CXCR3 expression was observed when CD16+ NK cells were incubated with IL-12 (data not shown). These results are in agreement with previous work demonstrating IL-12 mediated down-regulation of CXCR3 expression on total NK cells derived from peripheral blood.44 Remarkably, when peripheral CD16– NK cells were treated either with IL-15 or with a combination of all cytokines tested, the chemokine receptor expression levels obtained were very similar to those found on decidual CD16– NK cells (Figures 8F-G and 2C). Similar results were obtained following 24 and 36 hours of cytokine incubation (data not shown).
Expression of various chemokine receptors on cytokine-activated peripheral CD16– NK cells. PBLs were obtained from healthy donors and were either left untreated or incubated with various cytokines at a concentration of 100 ng/mL and then were quadruple stained for the expression of chemokine receptors on CD16– NK cells. (A) No cytokine treatment, (B) IL-8, (C) IL-10, (D) IL-12, (E) IL-15, (F) TGFβ1, and (G) IL-8 + IL-10 + IL-12 + IL-15 + TGFβ1. The indicated values represent the percentage of cells positive for a certain chemokine receptor of the total population. One representative experiment is shown of 3 performed.
Expression of various chemokine receptors on cytokine-activated peripheral CD16– NK cells. PBLs were obtained from healthy donors and were either left untreated or incubated with various cytokines at a concentration of 100 ng/mL and then were quadruple stained for the expression of chemokine receptors on CD16– NK cells. (A) No cytokine treatment, (B) IL-8, (C) IL-10, (D) IL-12, (E) IL-15, (F) TGFβ1, and (G) IL-8 + IL-10 + IL-12 + IL-15 + TGFβ1. The indicated values represent the percentage of cells positive for a certain chemokine receptor of the total population. One representative experiment is shown of 3 performed.
Discussion
The unique composition of the decidual lymphocyte population probably holds the key to proper fetomaternal immune tolerance. In addition, the fact that CD56brightCD16– NK cells constitute the vast majority of the decidual lymphocyte population (Figure 1) has brought up many question regarding how and why the decidual tissue is vigorously enriched with this NK subpopulation. Because it was recently shown, in mice models, that decidual NK cells are recruited from peripheral sites rather than self-renewal in the uterine mucosa,45 we tested whether the CD16– NK population in PBLs expresses a unique combination of chemokine receptors that enables the specific migration of this subset to the decidua.
NK cells that are part of the innate immunity system are able to quickly kill virus-infected and tumor cells.46-48 This finding suggests that at least some of the NK cells present in the peripheral blood should be equipped with a complete repertoire of chemokine receptors that will enable their rapid migration to various organs. Such cells might be the un-activated peripheral CD16– NK cells that express all of the chemokine receptor tested (Figure 2). In agreement with our results, a recent report49 has demonstrated different expression patterns of some chemokine receptors on peripheral CD16+ and CD16– NK cells. In contrast, only a few chemokine receptors are expressed on decidual NK cells (Figure 2). Furthermore, the expression of all of the chemokine receptors tested, except from CXCR3 whose ligands CXCL9 and CXCL10 are found in the decidua, were down-regulated following multiple cytokine activation (Figure 8). These results raised the intriguing hypothesis that suggests that once NK cells reach their target organ, the decidua, there is no need for further trafficking. NK cells, therefore, down-regulate the expression of all chemokine receptors except from those needed for their retention.
The expression, regulation, and function (including vascularization, adhesion molecule expression, and lymphocyte activation) of various local cytokines in the decidua have been characterized during the human menstrual cycle and pregnancy.35-39 The results presented here demonstrate an additional role for IL-15, as a major modulator of the chemokine receptor repertoire at the maternalfetal interface. In addition, IL-15 has been previously shown to induce the extensive proliferation of peripheral and decidual CD56brightCD16– NK cells.38,50 Therefore, the specific decidual enrichment with CD16–CXCR4+ NK cells derived from peripheral blood might further be potentiated by IL-15.
Throughout the comparative analysis performed in this study, consistently, there was no correlation between mRNA expression and levels of chemokine receptors detected by fluorescence-activated cell sorter (FACS) analysis on the surface of the various NK populations (activated and nonactivated). In addition, the mRNA expression of the various chemokine receptors is dependent on NK cells' growth conditions and treatment and does not always reflect proper surface expression.51,52 Therefore, to obtain a reliable monitoring of chemokine receptor on NK cells, expression should always be detected directly by using specific mAbs.
Both the CXCR3 and CXCR4 were the 2 dominant receptors expressed by unactivated decidual and peripheral blood CD16– NK cells (Figure 2). The fact that CXCR3 ligands are not expressed on EVTs, the relatively minor differences in CXCR3 expression levels between both peripheral NK subsets (Figure 2D), and the inability of CXCR3 ligands to induce preferential enrichment of CD16– NK cells (Figure 5) suggest that CXCR3 is probably less involved in recruiting peripheral CD16– NK cells to the decidua. Moreover, the fact that CXCR3 ligands are expressed by stromal cells rather than invasive trophoblasts43 and the ability of decidual NK cells to migrate in response to these chemokines might implicate CXCR3 in retention of the CD16– NK cells after they have migrated into the decidua. Supporting this hypothesis, a similar role for CXCL10 expressed on dendritic cells has been proposed in the retention of TH1 cells in secondary lymphoid organs.53
The main chemokine receptor shown here to be involved in the preferential enrichment of CD16– NK cells is CXCR4 (Figure 5). The level of CXCR4 expression on decidual NK cells is lower than that of CD16– NK cells derived from the peripheral blood (Figure 2). This lower expression might result from local cytokine stimulation of NK cells in the decidua (Figure 8), and/or from the interactions between the CXCR4 on the NK cells and its ligand CXCL12 on the EVTs (Figures 6, 7). Indeed, down-regulation of CXCR4 was previously observed after its interaction with CXCL12,52 and reduced levels of CXCR4 expression were observed in the migrated NK cells population tested after their migration to CXCL12 (data not shown). We show here that EVTs express CXCL12, but not CXCL9 or CXCL10 (Figure 6 and data not shown). These cells eventually perform endovascular invasion, acquire endothelial cell–like phenotype, and position themselves in direct contact with maternal blood flow3,4 and, thus, can attract the CD16– NK cells to the decidua because they maintain CXCL12 expression (Figure 7).
It was recently shown that CCL3 is also able to attract peripheral NK cells.15 On the basis of our chemokine receptor expression analysis (Figure 2), 17% of the peripheral CD16– NK cells and 3% of CD16+ NK cells express CCR5 (the receptor for CCL3). This difference in CCR5 expression cannot, however, explain the specific accumulation of CD16– NK cells in the deciduae, as there was no difference in the migration of CD16– NK cells over CD16+ NK cells to CCL3.15 Furthermore, mice deficient for CCR5 displayed normal recruitment of peripheral NK cells to the decidua.16 The CCR5 receptor probably plays a pivotal role in intradecidual organization and retention of various cell types in the decidua but is less important in the preferential migration of CD16– NK cells.
The model we propose is that CD16– NK cells are attracted during pregnancy from the peripheral blood to the decidua via the interactions between CXCR4 on their surface and CXCL12 on the invasive trophoblast cells lining maternal blood vessels in the decidua. The interaction between CXCR3 and CXCL10 or CXCL9 found in decidual stroma might lead to the retention of the NK cells that have already migrated into the decidua, and together with CXCL12 these chemokines may also be important for proper positioning and organization of decidual NK cells. In the decidua, because of the local microenvironment conditions, the CD16– NK cells probably acquire some unique characteristics noted in this article by the reduction of chemokine receptor expression. These special properties of the decidual NK cells, together with other reported properties such as the increased expression of CEACAM128 and other inhibitory NK receptors11-13 on these cells, probably enable proper fetal development.
Prepublished online as Blood First Edition Paper, May 1, 2003; DOI 10.1182/blood-2003-02-0517.
Supported by research grants from the Israel Cancer Research Foundation (O.M.), the Israel Science Foundation (O.M.), and the Charles H. Revson Foundation (no. 153/00) (O.M.), the Cancer Research Institute (O.M.), and the HWZOA Research Fund for Women's Health.
J.H. and O.W. contributed equally to the work presented.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Nabil Hanna for helpful discussions, Mally Sapir for assistance in pathology slide preparation, and Franceska Levi-Schaffer and Ariel Munitz for their help with flow cytometry experiments. We also thank Hanna Wald, Michal Dagan-Berger, Caryn Greenfield, Shira Natanson-Yaron, and Michael Berger for excellent assistance.