Abstract
Sustained high-level proviral expression is important for clinical applications of gene therapy. Genetic elements including the β-interferon scaffold attachment region (SAR) have been shown to improve transgene expression in hematopoietic cells. We hypothesized that SAR elements might improve expression and allow the preselection of successfully transduced cells. Thus, we transplanted green fluorescent protein (GFP)–selected cells, half of which had been transduced with either SAR or non–SAR-containing retrovirus vectors, into 3 animals. All animals showed delayed engraftment compared with historic controls (28 vs 15.5 days). GFP marking was seen at levels up to 8% but declined over the first 6 weeks. Importantly, fluorescence intensity was 2- to 9-fold increased in progeny of SAR versus non–SAR vector–modified cells in all hematopoietic lineages for the duration of follow-up (6-12 months). In conclusion, the use of SAR-containing vectors improved transgene expression in hematopoietic repopulating cells, which may obviate the need for multicopy integration to achieve high-level expression and reduce the risk for insertional mutagenesis.
Introduction
Attenuation of proviral expression is a frequent occurrence after oncoretroviral transduction of hematopoietic cells. Modifications in the design of the Moloney murine leukemia virus promoter/enhancer can improve transgene expression in the murine model1,2 but not necessarily in the nonhuman primate model.3 Alternative strategies to improve long-term expression in hematopoietic cells include the use of transcriptional modifiers such as scaffold attachment region (SAR) and insulator elements, or other posttranscriptional regulatory sequences.4-6 SAR elements constitute cis-regulatory elements that appear to create independent domains of transcription.7 Retrovirus vectors containing the SAR element of the human beta interferon (β-IFN) gene enhance the expression of heterologous reporter genes in T cells, macrophages, and thymocytes in vitro and in vivo.5,8-10
The current study investigates if the presence of the β-IFN SAR element in an oncoretrovirus vector improves proviral green fluorescent protein (GFP) fluorescence in a large-animal model and if these improvements may permit preinfusion selection of genetically modified hematopoietic cells to enhance posttransplantation gene marking levels. The latter strategy has been used successfully in murine stem cell gene transfer models11-13 but so far not with success in a large animal model.14
Study design
Animals
Retrovirus vectors
The retrovirus vectors encode the enhanced green fluorescent protein (EGFP; Clontech, Palo Alto, CA) or its yellow variant, EYFP, under the control of the 5′ modified viral long terminal repeat (LTR)2 and are otherwise identical. EGFP and EYFP genes were inserted into the MND-X retroviral vector plasmid17 to make the vectors MND-EGFP or MND-EYFP. The 800-bp human β-IFN SAR element from Bode et al7 was inserted between the EGFP or EYFP gene and the 3′ LTR to make MND-EGFP-SAR or MND-EYFP-SAR, respectively. Producer cell clones with similar titer (0.6-1.3 × 105 HT1080 TU/mL) were generated using Phoenix GALV producer cells as described.18
CD34 enrichment, gene transfer, and selection of GFP-positive baboon marrow cells
Following 48 hours of prestimulation, CD34-enriched cells (generated as previously described16 ) were transduced twice for 4 hours in the presence of CH-296 (2 μg/cm RetroNectin; Takara Shuzo, Otsu, Japan),16 interleukin-3 (IL-3), IL-6, recombinant human stem cell factor (rhSCF), recombinant human granulocyte colony-stimulating factor (rhG-CSF), flt3-ligand (Flt3-L), and megakaryocyte growth and development factor (MGDF) at 100 ng/mL. Cells (10 × 106) were exposed to a total of 60 mL volume of vector per flask resulting in a multiplicity of infection between 0.1 and 0.3 in all 3 animals. Cells were sorted by flow cytometry 36 hours after the last vector exposure according to EGFP or EYFP expression as previously described.3
Real-time polymerase chain reaction (PCR) assay
For PCR amplification of provirus, DNA (300 ng) was amplified in duplicate with sequence-specific primers and probes designed using Primer Express software (Perkin-Elmer Applied Biosystems, Foster City, CA). The following primers were used: EYFP, 5′-CTG CAC CAC CGG CAA-3′ and 5′-GTA GCG GGC GAA GCA CT-3′; probe, 5′-FAM-CCA CCT TCG GCT ACG GCC TG-TAMRA-3′ (Synthegen, Houston, TX); EGFP, 5′-CTG CAC CAC CGG CAA-3′ and 5′-GTA GCG GCT GAA GCA CTG-3′; probe, 5′-FAM-CCA CCC TGA CCT ACG GCG TG-TAMRA-3′; beta globin, 5′-CCT ATC AGA AAG TGG TGG CTG G -3′ and 5′-TTG GAC AGC AAG AAA GTG AGC TT-3′; probe, 5′-FAM-TGG CTA ATG CCC TGG CCC ACA AGT A-TAMRA-3′. Standards consisted of dilutions of DNA extracted from cell lines transduced with a single copy of the vector and DNA from control animal peripheral blood mononuclear cells. Reactions were run using the ABI master mix (Applied Biosystems, Branchburg, NJ) on the ABI Prism 7700 sequence detection system (Applied Biosystems) using the following thermal cycling conditions: 50°C for 2 minutes and 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute.
Results and discussion
Reinfusion of preselected, GFP-expressing CD34+ cells delays hematopoietic reconstitution
Table 1 contains details regarding enrichment, cell dose, purity, vector assignment, and engraftment. Results illustrate that preselection and infusion of GFP-expressing cells had a profound effect on the hematopoietic reconstitution and significantly delayed hematopoietic engraftment. The mean time to granulocyte engraftment in animals receiving preselected GFP-expressing cells was 28 days (standard deviation [SD] ± 1 day; range, 27-29 days) compared with 15.5 days (SD ± 2.8 days; range, 12-20 days; P = .01, Wilcoxon rank sum test) in 8 historic control animals.3,19 Consistent with data in other experimental models, the extended in vitro culture required for GFP expression prior to and the sorting manipulation itself may have prompted in vitro differentiation and compromised the repopulating ability.20-22
Preselection results in high marking levels in short-term repopulating cells
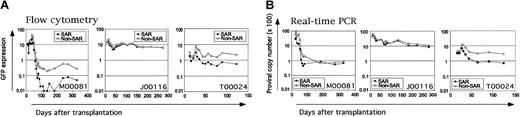
Gene transfer was variable among animals, and we found no evidence that the overall gene transfer rate or the ability to select for transduced stem cells is affected by the presence or absence of SAR elements or the GFP variant5,9 (Figure 1A-B). We determined overall marking at 4-, 13-, and 18-week follow-up time points by fluorescence-activated cell sorter (FACS) analysis to be as follows: M00081, 67%/0.3%/0.2%; J00116, 17%/23%/24%; and T00024, 11%/3.8%/2.7%. Long-term gene transfer rates in 2 of these animals (not J00116) appeared to range below those previously attained without preselection.16,18 This is unlikely to be accounted for by silencing or immunologic clearance given the correlation of transgene expression (marking by FACS, Figure 1A) and proviral persistence (marking by real-time PCR, Figure 1B). The PCR/FACS GFP marking ratios at given time points (1.9-2.8 on average per animal) were unaffected by fluorescence color (EGFP or EYFP) or the presence of SAR element within the vector. These ratios are within the limits of studies previously reported3 and consistent with large-animal studies by Rosenzweig et al.23
Frequency of genetically modified cells in the peripheral blood after transplantation. Serial determinations of GFP marking (expressed as percent cells marked in the peripheral blood) were made by flow cytometry (A) or PCR (average copy number × 100 shown) (B). Vector marking in cells transduced with SAR-containing vectors is shown in solid symbols, non–SAR-containing vectors in open symbols. Animal designations and fluorescence assignment are noted: EGFP indicates enhanced green fluorescent protein; EYFP, enhanced yellow fluorescent protein.
Frequency of genetically modified cells in the peripheral blood after transplantation. Serial determinations of GFP marking (expressed as percent cells marked in the peripheral blood) were made by flow cytometry (A) or PCR (average copy number × 100 shown) (B). Vector marking in cells transduced with SAR-containing vectors is shown in solid symbols, non–SAR-containing vectors in open symbols. Animal designations and fluorescence assignment are noted: EGFP indicates enhanced green fluorescent protein; EYFP, enhanced yellow fluorescent protein.
SAR elements enhance EGFP and EYFP fluorescence intensity in all hematopoietic lineages
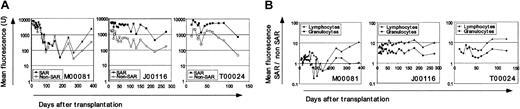
The most striking finding in this study is the improvement in proviral expression measured as mean EGFP (EYFP) fluorescence (mean fluorescence intensity [MFI]) at serial time points after transplantation (Figure 2A). Flow-cytometric analysis was performed separately in granulocytes and lymphocytes. The calculated ratio of MFI in granulocytes or lymphocytes for the SAR-containing over non–SAR-containing arm at a given time point (Figure 2B) exceeds 1 in all 3 animals, indicating greater fluorescence intensity of the SAR than non–SAR-containing vector regardless of EGFP (J00116) or EYFP (M00081 and T00024) fluorescence assignment. This difference in fluorescence intensity persists over time in the hematopoietic lineages analyzed, demonstrating predominant SAR-mediated effects on the magnitude of transgene expression.5,8,9 The gain in SAR-mediated fluorescence intensity was also apparent in red blood cells (analyzed in 2 animals at fewer time points; data not shown). Our observation that the SAR-mediated improvement in fluorescence intensity was relatively greater in T lymphocytes than granulocytes is supported by reports from other investigators in vitro and in the murine xenograft model.9,10
Improved fluorescence intensity from SAR-containing vectors. (A) MFI in peripheral blood leukocytes (PBLs) over time after transplantation. Fluorescence from cells transduced with SAR-containing vectors is shown in solid symbols, non–SAR-containing vectors in open symbols. (B) To calculate ratios of MFI in SAR over non–SAR-containing vector transduced cells we used mean fluorescence intensity (MFI) from forward and right-angle light scatter–gated granulocyte (circles) and lymphocyte (triangles) subsets using CELLQuest v3.1 (Becton Dickinson, San Jose, CA) as previously described. 3 A ratio of 1 implies equal fluorescence intensity between SAR versus non-SAR vectors. A ratio of 10 denotes fluorescence 10 times brighter in cells transduced with the SAR-containing vector. Animal designations are noted.
Improved fluorescence intensity from SAR-containing vectors. (A) MFI in peripheral blood leukocytes (PBLs) over time after transplantation. Fluorescence from cells transduced with SAR-containing vectors is shown in solid symbols, non–SAR-containing vectors in open symbols. (B) To calculate ratios of MFI in SAR over non–SAR-containing vector transduced cells we used mean fluorescence intensity (MFI) from forward and right-angle light scatter–gated granulocyte (circles) and lymphocyte (triangles) subsets using CELLQuest v3.1 (Becton Dickinson, San Jose, CA) as previously described. 3 A ratio of 1 implies equal fluorescence intensity between SAR versus non-SAR vectors. A ratio of 10 denotes fluorescence 10 times brighter in cells transduced with the SAR-containing vector. Animal designations are noted.
Findings of improved proviral expression from SAR-containing vectors are consistent with observations by Mielke et al who located highly expressing proviral integrants in the genome in close proximity to SAR elements.24 Thus, the use of SAR elements may provide for improved stable expression from a single copy proviral integrant. It is noteworthy that alternate positioning of the SAR element as described by Ramezani et al may have additional safety benefits while maintaining improvements in expression.25
In conclusion, we propose that SAR-mediated high-level transgene expression may provide an attractive strategy to improve long-term proviral expression, obviating the need for multicopy integration and thus reducing the risk of mutagenic insertion.26 SAR elements should be considered for the design of improved stem cell gene therapy vectors.
Prepublished online as Blood First Edition Paper, July 17, 2003; DOI 10.1182/blood-2003-03-0962.
Supported by grants HL54881, RR00166, DK47754, DK56465, and HL53750. H.-P.K. is a Markey Molecular Medicine Investigator.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are indebted to Hana Paik for her dedication and expertise as well as Dr Wendy Leisenring for statistical analysis and Dr Peter Horn for technical assistance. In addition we would like to acknowledge the contribution of research staff at the Washington State primate center, in particular Michael Gough and Dr Robert Andrews.