Abstract
Previously, we have shown that Fos/Jun transcription factor complexes function as positive modulators of myeloid differentiation. Fos, which is stably induced during normal myeloid differentiation, is not induced upon differentiation of M1 myeloblastic leukemia cells. Establishing M1 cells that express a β-estradiol-conditional FosER chimera, we show that in the absence of the differentiation inducer interleukin-6 (IL-6), Fos expression in M1 myeloblasts promoted apoptotic cell death, entailing cytochrome c release and caspase-9 activation. In contrast, in the presence of IL-6, Fos-mediated apoptosis was abrogated, and Fos promoted terminal differentiation, increasing the sensitivity of M1 cells to be induced for differentiation by IL-6. Fos-mediated apoptosis was accelerated by deregulated c-Myc. Furthermore, restoring Fos expression in M1 partially abrogated the block imparted by deregulated c-Myc on the myeloid differentiation program, increased the sensitivity of the cells to be induced for differentiation, and curtailed their leukemic phenotype. These data provide evidence that Fos/Jun transcription factor complexes play a role in modulating both myeloid cell survival and differentiation and suggest that genetic lesions that alter Fos expression may cooperate with deregulated c-Myc in leukemogenesis. (Blood. 2004;103:4259-4267)
Introduction
Evidence was obtained in this laboratory that myeloid differentiation primary response (MyD) genes and proto-oncogenes function as positive and negative regulators of terminal hematopoietic cell differentiation. This finding was achieved by genetically manipulating hematopoietic cells of the myeloid lineage, including both normal cells and differentiation-inducible leukemic cell lines.1,2
C-jun, JunB, and JunD, which are members of the Ap-1 family of leucine zipper transcription factors, were identified in this laboratory as MyD genes. They are stably induced during differentiation of M1 myeloblastic leukemia cells and normal myeloblasts,3,4 suggesting that they play key roles in the initiation, progression, and maintenance of the myelopoietic differentiation program. The AP-1 family of homodimeric and heterodimeric leucine zipper transcription factors also includes Fos and Fos-related proteins.3 It has been established that Jun proteins homodimerize and heterodimerize with each other, whereas Fos proteins need Juns to form stable AP-1 complexes.3 Intriguingly, c-Fos was found to be stably induced during differentiation of bone marrow (BM)-derived myeloblasts but not upon differentiation of the M1 myeloblastic leukemia cells.3,4 These observations have prompted us to investigate the role Fos plays in myeloid differentiation.
Previously, we have shown that enforced expression of very low levels of c-Fos (compared with what has been observed in differentiating bone marrow myeloblasts) in M1 myeloid progenitors resulted in both an increased propensity of the cells to differentiate in culture and a reduction in the aggressiveness of the M1 leukemic phenotype in nude mice. Blocking Fos/Jun expression using antisense oligomers was observed to impair myeloid differentiation of both normal and M1 myeloblasts, implicating Fos/Jun transcription complexes as positive modulators of hematopoietic differentiation. In the course of that work it became evident that M1 cells expressing c-Fos at levels comparable to what was observed in differentiating BM-derived myeloblasts could not be established.4
In other studies we have shown that deregulated expression of proto-oncogenes that play a role in cell cycle progression and whose expression is normally suppressed upon induction of terminal myeloid differentiation blocks the differentiation program.2,5-8 For example, deregulated c-Myc was found to block myeloid differentiation at an intermediate stage of the differentiation program.6
These findings have raised several interesting questions regarding the role c-Fos plays in the regulation of normal blood cell homeostasis. As indicated above, M1 cells that express c-Fos at levels comparable to what was observed in differentiating BM-derived myeloblasts could not be established. How physiologic levels of c-Fos effect M1 myeloblast cell proliferation, differentiation, and/or survival in the absence or presence of differentiation inducer remains to be addressed. Deregulated expression of c-Myc was shown to block the M1 differentiation program at an intermediate stage; however, M1 myeloblasts are devoid of Fos expression. It can be asked how the lack of Fos expression contributes to the block imparted by deregulated c-Myc on the M1 myeloid differentiation program.
To answer these questions we have generated M1 cell lines that conditionally express physiologic levels of c-Fos, using a transgene encoding for the chimeric FosER protein (M1FosER), whose activity is dependent on the presence of β-estradiol (βE2) in the culture media.3 Taking advantage of the M1FosER cells, it is shown that in the absence of the differentiation inducer IL-6, Fos expression resulted in apoptotic cell death of M1 myeloblasts, involving cytochromec release from the mitochondria and caspase-9 activation. In contrast, in the presence of the differentiation inducer interleukin-6 (IL-6), Fos-mediated apoptosis is shown to be abrogated and Fos promotes myeloid differentiation. Furthermore, it is shown that restoring Fos expression in M1 cells partially abrogates the block imparted by deregulated c-Myc on terminal myeloid differentiation and curtails their leukemic phenotype.
Taken together, these data provide evidence that Fos/Jun transcription factor complexes play an important role in modulating both myeloid cell survival and differentiation and suggests that genetic lesions that alter Fos expression cooperate with deregulated c-Myc in blocking myeloid differentiation and exacerbates the leukemic phenotype.
Materials and methods
Cells, cell culture, cytokines, and chemicals
Myeloblast-enriched BM cells were obtained from femurs of CD-1 mice (Charles River Laboratories, Wilmington, MA) injected intraperitoneally 3 days earlier with 3 mL 10% sodium caseinate (Difco, Detroit, MI) in phosphate buffered saline (PBS).4 M1 murine myeloblastic leukemia cell line competent for induction of terminal differentiation upon addition of physiologic factors such as IL-6, leukemia inhibitory factor (LIF), or lung-conditioned medium (LUCM, containing IL-6 and LIF) has been described previously.8 M1 cells were cultured in Dulbecco modified Eagle medium (DMEM; Gibco BRL, Gaithersburg, MD) supplemented with 10% heat-inactivated horse serum (Gibco BRL) and 1% penicillin and streptomycin (Gibco BRL). M1FosER were cultured in phenol red-free DMEM (Gibco BRL), 10% heat-inactivated horse serum, and 400 μg/mL G418 selection agent. Since the c-Myc and the bcl-2 transgenes carried a puromycin resistance selection marker, the M1FosER-Myc, M1myc, and the M1FosER-bcl-2 cell lines were grown in medium that was supplemented with 3.5 μg/mL puromycin (dissolved in 10% ethanol and 1 × PBS; Sigma, St Louis, MO), in addition to the G418 (400 μg/mL). Control cell lines expressing empty vector also were cultured with the antibiotic that pertained to the selection marker. All cells were maintained in a humidified atmosphere with 10% CO2 at 37°C. Serum-free conditioned medium from mouse lungs (LUCM) was prepared with LiCl8 and used at a concentration of 10% purified recombinant human IL-6 (rhuIL-6; a generous gift from L. Souza of Amgen, Thousand Oaks, CA). β-estradiol (βE2) was obtained from Sigma and used at 2 μM.
Establishment of genetically engineered cell lines
Freshly selected PA317 (American Type Culture Collection, Rockville, MD) viral packaging cells were transiently transfected overnight with 10 μg DNA using the CaPO4 method.9 Supernatant containing infectious retroviral pseudotypes was harvested 48 hours after transfection, pooled on ice, and filtered through a 45-μm membrane. Infection of exponentially growing M1, M1FosER, and M1Fos14 cells was performed in 10% CO2 atmosphere with 5 mL virus-enriched culture supernatant and 10 μg/mL polybrene (Sigma) per 100-mm dish. After 18 hours, the medium was changed to remove the polybrene, and the cells were allowed to recuperate for 24 hours. For selection, the infected cells were replated into 24-well plates at 100 cells/well, 500 cells/well, and 1000 cells/well in DMEM supplemented with the appropriate antibiotic (400 μg/mL G418 and/or 3.5 μg/mL puromycin). Individual clones were isolated and established as clonal cell lines. At least 3 clones were used for analysis and found to behave in a similar manner. Characteristics of control cell lines (M1-MSCV-puro, M1-MSCV-neo, M1-MSCV-puro/neo, and M1FosERMSCV-puro) were indistinguishable from that of their respective parental cells.
Leukemogenicity assay
Nude mice were intravenously injected (tail vein) with 105 cells prepared in 200 μL of 1 × PBS for each cell type. Control animals were injected with the same volume of 1 × PBS. Cells were treated for 5 days with or without IL-6 (100 ng/mL) prior to inoculation to mice. The number of mice that were leukemia-bearing or leukemia-free was evaluated statistically using standard Kaplan-Meier survival analysis method.
General recombinant DNA techniques and expression vectors
Plasmids and DNA probes were prepared as previously described.4,10 The retroviral plasmid expression vectors MSCV-neomycin (neo) and MSCV-puromycin (puro) were a gift from Dr Robert G. Hawley11 (University of Toronto, ON, Canada). pMV7-FosER/neo, kind gift from Dr Maniard Busslinger (Research Institute of Molecular Pathology, Vienna Biocenter, Vienna, Austria),12 was used for infection to generate the M1FosER cells, and MSCV-bcl-2/puro (constructed in this laboratory; B.G., unpublished data, October 1996) was used to generate the M1FosER-/bcl-2 cells. To construct the MSCV-myc/puro plasmid, full-length murine c-Myc cDNA (1.6 kilobase [kb]) was excised from the LK444-c-Myc plasmid using BAMHI and SALI and blunt-end ligated into the XhoI site of the 6.3-kb MSCV-puromycin plasmid.6
Analysis of cell morphology and cell viability
Cells were collected at indicated time points and following cytocentrifugation were stained with May-Grunwald-Giemsa. Morphologic differentiation was determined by counting 250 to 300 cells on stained cytospin smears and scoring the proportion of immature blast cells, cells at intermediate stages of differentiation, and mature macrophages.4,13 Immature blast cells are characterized by scant cytoplasm and round or oval nuclei; cells at the intermediate stage of differentiation are flattened, with a larger cytoplasm-to-nucleus ratio, and irregularly shaped nuclei with few interspersed vacuoles; mature macrophages are flattened, spread out, and interspersed with numerous vacuoles in a greatly enlarged cytoplasm. Cell viability was determined using the trypan blue dye exclusion method. At time points as indicated, cell aliquots were mixed 1:1 with trypan blue dye and percent viable cells was counted in a hemocytometer. All experiments were repeated at least 3 times.
Analysis of apoptotic DNA fragmentation
Cells were harvested, washed in 1 × PBS, and incubated overnight in TNE lysis buffer (10 mM Tris [tris(hydroxymethyl)aminomethane]-HCL, pH 8.0; 150 mM NaCl; and 10 mM EDTA [ethylenediaminetetraacetic acid], pH 8.0) supplemented with 200 μg/mL proteinase K and 0.5% Triton-X 100 at 55°C. Total high-molecular weight DNA was isolated by extracting twice with phenol (equilibrated in Tris and H2O saturated), followed by 2 extractions with equal volumes of 1:1 phenol/(1:24) chlorophorm/isoamyl alcohol, followed by 2 extractions of just 1:24 chlorophorm/isoamyl alcohol. The DNA was then precipitated, resuspended in 10 mM Tris, pH 8.0, 1 mM EDTA, and subjected to electrophoretic analysis in a 2% agarose gel and run overnight at 20 V. The gel was then stained with ethidium bromide for analysis.
RNA extraction, Northern blots, and probes
Total RNA was prepared from 5 to 10 × 106 cells using TRIzol reagent (Gibco BRL) as described in the manufacturer's specifications. Total RNA (10 μg per lane) was electrophoresed on a 1% agarose gel containing formaldehyde (0.7%), and the loading of equal amounts was confirmed by comparing intensity of ethidium bromide staining of ribosomal RNA bands. For Northern blot analysis, gels were denatured with 50 mM NaOH and 150 mM NaCl twice for 30 minutes; neutralized with 0.1 M Tris-HCl, pH 7.0 and 150 mM NaCl twice for 30 minutes; and then transferred onto Duralon-UV membranes (Stratagene, La Jolla, CA). The membranes were fixed by UV cross-linking in a Stratalinker (Stratagene) and baked at 80°C for 2 hours. Blots were then hybridized overnight at 42°C in 50% deionized formamide, 10% dextran sulfate, 1 M NaCl, 1% sodium dodecyl sulfate (SDS), and 100 μg/mL salmon sperm DNA. Blots were washed at room temperature in 2 × sodium citrate concentrate (SSC), 0.1% SDS for 5 minutes twice, at 60°C in 0.1 × SSC, 1% SDS for 30 minutes, and exposed to x-ray film at -80°C for 12 to 18 hours. To rehybridize, blots were stripped by washing at least 5 times with boiling stripping solution (0.1 × SSC and 0.1% SDS) for 4 minutes.14 Probes used include the 2.2-kb murine c-fos fragment isolated from an MTFos plasmid4 ; the c-Myc fragment (1.6 kb) cut from the LK444-myc plasmid using BamHI and SalI restriction enzymes; the lysozyme EcoRI fragment (600 base pair [bp]) cut from the pBS vector6 ; and a ferritin α fragment excised from pBS.6 Probes were labeled by random priming (RadPrime DNA labeling kit; Gibco-BRL) to a specific activity equal to or more than 109 cpm/μg.
Protein extraction and immunoblotting
Cells were collected, washed twice with PBS, and lysed on ice for 30 minutes in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 50 mM Tris-HCL, pH 8.0, 15 NP-40, 0.5% sodium deoxycolate, and 0.1% SDS) supplemented with a cocktail of protease inhibitors: 2 μg/mL leupeptin, 2 μg/mL aprotinin, 2 μg/mL benzamidine, 1 mM phenylmethylsulfonyl fluoride (PMSF) (Sigma), and 1 mM dithiothreitol (DTT). Cellular debris was removed from the soluble extract by centrifugation at 14 000g for 10 minutes, and protein concentration was determined with the Bio-Rad (Hercules, CA) protein assay kit. Then, 50 μg of each protein extract were electrophoresed on a denaturing polyacrylamide gel containing SDS (SDS-PAGE). Resolved proteins were transferred to hybond enhanced chemiluminescence (ECL) nitro-cellulose membranes in 10 mM cyclohexyl aminopropane sulfonic acid (CAPS)/10% methanol buffer (pH 11.0). Membranes were subsequently blocked with 5% nonfat dry milk in 1 × PBS and 0.05% Tween-20 for 1-2 hours at room temperature and probed with primary antibody overnight at 4°C. After washing with washing buffer (1 × PBS and 0.05% Tween-20) horseradish peroxidase (HRP)-linked anti-mouse IgG (1:5000 dilution; Amersham Life Sciences, Arlington Heights, IL) was added for 2 hours. The antibody was then detected using the Pierce chemiluminescence detection system (Amersham). Primary antibodies against cytochrome c and the Fas receptor were obtained from Pharmingen (San Diego, CA). Antibodies against murine bcl-2, c-Myc, were from Santa Cruz Biotechnology (Santa Cruz, CA). The antibody against the poly-(ADP [adenosine diphosphate]-ribose)-polymerase (PARP) substrate was from Boehringer Mannheim (Mannheim, Germany), and the antibodies used to detect caspase-8 and -9 cleavage were from Stressgen (Victoria, BC, Canada).
Flow cytometry analysis
For cell cycle analysis 2 × 106 cells were harvested by centrifugation, washed 2 times in 1 × PBS, and fixed in 70% ice-cold ethanol. The cells were subsequently treated for 30 minutes with RNAse A (180 μg/mL) and stained with propidium iodide (34 μg/mL in 7.6 mM sodium citrate; Sigma) prior to analysis using the Coulter epics elite system (Miami, FL). An appropriate window was chosen in the fluorescence-activated cell sorter (FACS) such that both living and apoptotic cells were analyzed. Cell cycle analysis was performed at least 3 times yielding similar results. For analysis of Fas receptor 2 × 106 cells were harvested by centrifugation and washed twice with 1 × PBS. To eliminate background, cells were resuspended in 1% BSA and allowed to incubate at room temperature for 30 minutes. The Fas receptor primary antibody (Pharmingen, San Diego, CA) was diluted 2 μg/mL in 1% bovine serum albumin (BSA), added 1:1 to tubes containing cells suspended in blocking solution, and allowed to incubate for 1 hour. Cells were further incubated with fluorescein isothiocyanate (FITC)-linked anti-mouse IgG secondary antibody (Santa Cruz Biotechnology). Cells were then washed 2 times, resuspended in 1 × PBS, and analyzed using the Coulter epic elite system for variation in signal.
Indirect immunofluorescent staining of cells
Cytospin preparation of cells were dried at room temperature for 15 minutes and fixed with 100% methanol at 4°C for at least 15 minutes (or left overnight). Cells were blocked with 3% BSA (in NP-40/PBS) for 30 minutes to 1 hour at room temperature. The blocking solution was aspirated and the slides were incubated for 3 hours with primary antibody (1 μg/mL in blocking solution) at room temperature in a moist chamber. Slides were washed 3 times with 1 × PBS and then incubated with a 1:500 dilution of secondary antibody conjugated with fluorescein (Santa Cruz Biotechnology). Slides containing the cells were then washed 3 times with 1 × PBS for 5 minutes per wash. The slides were mounted with mounting solution and analyzed directly under a fluorescent microscope. Primary antibodies against cytochrome c were obtained from Pharmingen (San Diego, CA).
Results
Fos modulates myeloid cell survival and differentiation
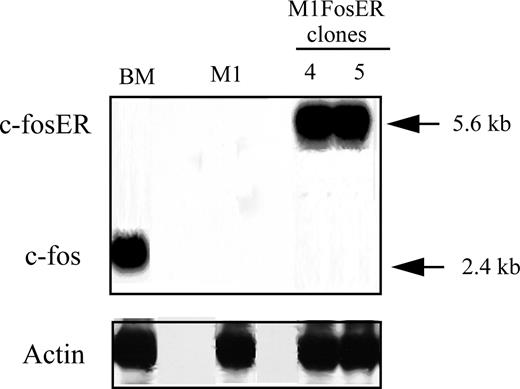
Fos, which is stably induced during normal myeloid differentiation, is not induced upon terminal differentiation of M1 myeloblastic leukemia cells.4 M1 myeloblasts that express physiologic levels of c-Fos mRNA, comparable to what is observed in myeloblast-enriched BM, could not be established.4 Thus, to further dissect the role Fos/Jun transcription complexes play in myeloid cell homeostasis, we have generated M1 cells that conditionally express functional Fos, using a transgene encoding for the chimeric FosER protein, whose activity is dependent on the presence of β-estradiol (βE2) in the culture media.12 M1 myeloblasts were infected with a retroviral vector harboring a chimeric foser transgene under control of the murine stem cell virus (MSCV) promoter.12 M1FosER cell lines were obtained where the expression of c-FosER mRNA (and protein; data not shown) is comparable to c-Fos expression in myeloblast-enriched BM cells (Figure 1). M1FosER clones no. 4 and no. 5 used in the experiments described in this section yielded similar data; data are shown for M1FosER clone no. 4.
Establishment of MlFosER cell lines. M1 cell lines that conditionally express c-Fos (MlFosER) levels comparable to myeloblasts cultured with LUCM for 3 days have been established by infection with a retroviral vector containing the chimeric foser transgene, under control of the MSCV promoter. The murine c-fos cDNA was ligated to the hormone-binding domain of the human estrogen receptor (ER), encoding for a chimeric FosER protein whose activity is dependent on the presence of β-estradiol. RNA expression was analyzed by hybridization of a c-fos probe to Northern blots, using total RNA (10 μg/lane).
Establishment of MlFosER cell lines. M1 cell lines that conditionally express c-Fos (MlFosER) levels comparable to myeloblasts cultured with LUCM for 3 days have been established by infection with a retroviral vector containing the chimeric foser transgene, under control of the MSCV promoter. The murine c-fos cDNA was ligated to the hormone-binding domain of the human estrogen receptor (ER), encoding for a chimeric FosER protein whose activity is dependent on the presence of β-estradiol. RNA expression was analyzed by hybridization of a c-fos probe to Northern blots, using total RNA (10 μg/lane).
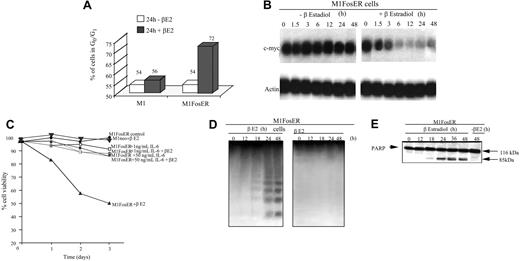
Activation of FosER in M1 myeloblasts was observed to retard cell growth. This was evident by the increase in the percentage of M1 myeloblasts in the G0/G1 phase of the cell cycle and decrease in the level of c-Myc mRNA (Figure 2A-B). Ultimately, M1 myeloblasts expressing functional Fos were observed to undergo apoptotic cell death (Figure 2C-E). As shown, treatment of M1FosER cells with βE2 decreased the percentage of viable cells (Figure 2C). This decrease in M1 viability was accompanied by a characteristic pattern of DNA fragmentation (Figure 2D) and cleavage of PARP (Figure 2E), downstream targets of the caspase cascade, which are activated by a variety of apoptotic stimuli.15 As a control, treatment of M1neo cells with βE2 did not affect cell viability (Figure 2C).
Activation of FosER in MlFosER cells results in growth retardation accompanied by suppression of c-Myc and apoptotic cell death that is abrogated by IL-6. (A) Untreated and estrogen-treated cells were collected 24 hours after initiation of treatment and subjected to FACS analysis as described in “Materials and methods.” Data presented are an average of at least 3 experiments, with each yielding similar results. (B). Cells were collected at the indicated time points after treatment with estrogen, and total RNA was extracted as described in “Materials and methods.” RNA (10 μg/lane) was resolved on a 1% agarose formaldehyde gel and transferred to a Duralon-UV membrane (Stratagene). RNA blots were hybridized to 32P-labeled c-Myc cDNA probe, washed, and subjected to autoradiography for 24 to 48 hours at -80°C as described in “Materials and methods.” (C) Cells were seeded at 0.1 × 106 cells/mL with estrogen, IL-6, or with both estrogen and IL-6, and viability was assessed by trypan blue dye exclusion as described in “Materials and methods.” Each time point represents the average of at least 3 experiments, with a standard deviation of up to 10% (ie, 40% + 4%). (D) For DNA fragmentation analysis, high-molecular weight genomic DNA was extracted from 1 × 107 cells at indicated time points and resolved on a 2% agarose gel using 10 μg/lane as described in “Materials and methods.” (E) Untreated and estrogen-treated cells were collected at indicated time points and lysed with RIPA buffer. Total protein extracts were resolved on a 7.5% SDS-PAGE gel using 50 μg/well, transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore; Billerica, Spain), and probed with anti-PARP antibody (0.5 μg/mL) in 5% milk. Signals were developed by using ECL Western blotting as described in “Materials and methods.”
Activation of FosER in MlFosER cells results in growth retardation accompanied by suppression of c-Myc and apoptotic cell death that is abrogated by IL-6. (A) Untreated and estrogen-treated cells were collected 24 hours after initiation of treatment and subjected to FACS analysis as described in “Materials and methods.” Data presented are an average of at least 3 experiments, with each yielding similar results. (B). Cells were collected at the indicated time points after treatment with estrogen, and total RNA was extracted as described in “Materials and methods.” RNA (10 μg/lane) was resolved on a 1% agarose formaldehyde gel and transferred to a Duralon-UV membrane (Stratagene). RNA blots were hybridized to 32P-labeled c-Myc cDNA probe, washed, and subjected to autoradiography for 24 to 48 hours at -80°C as described in “Materials and methods.” (C) Cells were seeded at 0.1 × 106 cells/mL with estrogen, IL-6, or with both estrogen and IL-6, and viability was assessed by trypan blue dye exclusion as described in “Materials and methods.” Each time point represents the average of at least 3 experiments, with a standard deviation of up to 10% (ie, 40% + 4%). (D) For DNA fragmentation analysis, high-molecular weight genomic DNA was extracted from 1 × 107 cells at indicated time points and resolved on a 2% agarose gel using 10 μg/lane as described in “Materials and methods.” (E) Untreated and estrogen-treated cells were collected at indicated time points and lysed with RIPA buffer. Total protein extracts were resolved on a 7.5% SDS-PAGE gel using 50 μg/well, transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore; Billerica, Spain), and probed with anti-PARP antibody (0.5 μg/mL) in 5% milk. Signals were developed by using ECL Western blotting as described in “Materials and methods.”
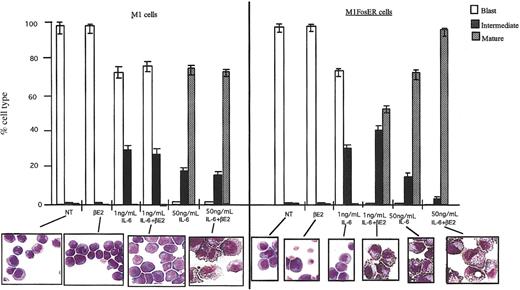
In sharp contrast, in the presence of the differentiation inducer IL-6, Fos-mediated apoptosis of M1 myeloblasts was completely abrogated, as indicated by cell survival and the lack of DNA fragmentation (Figure 2C; data not shown). Moreover, activation of FosER in the presence of IL-6 was observed to increase by 50-fold the sensitivity of M1 cells to be induced for terminal differentiation by IL-6 (Figure 3; 1 ng/mL IL-6 compared to 50 ng/mL). More than 50% of the M1FosER cells differentiated into mature macrophages upon treatment with βE2 and a suboptimal concentration of IL-6 (1 ng/mL), compared to no mature cells when treated with 1 ng/mL IL-6 only. βE2 had no effect on the differentiation of control M1 cells treated with 1 ng/mL IL-6. There was an increase in the percentage of mature cells and decrease in intermediate cells in M1FosER cells treated with βE2 + optimal IL-6 (50 ng/mL) compared to M1FosER cells treated with 50 ng/mL IL-6 only or M1 treated with IL-6 ± βE2.
Activation of FosER increases the propensity of M1 cells to be induced for differentiation by IL-6. Cells were treated as indicated, and after 3 days the morphology of at least 300 cells on May-Grunwald-Giemsa-stained cytospin smears was determined as detailed in “Materials and methods” (photomicrographs; original magnification, × 400). The proportion of immature blast cells, cells at intermediate monocyte stage, and mature macrophages was calculated. NT indicates not treated. Data presented are an average (± SD) of at least 3 experiments.
Activation of FosER increases the propensity of M1 cells to be induced for differentiation by IL-6. Cells were treated as indicated, and after 3 days the morphology of at least 300 cells on May-Grunwald-Giemsa-stained cytospin smears was determined as detailed in “Materials and methods” (photomicrographs; original magnification, × 400). The proportion of immature blast cells, cells at intermediate monocyte stage, and mature macrophages was calculated. NT indicates not treated. Data presented are an average (± SD) of at least 3 experiments.
Taken together, these observations indicate that in the absence of differentiation inducer (ie, IL-6), Fos expression promoted apoptosis of M1 myeloblasts. In the presence of differentiation inducer, Fos-mediated apoptosis was abrogated and Fos promoted myeloid differentiation.
Fos-induced apoptosis is mediated via cytochrome c release and caspase-9 activation
The data presented above indicated that Fos expression in M1 myeloblasts results in apoptosis. Thus, it was of interest to determine which of the known major apoptotic pathways is activated by Fos.
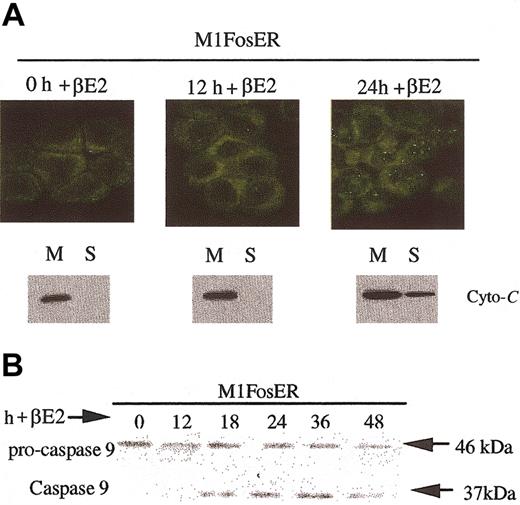
One archetypal apoptotic pathway induced by serum deprivation or environmental stress agents entails release of cytochrome c from mitochondria into the cytosol, where it associates with Apaf-1, which in turn results in cleavage and activation of procaspase-9.16 To determine if Fos uses this apoptotic pathway, cytochrome c localization was examined following activation of FosER. Cytospin smears of M1FosER cells, untreated and treated with βE2, were tested for intracellular localization of cytochrome c using indirect immunofluorescence with anti-murine cytochrome c primary antibody and FITC-conjugated secondary antibody. In addition to the data from the immunohistochemistry, Western blot analysis revealed that by 24 hours after activation of Fos with βE2, cytochrome c is released from mitochondria into the cytosol (Figure 4A). Consistent with the redistribution of cytochrome c, procaspase-9 was cleaved from the inactive 48-kDa protein species to the active 37-kDa form (Figure 4B). The protein band corresponding to the 37-kDa caspase-9 was first detected following 18 hours of treatment of M1FosER cells with estrogen, which roughly corresponds to the point in time when cytochrome c release was first detected (Figure 4; data not shown).
Fos-mediated apoptosis is mediated via the cytochrome c-caspase-9 pathway. (A) Analysis of cytochrome c release. Cytospin smears of untreated and βE2-treated MlFosER cells were analyzed for the redistribution of cytochrome c using immunohistochemistry with an anti-cytochrome c FITC-conjugated antibody (Stratagene) at 1 μg/mL (photomicrographs, original magnification × 400). In addition, at the indicated time points, 2 × 107 cells were collected and lysed. After pelleting of unlysed cells and nuclei, the mitochondrial fraction (M) and cytosolic supernatant fraction (S) were separated by centrifugation at 13 000g. For Western blot analysis, 10 μg mitochondrial and 20 μg supernatant fraction were resolved on a 15% SDS-PAGE gel, and an anti-cytochrome c antibody (murine; Pharmingen) was used to detect cytochrome c. (B) Analysis of procaspase-9 cleavage. Protein lysates were collected from cells at designated time points after treatment with estrogen and resolved on a 10% SDS-PAGE gel using 50 μg/well. Gels were transferred to PVDF membranes (Millipore) and probed with primary antibody recognizing the pro and cleaved form of caspase-9 (4 μg/mL; Stressgen). Signal was detected using an HRP-linked secondary antibody (Santa Cruz Biotechnology) and the Pierce detection system.
Fos-mediated apoptosis is mediated via the cytochrome c-caspase-9 pathway. (A) Analysis of cytochrome c release. Cytospin smears of untreated and βE2-treated MlFosER cells were analyzed for the redistribution of cytochrome c using immunohistochemistry with an anti-cytochrome c FITC-conjugated antibody (Stratagene) at 1 μg/mL (photomicrographs, original magnification × 400). In addition, at the indicated time points, 2 × 107 cells were collected and lysed. After pelleting of unlysed cells and nuclei, the mitochondrial fraction (M) and cytosolic supernatant fraction (S) were separated by centrifugation at 13 000g. For Western blot analysis, 10 μg mitochondrial and 20 μg supernatant fraction were resolved on a 15% SDS-PAGE gel, and an anti-cytochrome c antibody (murine; Pharmingen) was used to detect cytochrome c. (B) Analysis of procaspase-9 cleavage. Protein lysates were collected from cells at designated time points after treatment with estrogen and resolved on a 10% SDS-PAGE gel using 50 μg/well. Gels were transferred to PVDF membranes (Millipore) and probed with primary antibody recognizing the pro and cleaved form of caspase-9 (4 μg/mL; Stressgen). Signal was detected using an HRP-linked secondary antibody (Santa Cruz Biotechnology) and the Pierce detection system.
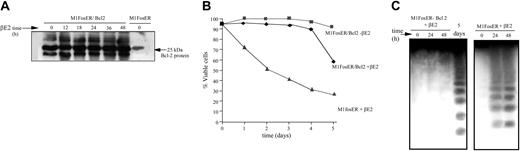
Bcl-2 is known to inhibit apoptosis by abrogating cytochrome c release from mitochondria.17,18 Thus, to further establish the role of cytochrome c in Fos-mediated apoptosis, M1FosER cells were genetically manipulated to express ectopic Bcl-2 (Figure 5A). Ectopic Bcl-2 significantly delayed the onset of c-Fos-induced apoptosis in M1 cells, as evidenced by the increase in cell viability and the delay in DNA fragmentation (Figure 5B-C).
Ectopic Bc1-2 protects MlFosER cells from Fos-mediated apoptosis. (A) MlFosER/Bcl-2 cell lines were established by infecting MlFosER cells with MSCV-Bcl-2/puro retroviral vectors followed by selection of cells in G418 and puromycin as described in “Materials and methods.” At designated time points after estrogen addition MlFosER/Bcl-2 cells (clone 2) were collected, lysed in RIPA buffer, resolved on a 12.5% SDS-PAGE gel, and analyzed for Bc1-2 expression by Western blotting. After blocking in 5% milk, blots were incubated with 1 μg/mL anti-Bcl-2 primary antibody (Santa Cruz Biotechnology; 1:5000), and the signal was detected using the ECL system. (B). Indicated cells were seeded at 0.1 × 106 cells/mL in the presence and absence of estrogen, and viability was assessed by trypan blue dye exclusion. Percent viability at each time point is an average of at least 3 experiments, with a standard deviation of up to 12% (ie, 30% ± 3.6%). (C) For analysis of DNA ladders, high-molecular weight DNA was extracted from 1 × 107 cells at indicated time points after treatment with estrogen (2 mM), and 10 μg/lane was resolved on a 2% agarose gel as described in “Materials and methods.”
Ectopic Bc1-2 protects MlFosER cells from Fos-mediated apoptosis. (A) MlFosER/Bcl-2 cell lines were established by infecting MlFosER cells with MSCV-Bcl-2/puro retroviral vectors followed by selection of cells in G418 and puromycin as described in “Materials and methods.” At designated time points after estrogen addition MlFosER/Bcl-2 cells (clone 2) were collected, lysed in RIPA buffer, resolved on a 12.5% SDS-PAGE gel, and analyzed for Bc1-2 expression by Western blotting. After blocking in 5% milk, blots were incubated with 1 μg/mL anti-Bcl-2 primary antibody (Santa Cruz Biotechnology; 1:5000), and the signal was detected using the ECL system. (B). Indicated cells were seeded at 0.1 × 106 cells/mL in the presence and absence of estrogen, and viability was assessed by trypan blue dye exclusion. Percent viability at each time point is an average of at least 3 experiments, with a standard deviation of up to 12% (ie, 30% ± 3.6%). (C) For analysis of DNA ladders, high-molecular weight DNA was extracted from 1 × 107 cells at indicated time points after treatment with estrogen (2 mM), and 10 μg/lane was resolved on a 2% agarose gel as described in “Materials and methods.”
Another major apoptotic pathway in mammalian cells entails signaling via death receptors such as Fas/CD95.19 To determine if death receptor signaling contributes to Fos-mediated apoptosis in M1 myeloblasts, flow cytometry was used to determine if the Fas receptor is expressed in M1FosER cells, untreated or following activation of Fos. It was previously reported that M1 cells do not express Fas receptors20 ; similarly, M1FosER cells, untreated or treated with βE2, do not express Fas receptors (data not shown), indicating that the Fas/CD95 pathway is not involved in c-Fos-mediated apoptosis. Caspase-8 is the initiator caspase not only in the Fas/CD95 pathway but also in other related death receptor (ie, TNFa/TNFR) pathways.15,19 To confirm that a death receptor pathway is not involved in c-Fos-mediated apoptosis, we assessed if procaspase-8 is cleaved and activated following activation of the FosER chimera in M1FosER cells. Procaspase-8 was not cleaved following βE2 treatment (data not shown). These observations indicated that neither the Fas/CD95 nor other death receptor pathways are involved in Fos-mediated apoptosis.
Taken together, these findings establish that cytochrome c release and activation of initiator procaspase-9 mediates Fos-induced apoptosis of M1 myeloblasts.
Fos partially abrogates the block imparted by deregulated c-Myc on myeloid differentiation and increases the sensitivity of deregulated M1myc cells to be induced for differentiation
Previously, we have shown that deregulated c-Myc in M1 cells6 (and normal BM20 ) blocks terminal differentiation. Thus, it was in our interest to determine the effect of Fos, which promotes myeloid differentiation, on the c-Myc block in terminal differentiation. Inappropriate expression of c-Myc under conditions that inhibit growth and down-regulate endogenous c-Myc expression is known to result in programmed cell death or acceleration of an ongoing apoptotic response.5 Fos expression in myeloid progenitors resulted in cell cycle arrest, accompanied by rapid down-regulation of c-Myc and apoptosis. It was thus also of interest to determine what effect deregulated c-Myc would have on Fos-mediated apoptosis of the M1 myeloblasts, and if having an effect, to ascertain if the same or a different apoptotic pathway is involved.
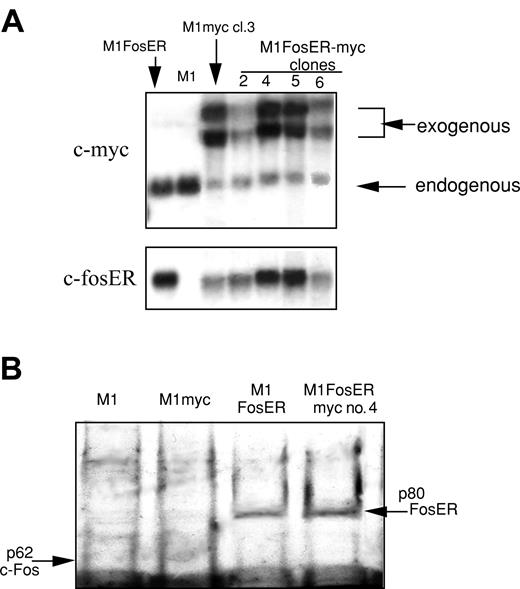
To this end, M1 and M1FosER clone 4 were transduced with an MSCV retroviral expression vector, with c-Myc under control of the MSCV retroviral promoter. M1FosER-myc clones 4 and 5 expressed ectopic c-Myc at levels comparable to the level of ectopic c-Myc in M1myc clone 3, and c-fosER at levels comparable to its level in parental M1FosER cells (Figure 6). Both clones used in subsequent experiments yielded similar data. Results obtained with M1FosER-myc clone 4 are presented.
Establishment of MlFosER cell lines that express deregulated c-Myc (MlFosER-myc). (A) MlFosER-myc and M1myc cells were established by infecting MlFosER and M1 cells, respectively, with the MSCV-myc/puro retroviral vector, followed by selection of cells in G418 + puro or puro as described in “Materials and methods.” Indicated clones were tested for c-Myc and c-FosER RNA expression using RNA blots. RNA (10 μg/lane) was resolved on a 1% agarose formaldehyde gel and transferred to nylon membranes (Duralon) for Northern blot analysis. Blots were hybridized with 32P-labeled c-Myc probe, then stripped and rehybridized with a c-fos probe (to detect c-fosER RNA; none of the clones expressed endogenous c-fos) as described in “Materials and methods.” (B) Western blot to test for FosER expression. Protein lysates were prepared from indicated cell lines, and 50 μg/well was run on a 10% SDS-PAGE gel. The gels were transferred onto a PVDF membrane (Millipore) and probed with a primary antibody specific for c-Fos (Santa Cruz Biotechnology). Detection of signal was done as described in “Materials and methods.”
Establishment of MlFosER cell lines that express deregulated c-Myc (MlFosER-myc). (A) MlFosER-myc and M1myc cells were established by infecting MlFosER and M1 cells, respectively, with the MSCV-myc/puro retroviral vector, followed by selection of cells in G418 + puro or puro as described in “Materials and methods.” Indicated clones were tested for c-Myc and c-FosER RNA expression using RNA blots. RNA (10 μg/lane) was resolved on a 1% agarose formaldehyde gel and transferred to nylon membranes (Duralon) for Northern blot analysis. Blots were hybridized with 32P-labeled c-Myc probe, then stripped and rehybridized with a c-fos probe (to detect c-fosER RNA; none of the clones expressed endogenous c-fos) as described in “Materials and methods.” (B) Western blot to test for FosER expression. Protein lysates were prepared from indicated cell lines, and 50 μg/well was run on a 10% SDS-PAGE gel. The gels were transferred onto a PVDF membrane (Millipore) and probed with a primary antibody specific for c-Fos (Santa Cruz Biotechnology). Detection of signal was done as described in “Materials and methods.”
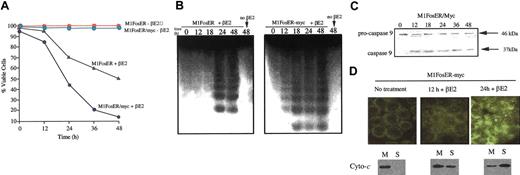
Fos activation in the presence of deregulated c-Myc was observed to accelerate apoptosis, determined by loss of viability, DNA cleavage (Figure 7A-B), activation of caspase-9, and release of cytochrome c from the mitochondria, compared with M1FosER cells (Figure 7C-D compared with Figure 4). No C95 receptor expression or caspase-8 activation was detected (data not shown). These data indicate that deregulated c-Myc interfaces with and amplifies the Fos apoptotic pathway in M1 myeloblasts.
Deregulated c-Myc enhances the c-Fos apoptotic pathway. (A) Cell viability. Cells were seeded at 0.1 × 106 cells/mL in the absence or presence of estrogen. At each time point cell viability was determined by trypan blue dye exclusion as described in “Materials and methods.” Data shown are representative of at least 3 experiments that were conducted using MlFosER-myc clones 4 and 5. (B) DNA fragmentation. At least 107 cells were harvested at indicated times after treatment with estrogen, and high-molecular-weight DNA was extracted and resolved on a 2% agarose gel using 10 μg/lane, as described in “Materials and methods.” Similar data were obtained using MlFosER-myc clones 4 and 5; results are shown for clone 4. (C) Analysis of procaspase-9 cleavage. Protein lysates were collected from cells at designated time points after treatment with estrogen and resolved on a 10% SDS-PAGE gel using 50 μg/well. Gels were transferred to PVDF membranes (Millipore) and probed with primary antibody recognizing both the pro and the cleaved forms of caspase-9 (4 μg/mL; Stressgen). Signal was detected using an HRP-linked secondary antibody (Santa Cruz Biotechnology) and the Pierce detection system. (D) Analysis of cytochrome c release. Cytospin smears of untreated and βE2-treated MlFosER cells were analyzed for the redistribution of cytochrome c using immunohistochemistry with an anti-cytochrome c FITC-conjugated antibody (Stratagene) at 1 μg/mL (photomicrographs, original magnification × 400). In addition, at the indicated time points, 2 × 107 cells were collected and lysed. After pelleting of unlysed cells and nuclei, the mitochondrial fraction (M) and cytosolic supernatant fraction (S) were separated by centrifugation at 13 000g. For Western blot analysis, 10 μg mitochondrial and 20 μg supernatant fraction were resolved on a 15% SDS-PAGE gel, and an anti-cytochrome c antibody (murine; Pharmingen) was used to detect cytochrome c.
Deregulated c-Myc enhances the c-Fos apoptotic pathway. (A) Cell viability. Cells were seeded at 0.1 × 106 cells/mL in the absence or presence of estrogen. At each time point cell viability was determined by trypan blue dye exclusion as described in “Materials and methods.” Data shown are representative of at least 3 experiments that were conducted using MlFosER-myc clones 4 and 5. (B) DNA fragmentation. At least 107 cells were harvested at indicated times after treatment with estrogen, and high-molecular-weight DNA was extracted and resolved on a 2% agarose gel using 10 μg/lane, as described in “Materials and methods.” Similar data were obtained using MlFosER-myc clones 4 and 5; results are shown for clone 4. (C) Analysis of procaspase-9 cleavage. Protein lysates were collected from cells at designated time points after treatment with estrogen and resolved on a 10% SDS-PAGE gel using 50 μg/well. Gels were transferred to PVDF membranes (Millipore) and probed with primary antibody recognizing both the pro and the cleaved forms of caspase-9 (4 μg/mL; Stressgen). Signal was detected using an HRP-linked secondary antibody (Santa Cruz Biotechnology) and the Pierce detection system. (D) Analysis of cytochrome c release. Cytospin smears of untreated and βE2-treated MlFosER cells were analyzed for the redistribution of cytochrome c using immunohistochemistry with an anti-cytochrome c FITC-conjugated antibody (Stratagene) at 1 μg/mL (photomicrographs, original magnification × 400). In addition, at the indicated time points, 2 × 107 cells were collected and lysed. After pelleting of unlysed cells and nuclei, the mitochondrial fraction (M) and cytosolic supernatant fraction (S) were separated by centrifugation at 13 000g. For Western blot analysis, 10 μg mitochondrial and 20 μg supernatant fraction were resolved on a 15% SDS-PAGE gel, and an anti-cytochrome c antibody (murine; Pharmingen) was used to detect cytochrome c.
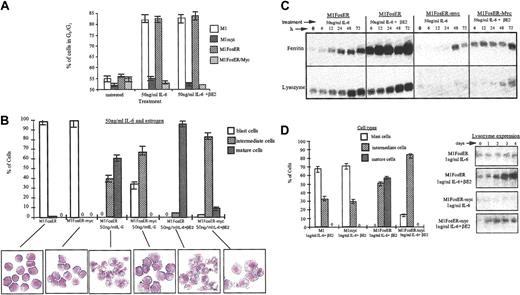
To assess what effect Fos has on the c-Myc block in terminal differentiation, M1, M1FosER, M1myc, and M1FosER-myc cells were treated with an optimal concentration of IL-6 (50 ng/mL6) with or without βE2, and after 3 days the cells were analyzed for cell cycle distribution and morphologic characteristics using FACS analysis and May-Grunwald-stained cytospin smears, respectively. As shown in Figure 8A, about 80% of M1 and M1 FosER cells, treated with either IL-6 only or IL-6 + βE2, arrested in the G0/G1 phase of the cell cycle. In contrast, M1myc and M1FosER-myc cells continued to cycle, regardless of treatment, with fewer than 55% of the cells in G1/G0 (Figure 8A). Nevertheless, a significantly higher percentage of the M1FosER-myc cells treated with IL-6 + βE2 differentiated into intermediate stage macrophages, compared to M1FosER-myc cells treated with IL-6 only (Figure 8B) or M1myc cells treated with IL6 + βE2 (data not shown). Moreover, reproducibly some (6%-8%) of the M1FosER-myc cells treated with IL-6 + βE2 displayed mature macrophage characteristics (Figure 8B). Consistent with these observations, expression of the myeloid differentiation markers ferritin and lysozyme was significantly higher in the M1FosER-myc cells treated with IL-6 + βE2, compared to the same cells treated with IL-6 only (Figure 8C) or M1myc cells treated with IL-6, with or without βE2 (data not shown). Furthermore, M1FosER-myc cells cultured with βE2 were more responsive to a suboptimal dose of IL-6 (1 ng/mL) compared to M1FosER-myc cells cultured without βE2 or M1myc cells cultured with IL-6 ± βE2 (data not shown), as evident by both the percentage of cells undergoing intermediate-stage differentiation and by lysozyme expression (Figure 8D-E).
Fos expression in M1 cells partially abrogates the c-Myc block in myeloid differentiation and increases propensity of the cells to be induced for differentiation by IL-6. (A) Cell cycle analysis. Indicated cell types, either untreated or 3 days after treatment, were collected and subjected to FACS analysis as described in “Materials and methods.” Data shown are representative of at least 4 experiments. (B) Morphologic characteristics of myeloid differentiation. May-Grunwald-Giemsa-stained cytospin smears of indicated cell types, untreated or treated for 3 days, were assessed for morphologic markers associated with various stages of differentiation. At least 300 cells were scored to obtain the proportion of immature blast cells, cells at intermediate monocyte stages of differentiation, and mature macrophages. Data are an average (± SD) of at least 3 experiments that yielded similar results. Representative photomicrographs (original magnification, × 400) of cytospin smears are shown. (C) Expression of the macrophage differentiation markers ferritin and lysozyme. RNA was extracted from cells collected at indicated times following treatments. RNA was resolved on a 1% agarose formaldehyde gel and transferred to nylon membranes (Duralon) for Northern blot analysis. Blots were hybridized with a 32P-labeled ferritin probe, then stripped and reprobed with a 32P-labeled lysozyme probe as described in “Materials and methods.” Hybridization to an actin probe was used to ensure equal loading of RNA, as shown for Figure 2. (D-E) Increased propensity of MlFosER-myc cells to be induced for differentiation by suboptimal (1 ng/mL) IL-6. (D) Cell morphology following treatment with estrogen and suboptimal dose (ng/mL) of IL-6. May-Grunwald-Giemsa-stained cytospin smears of cells treated for 3 days with 1 ng/mL IL-6 plus estrogen were analyzed for morphologic differentiation characteristics. At least 300 cells were scored. (E) Lysozyme expression following treatment with estrogen plus 1 ng/mL IL-6. RNA blots (10 μg RNA/lane) were prepared with total RNA obtained at indicated time points after treatment and hybridized with a lysozyme cDNA probe labeled with 32P.
Fos expression in M1 cells partially abrogates the c-Myc block in myeloid differentiation and increases propensity of the cells to be induced for differentiation by IL-6. (A) Cell cycle analysis. Indicated cell types, either untreated or 3 days after treatment, were collected and subjected to FACS analysis as described in “Materials and methods.” Data shown are representative of at least 4 experiments. (B) Morphologic characteristics of myeloid differentiation. May-Grunwald-Giemsa-stained cytospin smears of indicated cell types, untreated or treated for 3 days, were assessed for morphologic markers associated with various stages of differentiation. At least 300 cells were scored to obtain the proportion of immature blast cells, cells at intermediate monocyte stages of differentiation, and mature macrophages. Data are an average (± SD) of at least 3 experiments that yielded similar results. Representative photomicrographs (original magnification, × 400) of cytospin smears are shown. (C) Expression of the macrophage differentiation markers ferritin and lysozyme. RNA was extracted from cells collected at indicated times following treatments. RNA was resolved on a 1% agarose formaldehyde gel and transferred to nylon membranes (Duralon) for Northern blot analysis. Blots were hybridized with a 32P-labeled ferritin probe, then stripped and reprobed with a 32P-labeled lysozyme probe as described in “Materials and methods.” Hybridization to an actin probe was used to ensure equal loading of RNA, as shown for Figure 2. (D-E) Increased propensity of MlFosER-myc cells to be induced for differentiation by suboptimal (1 ng/mL) IL-6. (D) Cell morphology following treatment with estrogen and suboptimal dose (ng/mL) of IL-6. May-Grunwald-Giemsa-stained cytospin smears of cells treated for 3 days with 1 ng/mL IL-6 plus estrogen were analyzed for morphologic differentiation characteristics. At least 300 cells were scored. (E) Lysozyme expression following treatment with estrogen plus 1 ng/mL IL-6. RNA blots (10 μg RNA/lane) were prepared with total RNA obtained at indicated time points after treatment and hybridized with a lysozyme cDNA probe labeled with 32P.
Collectively, these data indicate that restoration of Fos expression in M1myc cells resulted in partial abrogation of the c-Myc block of myeloid differentiation and increased the sensitivity of the M1 cells expressing deregulated c-Myc to respond to IL-6.
Effect of restoring Fos expression in M1myc cells on IL-6-mediated suppression of M1 leukemogenicity
It has been shown that intravenous injection of untreated M1 cells into nude CD-1 mice results in the rapid development of a leukemic phenotype that is similar to acute myelogenous leukemia (AML) in human patients.2,4,21 All of the mice die as a direct result of the unregulated proliferation of M1 cells. If M1 cells were treated with IL-6 and induced for terminal differentiation prior to injection into the mice, then there was a complete absence of leukemia. Thus, IL-6 is able to suppress the leukemogenic potential of Ml cells in vivo. To better understand the relationship between blocks in differentiation and leukemogenicity, we examined the effects of deregulated c-Myc in M1 cells on the ability of IL-6 to suppress the leukemic phenotype in M1myc cells in nude mice, as well as the consequence of restoring c-Fos expression in these cells.
To this end, M1fos14 cells4 were transduced with the MSCV retroviral expression vector with c-Myc under control of the MSCV retroviral promoter. M1fos14-myc clones 2 and 7 expressed ectopic c-Myc at levels comparable to the level of ectopic c-Myc in M1myc clone 3, and c-Fos levels as shown before for M1fos14 cells4 (data not shown). Both clones used in subsequent experiments yielded similar data. Results obtained with M1myc7-fos14 are presented.
Ml, Mlmyc, M1fos14, and M1myc7-fos14 cells, either untreated or 5 days after treatment with IL-6, were intravenously injected into CD-I nu/nu mice. Whereas IL-6-treated Ml cells lost their leukemogenic potential, IL-6-treated M1myc cells retained their leukemogenicity, with 60% of the mice dead by 6 weeks and 100% dead by 10 weeks (Table 1). However, the leukemic phenotype of the IL-6-treated M1myc7-fos14 cells was significantly less aggressive compared to IL-6-treated Mlmyc cells. The evidence for this is that only 30% of the mice injected with IL-6-treated M1myc7-fos14 cells died of leukemia by 6 weeks and 50% by 10 weeks, compared to 60% mortality by 6 weeks and 100% mortality by 10 weeks in mice injected with IL-6-treated Mlmyc cells (Table 1, lines 7 and 9). Taken together, these data indicate that loss of c-Fos expression aggravates the leukemic phenotype of Mlmyc cells (see “Discussion”).
Discussion
Previously, we have shown that enforced expression of very low levels of c-Fos (compared to what has been observed in differentiating BM myeloblasts) in M1 myeloid progenitors resulted in an increased propensity of the cells to differentiate in culture. Blocking Fos/Jun expression, using c-fos antisense oligomers, was observed to impair myeloid differentiation of both normal and M1 myeloblasts, implicating Fos/Jun transcription complexes as positive modulators of hematopoietic differentiation. In the course of that work it became evident that M1 cells expressing c-Fos at levels comparable to what was observed in differentiating BM-derived myeloblasts could not be established.4 These observations raised the possibility that physiologic levels of c-Fos in hematopoietic progenitor cells may be antagonistic to cell growth and/or cell survival. Using M1 myeloblasts that express a βE2 conditional FosER chimera, we now have shown that Fos, in addition to promoting myeloid differentiation, also plays a role in modulating survival of myeloid cells.
Fos expression in M1 myeloid progenitors resulted in apoptotic cell death that was abrogated by the differentiation-inducing cytokine IL-6. To the best of our knowledge, this is the first instance where Fos has been documented to modulate myeloid progenitor cell survival dependent on the availability of differentiation inducer. These observations suggest that Fos expression following physiologic stress, such as low levels of differentiation/survival factors and/or following exposure of the organism to genotoxic stress,3,20 is a safeguard that keeps myeloid cell homeostasis in check to prevent fixation of mutations that may promote leukemogenesis. While our work was in progress, AP-1 had been implicated in the regulation of proliferation and survival of erythroid cells as well, suggesting that different AP-1 factors may play distinct roles in triggering apoptosis (JunB) verses protection from apoptosis (c-Jun).22
It was shown that cytochrome c release from mitochondria and activation of caspase-9 mediate, at least in part, Fos-induced apoptosis of M1 myeloblasts. It is noteworthy that this apoptotic pathway is the archetypal path that mediates physiologic and environmental stress-induced apoptosis, including apoptosis induced by survival factor withdrawal or by a variety of genotoxic stress agents including ionizing radiation-, UV-, and DNA-damaging chemicals.3,21 It has been suggested that p53 is an important target gene for AP-1 functions. However, since M1 myeloblasts are null for p53 expression, it is clear that Fos-mediated apoptosis in M1 cells is p53 independent.
We also have shown that in presence of the differentiation-inducing cytokine IL-6, Fos-mediated apoptosis was abrogated and instead Fos promoted myeloid differentiation and significantly increased the sensitivity of M1 cells to be induced for differentiation. The molecular nature of the mechanism by which IL-6 protects M1 cells from Fos-mediated apoptosis is currently unknown. Also not clear is the molecular nature of the mechanism that operates to switch the function of Fos from apoptosis to differentiation. Providing answers to these interesting questions is the subject for further investigations. Fos and Jun proteins heterodimerize and homodimerize to form functional Ap-1 transcription complexes. It has been established that Fos does not form homodimers. Thus, it is likely that Fos/Jun heterodimeric transcription complexes function both in apoptosis and myeloid differentiation. Determining the molecular nature of Fos/Jun transcription complexes that facilitate these activities is under way.
Since we have started our studies, other evidence has accumulated that implicates AP-1(Fos/Jun) transcription factors in the development of hematopoietic precursor cells along most, if not all, of the hematopoietic cell lineages, including the monocyte/macrophage, granulocyte, megakaryocyte, mastocyte, erythroid, and osteoclast lineages.3,23 Adding to these observations, recently it was shown that transgenic mice that lack JunB expression in the myeloid lineage develop a transplantable myeloproliferative disease, eventually progressing to blast crisis resembling human chronic myeloid leukemia. This suggests that JunB is a key transcriptional regulator of myelopoiesis and a potential tumor suppressor gene. It is possible, therefore, that Fos/JunB heterodimeric Ap-1 complexes are involved in mediating the differentiation promoting function of Fos. Since c-Fos-deficient mice do not display the phenotype observed for JunB-null mice, the possibility is raised that other AP-1 homodimeric and/or heterodimeric transcription complexes may compensate for the lack of Fos function.
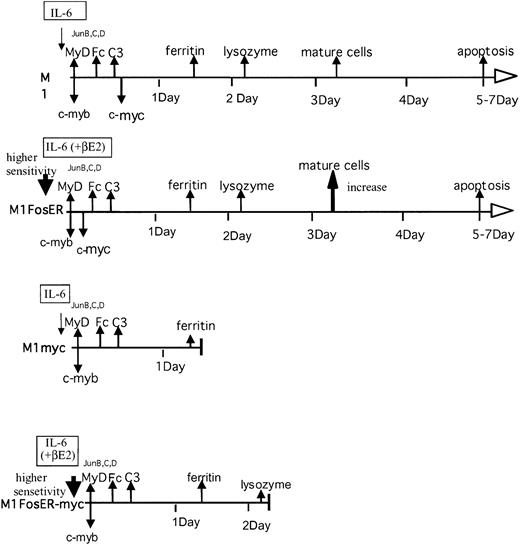
Previously, we have shown that deregulated c-Myc blocked M1 myeloid differentiation, where the cells kept proliferating at an intermediate differentiation stage. Since, unlike normal hematopoietic cells M1 cells are devoid of Fos expression, it would be interesting to know what effect Fos expression has on the block in differentiation due to deregulated c-Myc. We have shown that c-Fos partially abrogated the c-Myc block in M1 terminal differentiation and increased the sensitivity of M1myc cells to IL-6 (Figure 9).
Effect of c-Fos on the c-myc-mediated block in myeloid differentiation.
Inappropriate expression of c-Myc under conditions that inhibit growth and down-regulate endogenous c-Myc expression is known to result in programmed cell death or acceleration of an ongoing apoptotic response.5 Taking advantage of M1FosER cells that have been genetically altered to express a deregulated c-Myc transgene, we have shown that in the absence of differentiation inducer, deregulated c-Myc enhanced Fos-mediated apoptosis of M1 myeloblasts, including cytochrome c release and caspase-9 activation. The molecular nature of the mechanism by which deregulated c-Myc interfaces with and amplifies the Fos apoptotic pathway remains to be explored.
The development of leukemia is a multistage process, where mutations that alter normal expression and/or function of genes that participate in the regulation of blood cell homeostasis, including proliferation, terminal differentiation, and survival of hematopoietic cells cooperate in leukemia progression.24 Understanding of functional interactions between genes that regulate normal blood cell homeostasis and how their altered expression may contribute to leukemogenesis is, therefore, of paramount importance for rational drug design, as these proteins represent excellent targets for chemotherapeutic drug intervention. The data presented in this work increase this understanding, predicting that genetic lesions that abolish Fos expression may cooperate with deregulated c-Myc in leukemogenesis. Substantiation of this notion is provided by data that show that restoring Fos expression in M1myc cells curtailed the leukemic phenotype. It is possible that the status of c-Myc and Fos/Jun may effect the response of malignant cells to anticancer agents. Additional experimentation should enable us to correlate the in vitro differentiation potential of the genetically altered cell lines described in this study with their response to anticancer agents. Consistent with this notion are data showing that expression of c-Fos correlates with leukemogenicity and cytokine responsiveness in myeloid and lymphoid leukemias in humans.25-27 Thus, better understanding of the molecular nature of the interaction between c-Fos and c-Myc may have prognostic and predictive values for treatment of certain subtypes of myeloid leukemias involving alterations in the expression and/or function of these genes.
Prepublished online as Blood First Edition Paper, February 24, 2004; DOI 10.1182/blood-2002-09-2704.
Supported by National Institutes of Health grants 1 RO1 CA81168 (B.H.), 1 RO1 RO1 CA 59774 (D.A.L.), and R24 CA88261-03 (shared Resources for Cancer Research).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.