Abstract
Signal transducer and activator of transcription 5 (STAT5) plays a critical role in cytokine-induced survival of hematopoietic cells. One of the STAT5 target genes is pim-1, which encodes an oncogenic serine/threonine kinase. Here we demonstrate that Pim-1 inhibits STAT5-dependent transcription in cells responsive to interleukin-3, prolactin, or erythropoietin. Ectopic expression of Pim-1 in cytokine-dependent FDCP1 myeloid cells results in reduced tyrosine phosphorylation and DNA binding of STAT5, indicating that Pim-1 interferes already with the initial steps of STAT5 activation. However, the Pim-1 kinase does not directly phosphorylate or bind to STAT5. By contrast, Pim-1 interacts with suppressor of cytokine signaling 1 (SOCS1) and SOCS3 and potentiates their inhibitory effects on STAT5, most likely via phosphorylation-mediated stabilization of the SOCS proteins. Thus, both Pim and SOCS family proteins may be components of a negative feedback mechanism that allows STAT5 to attenuate its own activity.
Introduction
Interleukin-3 (IL-3) induces growth and differentiation of several types of cell populations, including multipotential hematopoietic stem cells, immature myeloid cells, as well as T- and pre–B-lymphoid cells. The IL-3 receptor along with the related receptors for IL-5 and granulocyte macrophage–colony-stimulating factor (GM-CSF) are composed of a ligand-binding alpha chain and a signal-transducing beta chain, which heterodimerize upon ligand binding.1 The membrane-proximal intracellular region of the beta chain is required for induction of proliferation-associated genes such as c-myc and pim-1.2,3 This region is also bound by multiple signal-transducing proteins such as Janus kinases (JAKs), signal transducers and activators of transcription (STATs), c-Src, and phosphotidylinositol-3 kinase.4
STAT5 with its 2 isoforms, STAT5A and STAT5B, plays a critical role in proliferation of hematopoietic cells in response to IL-2, IL-3, IL-5, GM-CSF, erythropoietin (EPO), prolactin (PRL), epidermal growth factor (EGF), and growth hormone.5-7 Activation of STAT5 by the oncogenic Bcr/Abl fusion protein is essential for its ability to transform hematopoietic cells,8 and STAT5 has been implicated in proliferation and survival of several types of cancer cells.9 Moreover, mutationally activated forms of STAT5 have been demonstrated to possess transforming capacity.10,11 After cytokine stimulation, STAT5 is phosphorylated on tyrosine by JAK2, dimerized, and translocated to the nucleus, where it induces expression of numerous proteins including cyclin D, Bcl-xL, Pim-1, c-Fos, c-Jun, and suppressors of cytokine signaling such as cytokine-inducible SH2-containing protein (CIS) and suppressor of cytokine signaling (SOCS) proteins.12-16 STAT5 is also regulated by serine phosphorylation by both mitogen-activated protein kinase (MAPK)–dependent and –independent pathways,17 but the exact mechanisms and kinases involved remain to be identified.
The pim-1 oncogene was originally identified as a common integration site for Moloney murine leukemia virus,18 and was shown to efficiently cooperate with c-myc, N-myc, or bcl-2 genes in lymphomagenesis.19,20 In the mouse, there are 2 major isoforms of the serine/threonine-specific Pim-1 kinase, 33 kDa and 44 kDa, the larger one being initiated from an upstream alternative translational initiation site.21 The expression of pim-1 is induced by several related cytokines including IL-2, IL-3, GM-CSF, EPO, and PRL,3,5,22,23 all of which also activate STAT5. Indeed, pim-1 is a STAT5 target gene. Moreover, it has been proposed that the IL-3 response mediated by STAT5 may be processed by the products of the STAT5 target genes c-fos and pim-1.12,16 More recently, pim-1 was shown to be up-regulated by Bcr/Abl via STAT5 and be required for Bcr/Abl-mediated cell survival and transformation.24 Indeed, Pim-1 can protect hematopoietic cells from apoptosis induced by cytokine withdrawal,25 glucocorticoids,26 or genotoxic stress.27 In addition, Pim-1 can stimulate activities of several transcription factors, such as c-Myb28 and NFATc1.29 Therefore, we hypothesized that it might be possible for Pim-1 to regulate the activity of STAT5 as well.
Suppressors of cytokine signaling such as SOCS and CIS proteins comprise a family of inhibitory molecules, whose expression is rapidly induced by cytokines. SOCS family proteins in turn down-regulate cytokine signal transduction and STAT activation by interacting with active JAKs or activated receptors.30 However, additional proteins may be needed to modify this negative feedback mechanism. Indeed, Pim family kinases Pim-1 and Pim-2 were recently reported to phosphorylate and stabilize the SOCS1 protein.31 Furthermore, coexpression of Pim-2 with SOCS1 enhanced inhibition of STAT6 activation by SOCS1. Here we report that Pim-1 inhibits STAT5-dependent transcription in several cytokine-responsive cell lines and that this inhibition is most likely mediated via SOCS family proteins.
Materials and methods
DNA constructs
Prokaryotic GST-Pim-1 and GST-Pim-1(K67M) fusion vectors and eukaryotic pLTR-pim-1, pLTR-pim-1(K67M), and pECFP-pim-1 expression vectors have been described previously,29 as have pCMV-pim-1-FLAG28 and p100-FLAG.32 GST-SOCS3, pCI-NEO-SOCS1-FLAG, and pCI-NEO-SOCS3-FLAG vectors were provided by Dr James Johnston (Queen's University Belfast, United Kingdom). The pME18S-based SOCS1, SOCS2, and SOCS3 expression vectors were provided by Dr Douglas Hilton (The Walter and Eliza Hall Institute of Medical Research, Parkville, Australia). The pSpi-LUC2 luciferase reporter construct33 containing the STAT5 binding site from the promoter of the 2.1 serine protease inhibitor gene, and mouse STAT5A expression vector were provided by Dr Timothy Wood (Affibody AB, Bromma, Sweden). The EPO receptor expression vector was provided by Dr James Ihle (St Jude Children's Research Hospital, Memphis, TN). The NFAT-LUC and AP-1-LUC reporters were from Dr Gerald Crabtree (Stanford University, CA). The region encoding amino acids 721-793 of STAT5A was cloned into pGEX-4T-1 vector (Amersham Biosciences, Uppsala, Sweden) to generate the GST-STAT5A TAD fusion construct with the transactivation domain (TAD) of STAT5. The pSV-β-GAL (galactosidase) and the pRLTK (Renilla luciferase) control reporters were from Promega (Madison, WI).
Antibodies
The following primary antibodies were used: antiphosphotyrosine antibody (clone 4G10; Upstate Biotechnology, Lake Placid, NY), anti-STAT5A antibody (Zymed Laboratories, San Francisco, CA), anti-FLAG M2 antibody (Sigma-Aldrich, St Louis, MO), and anti–green fluorescent protein (GFP) antiserum (Clontech Laboratories, Palo Alto, CA), which recognizes multiple GFP color variants including ECFP (enhanced cyan fluorescent protein).
Cell culture
COS-7 cells (American Type Culture Collection [ATCC], Manassas, VA) were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS), 2 mM l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. RPMI 1640 with equal supplements was used to grow murine myeloid FDCP1 cell lines expressing neomycin (FDCP1/Neo), murine full-length, 44-kDa Pim-1 protein (FDCP1/Pim44), or an N-terminally truncated NT81 mutant of Pim-1 (FDCP1/NT81).25 WEHI-conditioned medium (10%) was used as a source of IL-3 for the IL-3–dependent FDCP1 cells, or the cells were cultured with 2 U/mL murine IL-3 (PeproTech, London, United Kingdom). Mouse mammary epithelial cell line HC1134 was kindly provided by Dr Lars-Arne Haldosén (Karolinska Institutet, Huddinge, Sweden). HC11 cells were cultured in RPMI 1640 with 10% fetal bovine serum (FBS), 5 μg/mL insulin (Sigma-Aldrich), 10 ng/mL epidermal growth factor (EGF; Sigma-Aldrich), 50 μg/mL gentamycin (Biochrom KG, Berlin, Germany), l-glutamine, and antibiotics. Prior to hormone treatment, confluent HC11 cells were starved for 48 hours in medium lacking EGF, but containing 5 μg/mL insulin and 2% FBS. HeLa cells (ATCC) were cultured in minimum essential medium (MEM) supplemented with 10% FBS, l-glutamine, and antibiotics. For starvation of HeLa cells, 2% FBS was used.
Transfections and transactivation assays
For transient transfections by electroporation (GenePulser II; Bio-Rad Laboratories, Hercules, CA), FDCP1 cells were mixed with 1 μg pSpi-LUC2, NFAT-LUC, or AP-1-LUC; 0.5 μg pSV-β-GAL; and 2 μg pLTR-poly or pLTR-pim-1 vectors, pulsed at 260 V and 975 microfarad (μF) and grown further in the presence of IL-3. At 24 hours after transfection, cells were washed with phosphate-buffered saline (PBS), grown in the absence of IL-3 for 16 hours, and then stimulated with 10 U/mL IL-3 for 6 hours. For stimulation of NFAT or AP-1 activities, transfected cells were cultivated for 40 hours and then treated with 15 ng/mL PMA (phorbol 12-myristate 13-acetate; Sigma-Aldrich) and 1 μM ionomycin (Calbiochem, La Jolla, CA) for 6 hours. After treatments, cells were collected and analyzed for luciferase activities using the Labsystems luminometer (Labsystems, Helsinki, Finland). The transfection efficiencies were normalized against β-galactosidase activities. Transient transfections of cytokine- or serum-starved FDCP1, HC11, and HeLa cells with Fugene6 (Roche Diagnostics, Indianapolis, IN) or with ExGen 500 (MBI Fermentas, Hanover, MD) reagents were carried out according to manufacturer's instructions. FDCP1 cells were transfected with 1 μg pSpi-LUC2, 0.5 μg pRLTK, 1 μg pim-1, or 2 μg SOCS expression vectors. For HC11 and HeLa cells, 250 ng pSpi-LUC2, 50 ng to 100 ng pRLTK, 250 ng pim-1, and 50 ng to 100 ng STAT5A expression vectors were used. In addition, HeLa cells were cotransfected with 100 ng EPO receptor expression vector. In all cases, appropriate empty vectors were added to balance the total amounts of transfected DNA samples. After 16 hours incubation with transfection mixture, cells were stimulated with 10 U/mL IL-3, 5 μg/mL PRL (Sigma-Aldrich), or 10 U/mL EPO (Eprex; Janssen-Cilag, Schaffhausen, Switzerland) for 6 hours. Cells were then lysed and luciferase activities were measured using the Dual Luciferase Assay System (Promega), where the reporter luciferase activities were normalized to the activities of coexpressed Renilla luciferase. Shown in the figures are means and standard errors of representative examples of at least 3 independent experiments with duplicate or triplicate samples.
Electrophoretic mobility shift assays and nuclear extracts
Double-stranded β-casein (5′-AGATTTCTAGGAATTCAAATCC-3′) oligonucleotides were end-labeled with γ32P–adenosine 5′-triphosphate (ATP). Cells were incubated in the absence of IL-3 for 16 hours and then stimulated with IL-3 for 6 hours where indicated. Nuclear extracts were prepared from the lysates as previously described.35 Binding reactions were preformed in a total volume of 20 μL in binding buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA [ethylenediaminetetraacetic acid], 40 mM KCl (potassium chloride), 1 mM dithiothreitol [DTT], 10% glycerol, 2 μg poly dI-dC) with 10 μg nuclear extracts for 30 minutes at room temperature. Bound protein complexes were separated by 4% polyacrylamide gel electrophoresis (PAGE) and visualized by autoradiography. Competitions to confirm specificity of binding were carried out with unlabeled oligonucleotides. Relative intensities of distinct protein bands were determined by MCID M5+ image analysis software (Imaging Research, St Catharines, ON, Canada).
Immunoprecipitation and Western blotting assays
COS-7 cells were transfected by electroporation with 2 μg of indicated plasmids. Two days later, cells were collected and lysates prepared as previously described.29,36 Aliquots of protein (50 μg) were used as positive controls in Western blotting, whereas 250 μg to 500 μg aliquots were subjected to immunoprecipitation with specific antibodies at 4°C for 2 hours, followed by incubation with protein G–sepharose beads (Amersham Biosciences) at 4°C for 30 minutes. Immunoprecipitates were washed several times, dissolved in reducing Laemmli sample buffer, separated by sodium dodecyl sulfate (SDS)–PAGE and transferred to nitrocellulose membrane (Schleicher & Schuell, Dassel, Germany). Western blotting was performed using specific primary antibodies and biotinylated (Dako, Glostrup, Denmark) or streptavidin-biotin horseradish peroxidase (HRP)–conjugated (Amersham Biosciences) secondary antibodies. The proteins were visualized with ECL Plus reagents (Amersham Biosciences). Relative intensities of distinct protein bands were determined by MCID M5+ image analysis software.
In vitro kinase assays
Putative target proteins were phosphorylated in vitro as previously described29 and analyzed by SDS-PAGE followed by autoradiography. The amounts of proteins loaded were visualized by silver or Coomassie staining.
Results
The transcriptional activity of STAT5 is inhibited by Pim-1
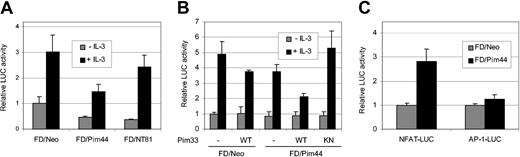
To examine the ability of the Pim-1 kinase to regulate STAT5 activity, we used IL-3–dependent murine myeloid FDCP1 cells as a model system. FDCP1 cells stably expressing either neomycin (FDCP1/Neo), the 44-kDa Pim-1 isoform (FDCP1/Pim44), or an N-terminally truncated, kinase-deficient mutant of Pim-1 (FDCP1/NT81) were transfected with a STAT5-dependent luciferase reporter construct containing a STAT5 binding site derived from the promoter of the 2.1 serine protease inhibitor gene. After transfection, IL-3 was withdrawn for 16 hours, after which half of the cell samples were stimulated with IL-3. As shown in Figure 1A, IL-3 efficiently activated STAT5, but the STAT5 activity was inhibited by nearly 50% in FDCP1/Pim44 cells as compared with FDCP1/Neo cells. By contrast, no significant inhibition was observed in FDCP1/NT81 cells, indicating that the kinase activity of Pim-1 was essential for its negative effects. The levels of endogenously expressed STAT5 protein remained unaffected in all 3 cell lines (Figure 3B), indicating that the Pim-1–induced reduction in STAT5 activity was not due to inhibition of STAT5 expression. Similar results were obtained when cells were cotransfected with expression vectors for either the 33-kDa Pim-1 isoform or a corresponding kinase-deficient mutant with a K67M point mutation in the ATP-binding pocket. The inactive Pim-1 mutant was able to rescue the Pim-1–dependent inhibition of STAT5 activity in FDCP1/Pim44 cells (Figure 1B). By contrast, the 33-kDa wild-type Pim-1 kinase further enhanced the inhibitory effects of the 44-kDa Pim-1, implying that the 2 isoforms of Pim-1 can inhibit STAT5 activity in a dose-dependent fashion (Figure 1B).
Pim-1 inhibits STAT5-dependent transcription in IL-3–stimulated FDCP1 cells. (A-B) FDCP1 cells stably expressing either neomycin (FD/Neo), the 44-kDa Pim-1 protein (FD/Pim44), or a kinase-deficient mutant of Pim-1 (FD/NT81) were transfected with the Spi-LUC2 reporter with or without wild-type (WT) or kinase-deficient (KN) 33-kDa Pim-1 protein (Pim33) as indicated. Cells were left unstimulated or stimulated with IL-3 as indicated. Luciferase activities from cell lysates were measured and normalized against cotransfected control vectors. (C) FD/Neo (▦) and FD/Pim44 (▪) cells were transfected with the NFAT-LUC or AP-1-LUC reporters as indicated and stimulated with PMA and ionomycin.
Pim-1 inhibits STAT5-dependent transcription in IL-3–stimulated FDCP1 cells. (A-B) FDCP1 cells stably expressing either neomycin (FD/Neo), the 44-kDa Pim-1 protein (FD/Pim44), or a kinase-deficient mutant of Pim-1 (FD/NT81) were transfected with the Spi-LUC2 reporter with or without wild-type (WT) or kinase-deficient (KN) 33-kDa Pim-1 protein (Pim33) as indicated. Cells were left unstimulated or stimulated with IL-3 as indicated. Luciferase activities from cell lysates were measured and normalized against cotransfected control vectors. (C) FD/Neo (▦) and FD/Pim44 (▪) cells were transfected with the NFAT-LUC or AP-1-LUC reporters as indicated and stimulated with PMA and ionomycin.
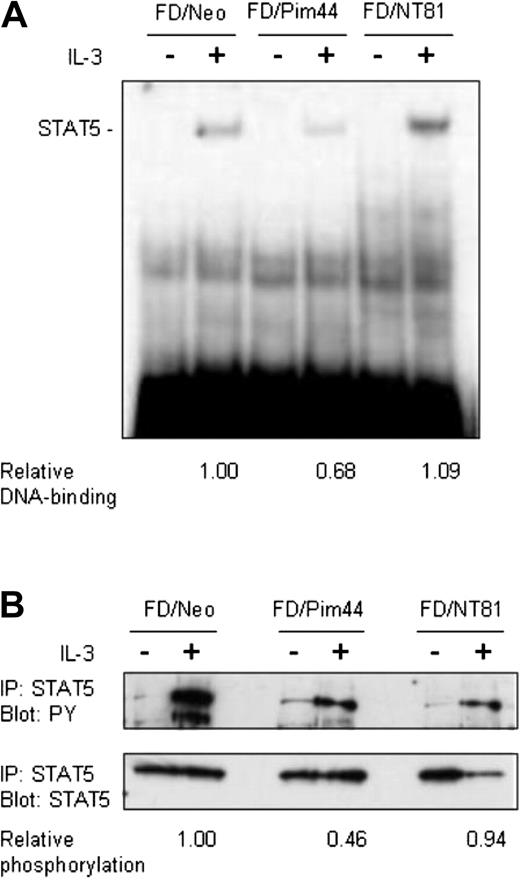
Pim-1 reduces DNA binding and tyrosine phosphorylation of STAT5. Starved FD/Neo, FD/Pim44, or FD/NT81 cells were left unstimulated or stimulated with IL-3 for 6 hours as indicated. (A) Aliquots of nuclear extracts (10 μg) prepared from cell lysates were subjected to electrophoretic mobility shift assay. Protein-DNA complexes were resolved by 4% nondenaturing PAGE and detected by autoradiography. Shown is the position of the specific STAT5-DNA complex. The relative DNA binding of STAT5 in FD/Pim44 or FD/NT81 cells as compared with FD/Neo control cells was quantitated by image analysis by measuring the intensities of specific versus nonspecific DNAcomplexes. (B) Endogenous STAT5 was immunoprecipitated (IP) from cell lysates and the samples were separated by SDS-PAGE, followed by immunoblotting with antiphosphotyrosine (PY) or anti-STAT5 antibodies. The relative phosphorylation levels of STAT5 in FD/Pim44 or FD/NT81 cells as compared with FD/Neo control cells were quantitated by image analysis by measuring the intensities of the upper (PY) versus lower (STAT5) protein bands.
Pim-1 reduces DNA binding and tyrosine phosphorylation of STAT5. Starved FD/Neo, FD/Pim44, or FD/NT81 cells were left unstimulated or stimulated with IL-3 for 6 hours as indicated. (A) Aliquots of nuclear extracts (10 μg) prepared from cell lysates were subjected to electrophoretic mobility shift assay. Protein-DNA complexes were resolved by 4% nondenaturing PAGE and detected by autoradiography. Shown is the position of the specific STAT5-DNA complex. The relative DNA binding of STAT5 in FD/Pim44 or FD/NT81 cells as compared with FD/Neo control cells was quantitated by image analysis by measuring the intensities of specific versus nonspecific DNAcomplexes. (B) Endogenous STAT5 was immunoprecipitated (IP) from cell lysates and the samples were separated by SDS-PAGE, followed by immunoblotting with antiphosphotyrosine (PY) or anti-STAT5 antibodies. The relative phosphorylation levels of STAT5 in FD/Pim44 or FD/NT81 cells as compared with FD/Neo control cells were quantitated by image analysis by measuring the intensities of the upper (PY) versus lower (STAT5) protein bands.
To demonstrate that the negative effects of Pim-1 were specific for the STAT5-dependent reporter and not a result of some general inhibition, we transfected the FDCP1-derived cell lines with luciferase constructs containing multiple NFAT or AP-1 sites. Two days later, cells were stimulated with the phorbol ester PMA and the calcium ionophore ionomycin. Very similarly to Jurkat T cells transiently expressing the 33-kDa Pim-1 protein,29 NFAT activity was up-regulated in FDCP1/Pim44 cells but AP-1 activity was hardly affected (Figure 1C). These results indicated that the effects on luciferase reporter activities that were shared by the 33 kDa and 44 kDa isoforms of Pim-1 specifically depended on the transcription factors involved.
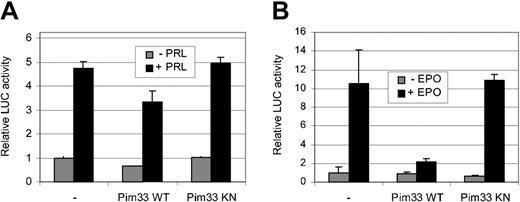
To rule out the possibility that the inhibition of STAT5 activity by Pim-1 is a cell- or stimulation-specific phenomenon, transactivation assays were carried out with transiently transfected HC11 cells and HeLa cells. For this purpose, cells were cotransfected with expression vectors for STAT5A and either wild-type or kinase-deficient Pim-1. In addition, an expression vector for EPO receptor was added to HeLa cells. Transfected HC11 and HeLa cells were then treated with PRL or EPO, respectively (Figure 2A-B). Ectopic expression of wild-type, but not the kinase-deficient mutant of Pim-1, reduced ectopic STAT5 activity in both of these cells, indicating that the observed inhibition is not specific for IL-3 signaling or FDCP1 cells, but that Pim-1 is capable of inhibiting STAT5 activity in other signaling pathways as well. However, the extent of the Pim-induced inhibition varied in a cell type–dependent fashion.
Pim-1 inhibits STAT5-dependent transcription in PRL-stimulated HC11 cells and in EPO-stimulated HeLa cells. HC11 (A) and HeLa (B) cells were transfected with the Spi-LUC2 reporter and STAT5A expression vector with or without WT or KN Pim33. EPO receptor expression vector was also transfected into HeLa cells. Starved cells were left unstimulated or stimulated with PRL (A) or EPO (B) as indicated.
Pim-1 inhibits STAT5-dependent transcription in PRL-stimulated HC11 cells and in EPO-stimulated HeLa cells. HC11 (A) and HeLa (B) cells were transfected with the Spi-LUC2 reporter and STAT5A expression vector with or without WT or KN Pim33. EPO receptor expression vector was also transfected into HeLa cells. Starved cells were left unstimulated or stimulated with PRL (A) or EPO (B) as indicated.
Pim-1 inhibits both DNA binding and tyrosine phosphorylation of STAT5
The activation of STAT5 can be measured both by its tyrosine phosphorylation and by its ability to bind to DNA. To determine whether these events upstream of transactivation are also affected by Pim-1, IL-3–starved or IL-3–stimulated FDCP1/Neo, FDCP1/Pim44, or FDCP1/NT81 cells were collected at several time points from 15 minutes up to 6 hours after stimulation. Nuclear extracts were prepared from cell lysates and analyzed in electrophoretic mobility shift assays with a radioactively labeled oligonucleotide containing a STAT5 binding site derived from the β-casein promoter. At any time point tested, the IL-3–induced DNA-binding activity of STAT5 was significantly reduced in FDCP1/Pim44 cells as compared with FDCP1/Neo or FDCP1/NT81 cells (Figure 3A and data not shown). These results indicated that Pim-1 inhibits DNA binding by STAT5.
To analyze the phosphorylation status of the endogenously expressed STAT5 protein, we immunoprecipitated STAT5 from the cell lysates with anti-STAT5A antibodies, separated the samples on SDS-PAGE, and subjected them to Western blotting with antiphosphotyrosine and anti-STAT5A antibodies. These analyses revealed that while the STAT5 protein levels were hardly affected, the degree of tyrosine phosphorylation of STAT5 was remarkably lower in FDCP1/Pim44 cells as compared with FDCP1/Neo or FDCP1/NT81 cells (Figure 3B). Thus, wild-type, but not kinase-deficient Pim-1, inhibits STAT5 already at the initial stage of activation.
Pim-1 does not phosphorylate or directly interact with STAT5
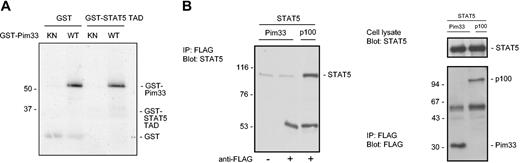
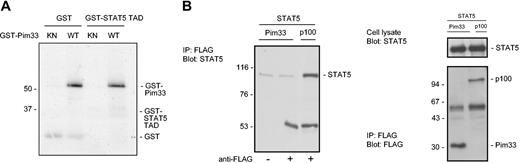
The C-terminal domains of STATs are indispensable for STAT transactivation function and contain phosphorylatable tyrosine and serine residues that are essential for optimal STAT activation. In the case of STAT5, phosphorylation of tyrosine 694 is required for its activation, but serines 725 and 779 have also been observed to become phosphorylated after IL-2 and PRL stimulation, respectively.37 However, the precise functions of these serine phosphorylations have remained unknown. Since Pim-1 is a serine/threonine kinase, one possible mechanism for inhibition of STAT5 would be its phosphorylation by Pim-1. To examine this possibility, we carried out in vitro kinase assays with bacterially expressed wild-type or kinase-deficient GST–Pim-1 fusion proteins and used full-length STAT5 protein expressed in COS-7 cells or STAT5 transactivation domain fused to GST (STAT5-TAD) as substrates. However, wild-type Pim-1 was clearly unable to phosphorylate either protein in vitro, even though it properly autophosphorylated itself (Figure 4A and data not shown).
Pim-1 does not phosphorylate STAT5 or interact with it. (A) GST or a GST-fusion protein containing the transactivation domain of STAT5 (GST-STAT5 TAD) was subjected to in vitro phosphorylation in the presence of wild-type (WT) or kinase-deficient (KN) GST-Pim33. Samples were analyzed by SDS-PAGE, followed by autoradiography. Shown are positions of the GST fusion proteins. (B) COS-7 cells were transfected with STAT5 together with either Pim33-FLAG or p100-FLAG expression vectors. Cell lysates were subjected to immunoprecipitation (IP) with or without anti-FLAG antibodies as indicated, followed by immunoblotting with anti-STAT5 (left and upper right panels) and anti-FLAG antibodies (lower right panel). Shown are positions of STAT5, p100, and Pim33 proteins. Note that only samples precipitated with anti-FLAG antibodies have been included in the right panels.
Pim-1 does not phosphorylate STAT5 or interact with it. (A) GST or a GST-fusion protein containing the transactivation domain of STAT5 (GST-STAT5 TAD) was subjected to in vitro phosphorylation in the presence of wild-type (WT) or kinase-deficient (KN) GST-Pim33. Samples were analyzed by SDS-PAGE, followed by autoradiography. Shown are positions of the GST fusion proteins. (B) COS-7 cells were transfected with STAT5 together with either Pim33-FLAG or p100-FLAG expression vectors. Cell lysates were subjected to immunoprecipitation (IP) with or without anti-FLAG antibodies as indicated, followed by immunoblotting with anti-STAT5 (left and upper right panels) and anti-FLAG antibodies (lower right panel). Shown are positions of STAT5, p100, and Pim33 proteins. Note that only samples precipitated with anti-FLAG antibodies have been included in the right panels.
To determine whether Pim-1 could bind to STAT5, full-length STAT5 was expressed in COS-7 cells together with FLAG-tagged Pim-1 or p100. p100-FLAG was used as a positive control, since it interacts with STAT5.36 Pim-1 and p100 were immunoprecipitated with anti-FLAG antibodies, after which the coprecipitating STAT5 proteins were detected by Western blotting with anti-STAT5A antibodies. As expected, p100 associated with STAT5 (Figure 4B). By contrast, no coprecipitation of STAT5 was observed with Pim-1. To exploit putative interactions of endogenously expressed proteins as well, coprecipitation assays were also carried out with untransfected FDCP1 cells. However, also in these assays, Pim-1 could not bind to endogenous STAT5 or vice versa (data not shown).
SOCS1 and SOCS3 cooperate with Pim-1 to inhibit STAT5 activity
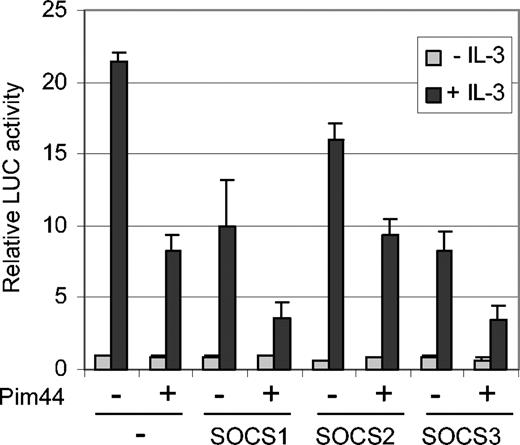
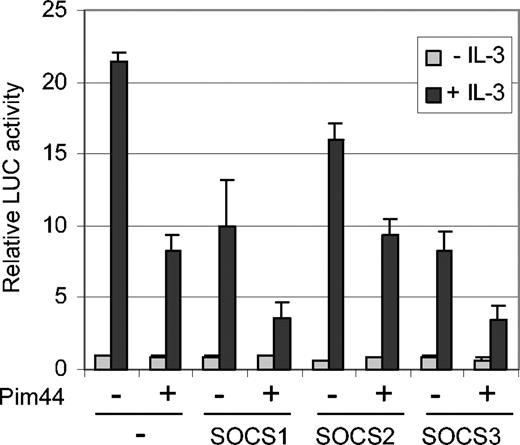
Since we were not able to detect any direct interaction between STAT5 and Pim-1, it seemed more likely that the inhibition of STAT5 activity by Pim-1 was mediated by another protein. Since Pim-1 was able to decrease STAT5 tyrosine phosphorylation and since Pim-1 has been reported to interact with SOCS1,31 we hypothesized that a member of the SOCS protein family might mediate STAT5 inhibition. To investigate this possibility, FDCP1/Neo and FDCP1/Pim44 cells were transfected with SOCS1, SOCS2, and SOCS3 expression vectors together with the STAT5-dependent luciferase reporter. In the absence of IL-3, no major differences were observed, but when the samples were treated with IL-3, all 3 SOCS proteins inhibited STAT5 activity in FDCP1/Neo cells in a dose-dependent fashion (Figure 5 and data not shown). However, SOCS1 and SOCS3 were more efficient inhibitors than SOCS2. Furthermore, only these 2 SOCS family members were able to synergize with Pim-1 to inhibit STAT5 activity in FDCP1/Pim44 cells in a more profound fashion (Figure 5). These results suggest specific cooperation between Pim-1 and SOCS1 or SOCS3.
Pim-1 cooperates with SOCS1 and SOCS3 to inhibit STAT5-dependent transcription in IL-3–stimulated FDCP1 cells. FD/Neo and FD/Pim44 cells were transfected with the Spi-LUC2 reporter with or without expression vectors for SOCS1, SOCS2, or SOCS3. Cells were left unstimulated or stimulated with IL-3 as indicated.
Pim-1 cooperates with SOCS1 and SOCS3 to inhibit STAT5-dependent transcription in IL-3–stimulated FDCP1 cells. FD/Neo and FD/Pim44 cells were transfected with the Spi-LUC2 reporter with or without expression vectors for SOCS1, SOCS2, or SOCS3. Cells were left unstimulated or stimulated with IL-3 as indicated.
SOCS3 is a novel interaction partner and phosphorylation target for Pim-1
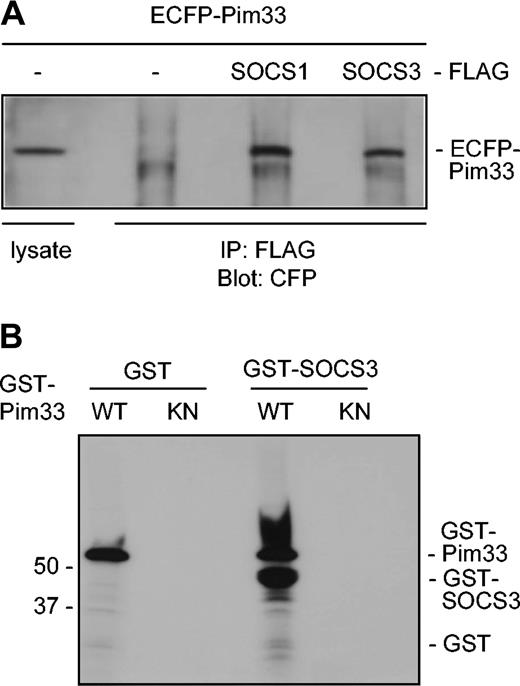
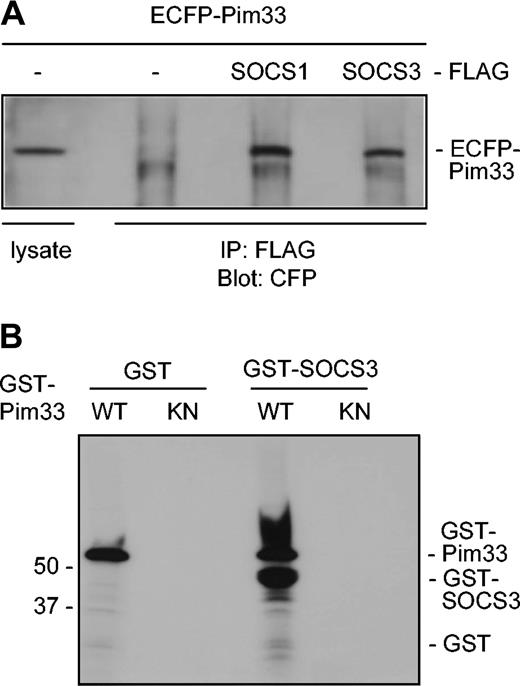
It has previously been demonstrated that Pim kinases phosphorylate SOCS1 and thereby stabilize it.31 Since SOCS1 and SOCS3 share the greatest sequence homology within the SOCS family, it seemed likely that Pim-1 interacted also with SOCS3. To investigate this possibility, Pim-1 and either SOCS1 or SOCS3 proteins were expressed in COS-7 cells as ECFP- and FLAG-tagged proteins, respectively. Indeed, Pim-1 was able to coprecipitate with both SOCS proteins to a similar extent, whereas it did not bind to the FLAG-tagged only control or to some irrelevant tagged proteins (Figure 6A and data not shown). In addition, ECFP did not bind to any of the FLAG-tagged proteins. Western blotting of both cell lysates and immunoprecipitates with FLAG antibodies further confirmed that SOCS1 and SOCS3 proteins were expressed to an equivalent extent (data not shown). To determine whether Pim-1 could phosphorylate SOCS3, bacterially expressed GST–Pim-1 fusion protein and the corresponding K67M mutant were subjected to in vitro kinase assays using GST-SOCS3 fusion protein as a substrate. The kinase-deficient Pim-1 mutant was not able to phosphorylate SOCS3, whereas wild-type Pim-1 phosphorylated both itself and SOCS3 (Figure 6B). These results thus suggested that SOCS3 also is a direct substrate for Pim-1.
Pim-1 interacts with both SOCS1 and SOCS3 in vivo and phosphorylates SOCS3 in vitro. (A) COS-7 cells were transfected with expression vectors encoding for ECFP or ECFP-Pim33 and SOCS1-FLAG, SOCS3-FLAG, or just the FLAG-tag. Part of the cell lysates were subjected to immunoprecipitation with anti-FLAG antibodies. Samples were separated on SDS-PAGE, followed by immunoblotting with anti-GFP antibodies, which also recognize CFP. Shown is the position of ECFP-Pim33 protein. (B) GST or GST-SOCS3 proteins were subjected to in vitro phosphorylation in the presence of wild-type (WT) or kinase-deficient (KN) GST-Pim33. Shown are positions of the GST fusion proteins.
Pim-1 interacts with both SOCS1 and SOCS3 in vivo and phosphorylates SOCS3 in vitro. (A) COS-7 cells were transfected with expression vectors encoding for ECFP or ECFP-Pim33 and SOCS1-FLAG, SOCS3-FLAG, or just the FLAG-tag. Part of the cell lysates were subjected to immunoprecipitation with anti-FLAG antibodies. Samples were separated on SDS-PAGE, followed by immunoblotting with anti-GFP antibodies, which also recognize CFP. Shown is the position of ECFP-Pim33 protein. (B) GST or GST-SOCS3 proteins were subjected to in vitro phosphorylation in the presence of wild-type (WT) or kinase-deficient (KN) GST-Pim33. Shown are positions of the GST fusion proteins.
Discussion
Here we report that the Pim-1 kinase inhibits STAT5-mediated signaling in response to IL-3, PRL, and EPO and most likely also other STAT5-activating cytokines. These negative effects shared by both the 33-kDa and 44-kDa Pim-1 isoforms are independent of the cellular context, since dose-dependent inhibition of STAT5 activity was observed not only in naturally cytokine-responsive cells such as FDCP1 and HC11 cells, but also in HeLa cells ectopically expressing the EPO receptor. By contrast, the effects of Pim-1 are dependent on its kinase activity, since coexpression of a kinase-deficient mutant was able to rescue the inhibition, most likely due to its ability to sequester wild-type Pim-1 into inactive oligomers, as suggested by our previous studies.29 Furthermore, the negative effects of Pim-1 are transcription factor–specific, since in parallel experiments with FDCP1 cells stably overexpressing Pim-1, only STAT5 activity was inhibited, whereas NFAT activity was upregulated and AP-1 activity remained unaffected. These results on NFAT and AP-1 activities correlated well with those previously obtained from Jurkat T cells,29 again confirming that the positive or negative effects of Pim-1 are not dependent on the cell type, but on the transcription factors involved.
Tyrosine phosphorylation and consequently also DNA binding of STAT5 are reduced in Pim-1–expressing cells, indicating that Pim-1 inhibits STAT5-dependent transcription by interfering with the initial events involved in the activation of STAT5. Yet we could not detect a direct interaction between Pim-1 and STAT5, and Pim-1 was unable to phosphorylate STAT5, although kinase activity was required for all of its effects. Thus we assumed that another protein cooperates with Pim-1 in the inhibition of STAT5 activity. According to our results, SOCS1 and SOCS3 proteins are the most likely candidates, since both of them further enhance the negative effects of Pim-1 on STAT5 activity. Moreover, we demonstrate that Pim-1 can interact with both SOCS1 and SOCS3 under in vivo conditions and that SOCS3 is a novel substrate for Pim-1. Via phosphorylation, Pim-1 is likely to stabilize SOCS3 and thereby potentiate its inhibitory effects, as recently reported for SOCS1 phosphorylated by Pim family kinases.31
Our results correlate well with the abilities of the closely related SOCS1 and SOCS3 proteins, but not SOCS2, to down-regulate IL-3, EPO, and PRL responses.38,39 SOCS1 and SOCS3 also inhibit STAT5 activation in response to several other cytokines, including IL-2.40,41 However, this does not necessarily result in decreased cell proliferation or survival. By contrast, SOCS3 has been shown to support IL-2–induced signaling via the Ras/MAPK pathway by binding the Ras inhibitor RasGAP and probably targeting it to proteasome-mediated degradation.42,43 Interestingly, both STAT5 and Ras contribute to Bcr/Abl-induced transformation of hematopoietic cells by having both cooperative and redundant functions.44
Also, Pim-1 has recently been reported to play a critical role in Bcr/Abl-mediated leukemogenesis.24 Since pim-1 is a STAT5 target gene and since Pim-1 appears to act downstream of Ras to stimulate activities of Myb and NFATc family transcription factors,28,29 Pim-1 may provide an important link between the STAT5 and Ras signaling pathways by maintaining a balance possibly mediated by SOCS proteins to protect cells from the adverse effects of too-strong stimuli. Thus, STAT5 may attenuate its own activation via up-regulation of both pim and SOCS family genes. Since Pim-1 does not completely abolish STAT5-mediated signaling, we propose that the observed inhibition is a fine-tuning mechanism to ensure an optimal level of STAT5 activity. In nontransformed cells, this negative feedback loop, which will also eventually shut down pim-1 expression, may be required during cellular differentiation to turn proliferative signals down. It may also be essential for cellular homeostasis, as evidenced by the critical roles of SOCS1 and SOCS3 during embryogenesis.39 By contrast, in transformed cells constitutively overexpressing pim-1, signals restricting growth or survival may have been overridden by additional activated oncogenes, such as the gfi-1 family members which can cooperate with pim and myc genes to accelerate lymphomagenesis.45-47 Interestingly, in contrast to Pim-1, Gfi-1B reduces SOCS activity by specifically repressing transcription of SOCS1 and SOCS3, but not SOCS2 genes.48 By promoting activities of SOCS1 and SOCS3, Pim-1 is likely to affect also other STATs than STAT5, as supported by the observation that Pim-2 is able to inhibit IL-4–induced STAT6 activation in cooperation with SOCS1.31 Furthermore, since SOCS family proteins have been suggested to have also STAT-independent functions,49 Pim kinases may stimulate those as well.
In conclusion, our data indicate that Pim-1 can cooperate with SOCS1 and SOCS3 to inhibit STAT5 activity in response to several cytokines. Pim and SOCS proteins may therefore comprise a negative feedback loop that fine-tunes STAT5 activation and is needed to regulate normal cell growth, survival, and differentiation. Constitutive coexpression of Pim-1 with other oncoproteins (eg, in Bcr/Abl-transformed cells) may impair this fine-tuning mechanism and thereby result in hyperproliferation and eventually in malignant tumors.
Prepublished online as Blood First Edition Paper, February 5, 2004; DOI 10.1182/blood-2003-09-3126.
Supported by the Academy of Finland, the Turku Graduate School of Biomedical Sciences, Paulo Foundation, Maud Kuistila Foundation, Finnish Cultural Foundation, Finnish Cancer Organizations, Biomedicum Helsinki Foundation, Ida Montin Foundation, Sigrid Juselius Foundation, and the Medical Research Fund of Tampere University Hospital.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs James Johnston, Douglas Hilton, Timothy Wood, James Ihle, Gerald Crabtree, and Lars-Arne Haldosén for reagents, and Jouko Sandholm, Kaija-Liisa Laine, and Anne Sovikoski-Georgieva for expert technical assistance.