Abstract
CCAAT enhancer binding protein epsilon (C/EBPϵ) is a myeloid specific transcription factor that is essential for terminal granulocytic differentiation. Retinoblastoma (Rb) and E2F1 are critical cell cycle regulators that also have been implicated in several differentiation systems. Here, we demonstrate that C/EBPϵ interacts with Rb and E2F1 during granulocytic differentiation in NB4 and U937 human myeloid cells and in 32Dcl3 murine myeloid precursor cells. The interaction between C/EBPϵ and Rb enhances C/EBPϵ-mediated transcription of myeloid specific genes both in reporter assays and endogenously. The C/EBPϵ-E2F1 interaction results in repression of E2F1-mediated transcriptional activity. Finally, overexpression of C/EBPϵ in human myeloid cells leads to down-regulation of c-Myc. We propose that the interactions between C/EBPϵ, a tissue-specific transcription factor, and the broad-spectrum proteins, Rb and E2F1, are important in C/EBPϵ-induced terminal granulocytic differentiation.
Introduction
In hematopoiesis, stem cells generate mature blood cells of specific lineages, in a process that is tightly regulated by transcription factors. CCAAT enhancer binding protein epsilon (C/EBPϵ) is a member of the C/EBP family of transcription factors that share a highly conserved basic region and a leucine zipper domain (bZIP).1 C/EBPϵ is expressed almost exclusively in myeloid cells and activates the transcription of a subset of myeloid specific genes.2,3 Mice and humans with genetic deletion of C/EBPϵ have a block in granulocytic differentiation and often have either severe or fatal chronic bacterial infections.4-6 Likewise, induction of neutrophil differentiation in promyelocytic leukemia lines is associated with C/EBPϵ expression, and forced expression of C/EBPϵ in these cells can induce granulocytic differentiation.7-9 These findings indicate that C/EBPϵ is a critical regulator of terminal granulopoiesis.
Recent studies have emphasized the important role that protein-protein interactions play in the ability of C/EBPs to regulate differentiation and cell growth.10 Interestingly, 2 key regulators of the cell cycle, retinoblastoma (Rb) and E2F1, have been linked to differentiation and growth suppression events that are mediated by members of the C/EBP family. The tumor suppressor gene Rb has a well-established role in the regulation of cell cycle progression.11 In G1, hypophosphorylated Rb sequesters the E2F transcription factors whose target genes are necessary for the G1/S transition. In recent years, evidence has been accumulating that Rb also is involved in cellular differentiation.12,13 Transgenic mice with inactivated Rb show defective differentiation of hematopoietic and neuronal tissues and die after 14 to 15 days of gestation.14-16 In addition, hypophosphorylation of Rb correlates with the differentiation of normal and leukemic hematopoietic cells in vitro.17,18 Rb binds and activates C/EBPβ, and this interaction has been found to be involved in differentiation of adipocytes and monocytes.19,20 C/EBPα recently was shown to inhibit cellular growth and induce adipogenesis and granulopoiesis through direct repression of E2F-mediated transcription.21-23
The use of knockout models and conditionally expressing cell lines have demonstrated that C/EBPϵ plays a key role in terminal granulocytic differentiation. However, the molecular mechanisms involved in this process are not completely understood. In this report, we demonstrate that C/EBPϵ interacts with Rb during granulocytic differentiation and that this interaction activates C/EBPϵ transcription of myeloid specific genes. We also show that C/EBPϵ interacts in vivo with E2F1 and represses E2F1 transcriptional activity. Finally, we show that overexpression of C/EBPϵ leads to down-regulation of c-Myc in myeloid cells.
Materials and methods
Cell culture and transfections
COS-1 (monkey kidney), NB4 (acute promyelocytic leukemia), U937 (myelomonoblastic), Saos-2 (osteosarcoma), and NIH 3T3 (murine fibroblast) cell lines were obtained from the American Type Culture Collection (Manassas, VA) and grown in the recommended medium and conditions. The murine myeloid precursor cell line 32Dcl3 was a kind gift from A. D. Friedman (Johns Hopkins University, Baltimore, MD). The 32Dcl3 cells were maintained in Iscove modified Dulbecco medium (IMDM) medium supplemented with 10% fetal calf serum (FCS) and 10% WEHI-conditioned medium as a source of interleukin-3 (IL-3). To induce C/EBPϵ expression in NB4 and U937 cells, the cells were treated with all-trans retinoic acid (ATRA, 1 × 10–7 M). For induction of C/EBPϵ expression in 32Dcl3 cells, the WEHI-conditioned medium was removed, and the cells were treated with granulocyte colony-stimulating factor (G-CSF, 30 ng/mL). U937 human myeloid stable cell lines (U937pMTϵ and U937pMT) were previously described.8 The 32Dϵ stable cell line was generated by infecting 32Dcl3 with a modified pBabe-C/EBPϵ expression vector followed by selection in puromycin (2 μg/mL, E. Williamson and H.P.K., unpublished data, June 2003). To generate the 3T3pMTϵ and the 3T3pMTα stable cell lines, NIH 3T3 cells were transfected with zinc-inducible C/EBPϵ8 and C/EBPα24 expression vectors using the GenePORTER transfection Reagent (GTS Inc, Greenbelt, MD) followed by selection in G418 (700 μg/mL). C/EBPϵ and C/EBPα expression were induced by adding ZnSO4 (100 μM) to the medium. Transient transfections were performed with various expression vectors (C/EBPϵ,24 Rb,25 and E2F126 ) or a control empty vector (pcDNA3; Invitrogen, Carlsbad, CA) using the GenePORTER transfection reagent.
Western analysis
Cells were washed twice with phosphate-buffered saline (PBS) and lysed on ice with lysis buffer (50 mM Tris[tris(hydroxymethyl)aminomethane]-HCl pH 7.4,150 mM NaCl, 0.5% NP-40). Cell lysates were subsequently resolved on 4%-15% gradient sodium dodecyl sulfate–polyacrylamide gels (SDS-PAGE) and transferred to nitrocellulose membranes (Sigma, St Louis, MO). Immunoblots were incubated with various primary antibodies followed by incubation with appropriate antirabbit or antimouse secondary immunoglobulin G antibody conjugated with horseradish peroxidase (Amersham Pharmacia Biotech, Piscataway, NJ). SuperSignal West Pico and West Dura Chemiluminescent substrates (Pierce, Rockford, IL) were used for detection. The following primary antibodies were used: anti-C/EBPϵ (sc-158), anti-Rb (sc-102), anti-E2F1 (sc-251), anti–c-Myc (sc-764) from Santa Cruz Biotechnology (Santa Cruz, CA), and anti-GAPDH from Research Diagnostics (Flanders, NJ). Western blots were stripped between hybridizations with stripping buffer (10 mM Tris-HCl pH 2.3, 150 mM NaCl).
Immunoprecipitation
Protein extracts were incubated with either a Rb antibody (sc-102AC) or an E2F1 antibody (sc-251AC) at 4°C for 16 hours. The precipitated proteins were washed 3 times with PBS, eluted with SDS sample buffer, and subjected to Western blot analysis as described for Western analysis.
GST pull-down assays
C/EBPϵ and C/EBPϵ cDNA deletion mutations were generated by reverse transcriptase–polymerase chain reaction (RT-PCR) and cloned into the pGEX-5X-2 vector (Pharmacia Biotech) to produce glutathione-S-transferase (GST) fusion proteins. GST-Rb (amino acids [aa's] 373-928 of Rb) was made by RT-PCR followed by cloning into the pGEX-2T vector (Pharmacia Biotech). The fusion proteins were expressed in Escherichia coli and purified by glutathione sepharose 4B (Amersham Pharmacia Biotech). Proteins were quantified by SDS-PAGE and Coomassie blue staining. NB4 lysates were incubated with GST fusion proteins at 4°C for 16 hours. GST complexes were washed 3 times with PBS, eluted with SDS sample buffer, and analyzed by Western blot as described above.
Protein in vitro transcription-translation
C/EBPϵ and E2F1 proteins were made by using 2 μg of pCMVSPORT-C/EBPϵ24 and pRC-E2F126 as template for in vitro transcription-translation (TNT Coupled Reticulocyte Lysate System; Promega, Madison, WI). For in vitro binding assays, equal amounts of GST proteins were incubated with 5 μL in vitro–translated proteins at 4°C for 16 hours. GST complexes were washed 3 times with PBS, eluted with SDS sample buffer, and analyzed by Western blot as described for Western analysis.
Luciferase reporter assays
NIH 3T3 and Soas-2 cells were transiently cotransfected with 1 μg of one of the following promoter reporter constructs: pGL3B/granulocyte colony-stimulating factor receptor (pTK81G/G-CSFR, generous gift from D. G. Tenen, Harvard Institutes of Medicine, Boston, MA), pGL3B/mim-1 (generous gift from A. Leutz, Max-Delbruck-Centre for Molecular Medicine, Germany), or pGL3B/3 × E2F and different expression plasmids as indicated. Lysates were harvested 24 hours after transfection, and luciferase activity was measured with the dual-luciferase reporter 1000 assay system (Promega). Transfection efficiency was normalized using 0.2 μg pRL-SV40. Results represent the mean of triplicate transfections. The experiments were repeated 3 times.
Real-time RT-PCR
Two micrograms of total RNA was converted into cDNA using Moloney murine leukemia virus (MMLV) reverse transcriptase (Invitrogen). B9 and 18S (for endogenous reference) expression levels were determined with specific primers and probes using Taqman PCR mastermix (Applied Biosystems, Foster City, CA). PCR conditions were as follows: 2 minutes at 50°C, 10 minutes at 95°C, followed by 45 cycles of 95°C for 15 seconds and 60°C for 1 minute. c-Myc and C/EBPϵ expression levels were determined with specific primers using HotMaster Taq DNA Polymerase (Eppendorf, Westbury, NY) and SYBRGreen I (Molecular Probes, Eugene, OR). PCR conditions were as follows: 2 minutes at 94°C, followed by 45 cycles of 94°C for 20 seconds, 60°C for 10 seconds, 65°C for 25 seconds, and fluorescence determination at the melting temperature of the product for 20 seconds. Specificity of PCR products was checked on agarose gel. All reactions were performed in triplicates in an iCycler iQ system (Biorad, Hercules, CA). For each sample, the amount of the target gene and 18S was determined from a standard curve. The results are expressed in arbitrary units as a ratio of the target gene transcripts/18S transcripts (each value represents the mean of 3 measurements of the sample).
Electrophoretic mobility shift assay
The mim-1 oligonucleotide sequence containing a C/EBP site (underlined) used in electrophoretic mobility shift assay (EMSA) was 5′-ACTGATTGGCCAACAC AACAG-3′. Double-stranded oligonucleotides were end-labeled with γ-32P-ATP by T4 polynucleotide kinase. Nuclear extracts from both U937 cells and transfected COS-1 cells were prepared with the CelLytic Nuclear extraction Kit (Sigma). Nuclear extract proteins (10 μg) were incubated with 20 000 cpm of labeled oligonucleotides. Binding reactions were performed for 30 minutes on ice and then analyzed on 4% polyacrylamide gels. When cold competitor (100-fold excess) or antibodies (anti-C/EBPϵ, sc-158X, anti-Rb, and sc-50; Santa Cruz Biotechnology) were used, they were added to the reactions 20 minutes prior to the labeled probe.
Results
C/EBPϵ interacts with Rb
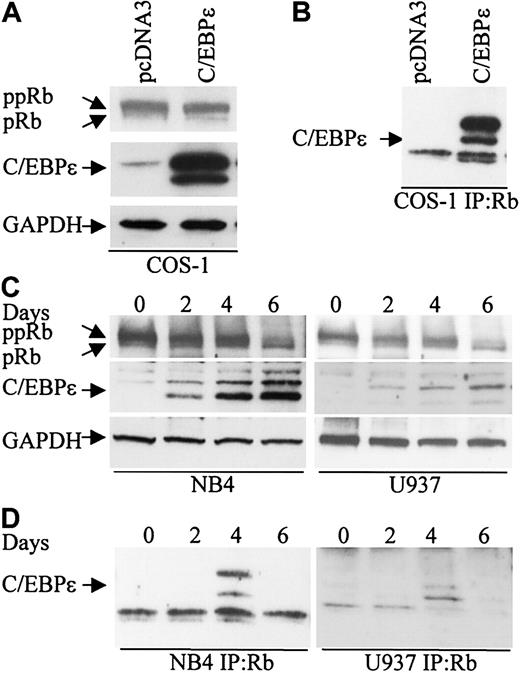
To explore the possibility that Rb interacts with C/EBPϵ, COS-1 cells were transfected with a C/EBPϵ expression vector that expresses the p32 and p30 isoforms of C/EBPϵ (Figure 1A). Cell extracts were immunoprecipitated with an Rb antibody followed by Western analysis with C/EBPϵ antibody. Both the p32 and p30 isoforms of C/EBPϵ were found in complexes immunoprecipitated with the Rb antibody (Figure 1B).
C/EBPϵ and Rb interact in vivo. (A) COS-1 cells were transfected either with an empty vector or a C/EBPϵ expression vector. Western blot analysis was done to detect Rb and C/EBPϵ expression. A GAPDH antibody was used as control for equal loading. (B) COS-1 cell lysates were immunoprecipitated with an Rb antibody and analyzed by immunoblotting with a C/EBPϵ antibody. (C) NB4 and U937 myeloid cells were treated with ATRA for 0, 2, 4, and 6 days and analyzed by Western blot for Rb and C/EBPϵ expression. The blots were stripped and rehybridized with a GAPDH antibody. (D) NB4 and U937 lysates were immunoprecipitated with a Rb antibody followed by Western analysis with a C/EBPϵ antibody.
C/EBPϵ and Rb interact in vivo. (A) COS-1 cells were transfected either with an empty vector or a C/EBPϵ expression vector. Western blot analysis was done to detect Rb and C/EBPϵ expression. A GAPDH antibody was used as control for equal loading. (B) COS-1 cell lysates were immunoprecipitated with an Rb antibody and analyzed by immunoblotting with a C/EBPϵ antibody. (C) NB4 and U937 myeloid cells were treated with ATRA for 0, 2, 4, and 6 days and analyzed by Western blot for Rb and C/EBPϵ expression. The blots were stripped and rehybridized with a GAPDH antibody. (D) NB4 and U937 lysates were immunoprecipitated with a Rb antibody followed by Western analysis with a C/EBPϵ antibody.
During granulocytic differentiation of human myeloid NB4 and U937 cells after exposure to all-trans retinoic acid (ATRA), C/EBPϵ is rapidly induced and Rb is hypophosphorylated (Park et al,8 Brooks et al,27 and Figure 1C). To test whether Rb interacts with C/EBPϵ during this differentiation process, protein extracts were made from NB4 and U937 cells at days 0, 2, 4, and 6 after ATRA treatment, immunoprecipitated with an Rb antibody, and analyzed by Western blot with C/EBPϵ antibody. A definite interaction between C/EBPϵ (p32 and p30 isoforms) and Rb was detected by day 4 (Figure 1D). By day 6 of ATRA exposure, the interaction was no longer evident. This suggests that the Rb-C/EBPϵ interaction is transient and occurs only in a defined window during differentiation.
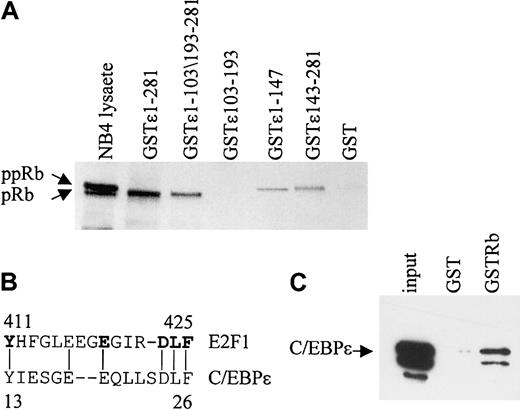
In vitro GST pull-down assays were performed to determine the regions of C/EBPϵ that are important for the Rb-C/EBPϵ interaction. By using GST-C/EBPϵ deletion mutants, 2 regions, one in the C-terminal and another in the N-terminal portion of C/EBPϵ were shown to interact with Rb (Figure 2A). A deletion mutant encoding only the repression domain of C/EBPϵ (aa's 103-193) failed to bind Rb. The N-terminal portion of C/EBPϵ contains a sequence (aa's to 13-27) that may be involved in the C/EBPϵ-Rb interaction because it is similar to a sequence used by E2F1 to interact with Rb28 (Figure 2B). Similar sequences also are found in C/EBPα, C/EBPβ, and C/EBPδ.20 To test whether C/EBPϵ interacts directly with Rb, in vitro–translated C/EBPϵ was incubated with a GST-Rb fusion protein. Results from pull-down assays showed that in vitro–translated C/EBPϵ binds the GST-Rb fusion (Figure 2C), demonstrating that the C/EBPϵ-Rb interaction is direct.
Two domains in C/EBPϵ interact with Rb. (A) NB4 lysates were incubated with either various GST-C/EBPϵ fusion proteins or GST alone as indicated in the figure. Bound Rb was detected by Western blot analysis. NB4 lysate (10% of the input) was used as control for Rb expression. (B) A deduced Rb binding sequence of C/EBPϵ is similar to that of E2F1. The amino acids critical for the E2F1-Rb binding are shown in bold. (C) In vitro–translated C/EBPϵ (input) was incubated with either GST or GST-Rb followed by Western blot analysis with C/EBPϵ antibody.
Two domains in C/EBPϵ interact with Rb. (A) NB4 lysates were incubated with either various GST-C/EBPϵ fusion proteins or GST alone as indicated in the figure. Bound Rb was detected by Western blot analysis. NB4 lysate (10% of the input) was used as control for Rb expression. (B) A deduced Rb binding sequence of C/EBPϵ is similar to that of E2F1. The amino acids critical for the E2F1-Rb binding are shown in bold. (C) In vitro–translated C/EBPϵ (input) was incubated with either GST or GST-Rb followed by Western blot analysis with C/EBPϵ antibody.
Rb enhances transcriptional activation by C/EBPϵ
To determine the functional significance of the interaction between Rb and C/EBPϵ, the effect of Rb expression on C/EBPϵ-mediated transcriptional activation of myeloid specific genes was examined. The Saos-2 osteosarcoma cell line, which expresses a truncated, nonfunctional Rb, was used in luciferase reporter assays because it provides a background on which the effects of Rb expression can be measured. Expression of C/EBPϵ alone increased by 3.4-fold the luciferase activity from a granulocyte colony-stimulating factor receptor (G-CSFR) promoter reporter construct (Figure 3A). Coexpression of C/EBPϵ and Rb resulted in an additional 3.3-fold increase of luciferase activity. An Rb mutant that is constitutively active (Rb5C) also increased luciferase activity by 3.7-fold, whereas an inactive Rb mutant (Rb16) did not enhance C/EBPϵ transcription activity. Similarly, the wild-type Rb and the constitutively active Rb enhanced C/EBPϵ transcriptional activation of the myeloid specific promoter mim-1 reporter construct by 2.9- and 3.1-fold, respectively (Figure 3B). Expression of Rb itself had no effect on luciferase activity.
Rb enhances C/EBPϵ transactivation. (A) Soas-2 cells were cotransfected with a G-CSFR promoter reporter vector (pTK81G/G-CSFR, 1 μg) and one or more of 4 constructs (1 μg each) that express C/EBPϵ, wild-type Rb (Rbwt), a constitutively active mutant Rb (Rb5C), or an inactive mutant Rb (Rb16). (B) Soas-2 cells were cotransfected with a mim-1 promoter reporter vector (pGL3B/mim-1, 1 μg) and the various expression constructs as described for panel A. All transfections included the pRL-SV40 vector (0.2 μg) that served as internal control for transfection efficiency. Results represent the mean of triplicate transfections. The experiments were repeated 3 times. (C) NIH 3T3 cells stably transfected with either an inducible C/EBPϵ gene (pMTϵ) or C/EBPα gene (pMTα) were incubated either without (–) or with (+) zinc (16 hours) and analyzed by Western blot using antibodies to C/EBPϵ and C/EBPα. The blots were stripped and rehybridized with a GAPDH antibody. (D) The pMTϵ and pMTα cells were transiently transfected with either an empty vector (ev) or a Rb expression vector (Rb) and treated with zinc for 16 hours. cDNA from these cells was subjected to real-time RT-PCR with specific primers and probes for B9 and 18S. The results are expressed in arbitrary units as a ratio of either B9 transcripts/18S transcripts (each value represents the mean of 3 measurements of the sample).
Rb enhances C/EBPϵ transactivation. (A) Soas-2 cells were cotransfected with a G-CSFR promoter reporter vector (pTK81G/G-CSFR, 1 μg) and one or more of 4 constructs (1 μg each) that express C/EBPϵ, wild-type Rb (Rbwt), a constitutively active mutant Rb (Rb5C), or an inactive mutant Rb (Rb16). (B) Soas-2 cells were cotransfected with a mim-1 promoter reporter vector (pGL3B/mim-1, 1 μg) and the various expression constructs as described for panel A. All transfections included the pRL-SV40 vector (0.2 μg) that served as internal control for transfection efficiency. Results represent the mean of triplicate transfections. The experiments were repeated 3 times. (C) NIH 3T3 cells stably transfected with either an inducible C/EBPϵ gene (pMTϵ) or C/EBPα gene (pMTα) were incubated either without (–) or with (+) zinc (16 hours) and analyzed by Western blot using antibodies to C/EBPϵ and C/EBPα. The blots were stripped and rehybridized with a GAPDH antibody. (D) The pMTϵ and pMTα cells were transiently transfected with either an empty vector (ev) or a Rb expression vector (Rb) and treated with zinc for 16 hours. cDNA from these cells was subjected to real-time RT-PCR with specific primers and probes for B9 and 18S. The results are expressed in arbitrary units as a ratio of either B9 transcripts/18S transcripts (each value represents the mean of 3 measurements of the sample).
Previous studies showed that overexpression of C/EBPϵ in NIH 3T3 fibroblasts induced endogenous expression of the neutrophil specific genes such as neutrophilic-granule-protein (Ngp/B9), neutrophil-gelatinase–associated lipocalin (N-GAL), and murine cathelinlike peptide (MCLP/CNLP).24 We therefore used these cells to test whether Rb enhances C/EBPϵ transcriptional activation of endogenously expressed genes. NIH 3T3 cells were stably transfected with a C/EBPϵ gene under the control of a zinc-inducible metallothionein promoter (3T3pMTϵ, Figure 3C). The 3T3pMTϵ cell line was transiently transfected with either an Rb expression vector or a control empty vector and treated with zinc to induce C/EBPϵ expression. Expression levels of B9 were measured by real-time PCR with specific primers and probe. The results showed that overexpression of Rb increased B9 induction by 3-fold, compared to cells expressing C/EBPϵ alone (Figure 3D). Since overexpression of C/EBPα in NIH 3T3 fibroblasts also induces the expression of neutrophil specific genes,24 we performed a similar experiment with NIH 3T3 overexpressing C/EBPα (3T3pMTα, Figure 3C). The results showed that Rb enhanced B9 induction by 3.1-fold compared to cells expressing C/EBPα alone (Figure 3D). Similarly, Rb increased the expression of N-GAL in the 3T3pMTϵ and the 3T3pMTα cell lines (data not shown). These results suggest that both the C/EBPϵ-Rb and the C/EBPα-Rb interactions are important for induction of neutrophil genes.
To determine if Rb is present in complex with C/EBPϵ as C/EBPϵ binds to its cognate DNA site, we performed EMSA using a consensus C/EBP binding site from the mim-1 promoter and nuclear extracts from U937 cells.
The results showed that a complex from U937 cells treated with ATRA could bind the mim-1 probe (Figure 4A, lane 2). The binding was specific, as it was competed by unlabeled consensus oligonucleotides and addition of a C/EBPϵ antibody completely super-shifted the protein complex (Figure 4A, lanes 5 and 3, respectively). Addition of an Rb antibody also led to a supershift of the C/EBPϵ complex, although not all the complex was shifted (Figure 3A, lane 4). These results demonstrate that Rb is present in the C/EBPϵ protein complexes that bind the consensus C/EBP site. EMSA also was performed in the presence of in vitro–translated C/EBPϵ incubated with nuclear extracts from COS-1 cells overexpressing Rb. Results showed that in vitro–translated C/EBPϵ was able to bind the C/EBP consensus site (Figure 4B, lane 2). Addition of a C/EBPϵ antibody completely supershifted the protein complex, while addition of an Rb antibody resulted in partial supershift of the complex (Figure 4B, lanes 3 and 4, respectively). These results further support the finding that C/EBPϵ can directly interact with Rb and that the Rb-C/EBPϵ complex is found at the C/EBP DNA binding site.
Rb is found in C/EBPϵ binding complexes in myeloid cells. (A) EMSA was performed using 10 μg of nuclear extract proteins from either untreated U937 cells (U937) or U937 cells treated with ATRA for 4 days (U937A). Extracts were incubated with 32P-labeled oligonucleotides containing the C/EBP site from the mim-1 promoter. C/EBPϵ antibody (anti-ϵ), Rb antibody (anti-Rb), and unlabeled competitor (competitor) were added to the reactions as indicated. (B) Nuclear extract proteins (10 μg) from COS-1 cells transfected with an Rb expression vector were incubated with the mim-1 probe. In vitro–translated C/EBPϵ protein, C/EBPϵ antibody (anti-ϵ), and Rb antibody (anti-Rb) were added to the reactions as indicated. SS C/EBPϵ indicates the complex supershifted by the C/EBPϵ and the Rb antibodies.
Rb is found in C/EBPϵ binding complexes in myeloid cells. (A) EMSA was performed using 10 μg of nuclear extract proteins from either untreated U937 cells (U937) or U937 cells treated with ATRA for 4 days (U937A). Extracts were incubated with 32P-labeled oligonucleotides containing the C/EBP site from the mim-1 promoter. C/EBPϵ antibody (anti-ϵ), Rb antibody (anti-Rb), and unlabeled competitor (competitor) were added to the reactions as indicated. (B) Nuclear extract proteins (10 μg) from COS-1 cells transfected with an Rb expression vector were incubated with the mim-1 probe. In vitro–translated C/EBPϵ protein, C/EBPϵ antibody (anti-ϵ), and Rb antibody (anti-Rb) were added to the reactions as indicated. SS C/EBPϵ indicates the complex supershifted by the C/EBPϵ and the Rb antibodies.
C/EBPϵ interacts with E2F1
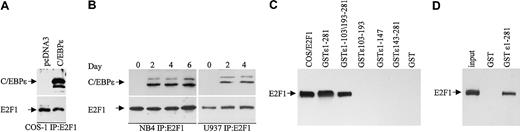
To test whether C/EBPϵ can bind E2F1, COS-1 cells were transfected with a C/EBPϵ expression vector, and cell extracts were immunoprecipitated with an E2F1 antibody followed by Western analysis with a C/EBPϵ antibody. The results showed that the p32 and p30 forms of C/EBPϵ were found in complexes immunoprecipitated with the E2F1 antibody (Figure 5A). We next examined whether E2F1 can interact with endogenously expressed C/EBPϵ. NB4 cells were treated with ATRA for 0, 2, 4, and 6 days; U937 cells were treated for 0, 2, and 4 days to induce C/EBPϵ expression; and protein extracts were analyzed as before. The results show that upon C/EBPϵ induction (by day 2), it could be detected in complexes immunoprecipitated by the E2F1 antibody (Figure 5B).
C/EBPϵ and E2F1 interact in vivo and in vitro. (A) COS-1 cells were transfected with either an empty vector or a C/EBPϵ expression vector. Lysates were immunoprecipitated with an E2F1 antibody and analyzed by immunoblotting with C/EBPϵ antibody. (B) Protein lysates from NB4 and U937 cells treated with ATRA as indicated were immunoprecipitated with an E2F1 antibody followed by Western blot with a C/EBPϵ antibody. The blots were stripped and rehybridized with an E2F1 antibody. (C) NB4 and U937 lysates were incubated with either various GST-C/EBPϵ fusion proteins or GST alone as indicated. Bound E2F1 was detected by Western blot analysis. Protein lysate from COS-1 cells transfected with an E2F1 expression vector was used as control for E2F1 expression. (D) In vitro–translated E2F1 (input) was incubated with either GST or GST-C/EBPϵ, followed by Western blot analysis with E2F1 antibody.
C/EBPϵ and E2F1 interact in vivo and in vitro. (A) COS-1 cells were transfected with either an empty vector or a C/EBPϵ expression vector. Lysates were immunoprecipitated with an E2F1 antibody and analyzed by immunoblotting with C/EBPϵ antibody. (B) Protein lysates from NB4 and U937 cells treated with ATRA as indicated were immunoprecipitated with an E2F1 antibody followed by Western blot with a C/EBPϵ antibody. The blots were stripped and rehybridized with an E2F1 antibody. (C) NB4 and U937 lysates were incubated with either various GST-C/EBPϵ fusion proteins or GST alone as indicated. Bound E2F1 was detected by Western blot analysis. Protein lysate from COS-1 cells transfected with an E2F1 expression vector was used as control for E2F1 expression. (D) In vitro–translated E2F1 (input) was incubated with either GST or GST-C/EBPϵ, followed by Western blot analysis with E2F1 antibody.
To determine the regions of C/EBPϵ that are important for the C/EBPϵ-E2F1 interaction, we used GST-C/EBPϵ deletion mutants. Results from GST pull-down assays demonstrated that both the C-terminal (aa's 1-147) and the N-terminal (aa's 143-281) domains but not the repression domain (aa's 103-193) of C/EBPϵ are necessary for the C/EBPϵ-E2F1 interaction (Figure 5C). To test whether C/EBPϵ interacts directly with E2F1, an in vitro–translated E2F1 was incubated with a GST-C/EBPϵ fusion protein. Results of pull-down assays showed that in vitro–translated E2F1 interacted with GST-C/EBPϵ (Figure 5D), indicating that the E2F1 binds directly to C/EBPϵ.
C/EBPϵ represses transcriptional activation by E2F1
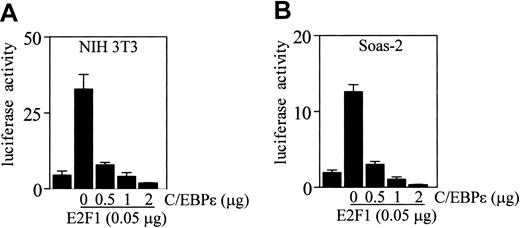
Recent studies demonstrated that C/EBPα suppresses transcription mediated by E2F in several cell lines including NIH 3T3.22,23 To determine if the binding of C/EBPϵ to E2F1 also results in repression of E2F1 transcription activity, we cotransfected NIH 3T3 cells with an E2F site-responsive promoter reporter construct (that contains a concatamer of 3 E2F binding sites in the pGL3 vector, pGL3B/3 × E2F) along with E2F1 and C/EBPϵ expression vectors. The results show that increasing amounts of C/EBPϵ led to a linear decrease of luciferase activity, demonstrating that C/EBPϵ is able to inhibit the E2F1-mediated transcription in a dose-dependent manner (Figure 6A).
Direct repression of E2F1-mediated transcription by C/EBPϵ. NIH 3T3 cells or Soas-2 cells were either transfected with an E2F-responsive reporter vector (pGL3B/3 × E2F, 1 μg) or cotransfected with an E2F-responsive reporter vector (1 μg), along with an E2F1 expression vector (0.05 μg). Where indicated, a C/EBPϵ expression vector was added to the transfections. All transfections included the pRL-SV40 vector (0.2 μg). Results represent the mean of triplicate transfections. The experiments were repeated 3 times.
Direct repression of E2F1-mediated transcription by C/EBPϵ. NIH 3T3 cells or Soas-2 cells were either transfected with an E2F-responsive reporter vector (pGL3B/3 × E2F, 1 μg) or cotransfected with an E2F-responsive reporter vector (1 μg), along with an E2F1 expression vector (0.05 μg). Where indicated, a C/EBPϵ expression vector was added to the transfections. All transfections included the pRL-SV40 vector (0.2 μg). Results represent the mean of triplicate transfections. The experiments were repeated 3 times.
Regulation of E2F1 transcriptional activity is tightly controlled through the E2F1-Rb association that results in repression of E2F-regulated genes. Since C/EBPϵ binds both Rb and E2F1, we tested the possibility that C/EBPϵ repression of E2F1 activity requires Rb. Soas-2 cells were cotransfected with the E2F-responsive promoter reporter construct, E2F1, and increasing amounts of C/EBPϵ. Results showed that C/EBPϵ also inhibited transcription mediated by the E2F1 in a dose-dependent manner in these Rb-negative cells (Figure 6B). This indicates that Rb is not required for the C/EBPϵ repression of E2F transcription.
C/EBPϵ represses c-Myc expression
A recent study showed that down-regulation of c-Myc by C/EBPα is mediated through an E2F site in the c-Myc promoter, and this down-regulation is critical for granulopoiesis in U937 cells.29 We have previously shown that U937 cells stably transfected with a zinc inducible C/EBPϵ gene (U937pMTϵ) undergo granulocytic differentiation upon induction of C/EBPϵ expression. To test whether C/EBPϵ regulates c-Myc expression, RNA was isolated from the U937pMTϵ cell line and the control U937pMT cell line (stably transfected with the empty vector) at various time points after treatment with zinc. c-Myc and C/EBPϵ expression levels were analyzed by real-time PCR with specific primers. Following induction of C/EBPϵ expression, the level of c-Myc RNA dramatically decreased by 70% at 6 hours of zinc treatment (Figure 7A). The level of c-Myc remained the same in the U937pMT cell line.
C/EBPϵ represses c-Myc expression. (A) U937 cells stably transfected with either empty vector (pMT) or an inducible C/EBPϵ vector (pMTϵ) were treated with zinc for the indicated times. cDNA from these cells was used for real-time RT-PCR with c-Myc, C/EBPϵ, and 18S specific primers. The results are expressed in arbitrary units as a ratio of either c-Myc or C/EBPϵ transcripts/18S transcripts (each value represents the mean of 3 independent measurements of the sample). (B) Stably transfected U937 cells (pMT, empty vector, and pMTϵ, inducible-C/EBPϵ) were incubated either without (–) or with (+) zinc for 24 hours, followed by Western blot analysis with a c-Myc antibody. The blot was stripped and rehybridized sequentially with C/EBPϵ antibody and GAPDH antibodies. (C) Protein lysates were made from U937 cells stably transfected with empty vector (pMT) and U937 cells stably transfected with an inducible C/EBPϵ vector (pMTϵ) following 24 hours of exposure to zinc. Lysates were immunoprecipitated with an E2F1 antibody followed by Western analysis with a C/EBPϵ antibody. The blot was stripped and rehybridized with an E2F1 antibody.
C/EBPϵ represses c-Myc expression. (A) U937 cells stably transfected with either empty vector (pMT) or an inducible C/EBPϵ vector (pMTϵ) were treated with zinc for the indicated times. cDNA from these cells was used for real-time RT-PCR with c-Myc, C/EBPϵ, and 18S specific primers. The results are expressed in arbitrary units as a ratio of either c-Myc or C/EBPϵ transcripts/18S transcripts (each value represents the mean of 3 independent measurements of the sample). (B) Stably transfected U937 cells (pMT, empty vector, and pMTϵ, inducible-C/EBPϵ) were incubated either without (–) or with (+) zinc for 24 hours, followed by Western blot analysis with a c-Myc antibody. The blot was stripped and rehybridized sequentially with C/EBPϵ antibody and GAPDH antibodies. (C) Protein lysates were made from U937 cells stably transfected with empty vector (pMT) and U937 cells stably transfected with an inducible C/EBPϵ vector (pMTϵ) following 24 hours of exposure to zinc. Lysates were immunoprecipitated with an E2F1 antibody followed by Western analysis with a C/EBPϵ antibody. The blot was stripped and rehybridized with an E2F1 antibody.
Western blot analysis demonstrated that c-Myc protein level also decreased in the U937pMTϵ cells compared to U937pMT cells following a 24-hour exposure to zinc (Figure 7B). We further examined whether C/EBPϵ expressed in the U937pMTϵ cells could be found in complexes with E2F1. U937pMT and U937pMTϵ cell extracts were immunoprecipitated with an E2F1 antibody followed by Western analysis with C/EBPϵ antibody. C/EBPϵ was easily detected in E2F1 complexes from U937pMTϵ cells (Figure 7C). Thus, down-regulation of c-Myc in U937pMTϵ cells temporally correlated with the interaction of C/EBPϵ with E2F1.
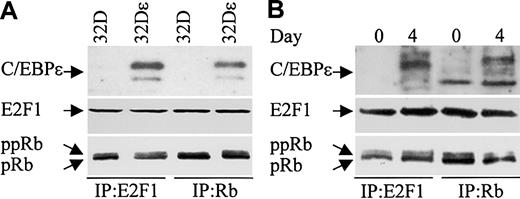
C/EBPϵ, Rb, and E2F1 can be found in complexes in myeloid cells. We next examined whether C/EBPϵ interacts with Rb and/or E2F1 in the murine myeloid precursor cell line 32Dc13. During granulocytic differentiation of these cells, C/EBPϵ is induced and C/EBPϵ target genes are transcribed.14,29 Furthermore, studies previously showed that overexpression of C/EBPϵ in 32Dcl3 cells facilitated their differentiation to granulocytes (Nakajima and Ihle29 and our unpublished data). We therefore used a 32Dcl3 stable cell line to detect the C/EBPϵ-Rb and the C/EBPϵ-E2F1 interactions. 32Dϵ (stably transfected with a constitutively expressed C/EBPϵ gene, E. Williamson and H.P.K., unpublished data, July 2003), and nontransfected 32Dcl3 cell extracts were immunoprecipitated with either E2F1 or Rb antibodies followed by Western analysis with C/EBPϵ antibody. C/EBPϵ was easily detected in E2F1 and Rb complexes from cells that overexpress C/EBPϵ (Figure 8A). These data further suggest that the C/EBPϵ-Rb and the C/EBPϵ-E2F1 interactions play an important role in myeloid cells.
C/EBPϵ, Rb, and E2F1 can be found in complexes together in 32Dcl3 cells. (A) Protein lysates were made from 32Dcl3 parental cells (32D) and 32Dcl3 cells stably transfected with a constitutively expressed C/EBPϵ gene (32Dϵ). Lysates were immunoprecipitated with either E2F1 or Rb antibodies followed by Western analysis with a C/EBPϵ antibody. (B) Protein lysates from 32Dcl3 cells untreated (day 0) or treated with G-CSF (day 4) were immunoprecipitated with either E2F1 or Rb antibodies followed by Western analysis with a C/EBPϵ antibody. The blots (A, B) were stripped and rehybridized with E2F1 and Rb antibodies.
C/EBPϵ, Rb, and E2F1 can be found in complexes together in 32Dcl3 cells. (A) Protein lysates were made from 32Dcl3 parental cells (32D) and 32Dcl3 cells stably transfected with a constitutively expressed C/EBPϵ gene (32Dϵ). Lysates were immunoprecipitated with either E2F1 or Rb antibodies followed by Western analysis with a C/EBPϵ antibody. (B) Protein lysates from 32Dcl3 cells untreated (day 0) or treated with G-CSF (day 4) were immunoprecipitated with either E2F1 or Rb antibodies followed by Western analysis with a C/EBPϵ antibody. The blots (A, B) were stripped and rehybridized with E2F1 and Rb antibodies.
The finding that C/EBPϵ binds Rb and E2F1, coupled with the well-documented Rb-E2F1 interaction, raises the question as to whether the C/EBPϵ-Rb and/or the C/EBPϵ-E2F1 interactions are mutually exclusive or whether all 3 proteins can be found complexed together. To answer this question, we probed Rb and E2F1 immune complexes obtained from lysates of 32Dϵ cells for the presence of E2F1 and Rb, respectively. The results showed that E2F1 is found in complexes precipitated with the Rb antibody, and Rb is found in complexes precipitated with the E2F1 antibody (Figure 8A). This demonstrates that the C/EBPϵ-Rb and the C/EBPϵ-E2F1 interactions do not disrupt the Rb-E2F1 interaction.
We next examined whether endogenously expressed C/EBPϵ from 32Dcl3 cells can be found in complexes with E2F1 and Rb. 32Dcl3 cells were treated with granulocyte colony-stimulating factor (G-CSF) to induce differentiation. Protein extracts made from untreated (day 0) and treated (day 4) cells were immunoprecipitated with either E2F1 or Rb antibodies and analyzed by Western blot with C/EBPϵ antibody. Results show that following G-CSF treatment, C/EBPϵ could be detected in E2F1 and Rb complexes (Figure 8B). Reprobing the blot with E2F1 and Rb antibodies demonstrated that E2F1 could be detected in the Rb complexes and Rb could be detected in the E2F1 complexes.
Discussion
The most recently cloned member of the C/EBP family, C/EBPϵ, acts downstream of C/EBPα in myeloid differentiation. To gain insight into the molecular mechanisms involved in this process, we explored the possibility that C/EBPϵ interacts with Rb and E2F1. Previous studies showed that interactions between those 2 proteins and C/EBP family members play critical roles in several differentiation systems where C/EBPs have been implicated.10,19-23
Rb is best known for its function in the control of cell cycle progression by negatively regulating the E2F transcription factors. In recent years, however, a new role for this protein has emerged as Rb was shown to activate transcription factors during terminal differentiation in a number of tissues and cell types. Previous studies showed that Rb interacts with C/EBPβ in U937 cells that were induced to differentiate along the monocyte-macrophage lineage and in 3T3-L1 fibroblasts that were induced to differentiate terminally into adipocytes.19,20 In those studies, Rb enhanced both the binding of C/EBPβ to cognate DNA sequences in vitro and the transactivation by C/EBPβ of a C/EBPβ-responsive promoter in cells. Although Rb was also shown to bind and activate C/EBPα and C/EBPδ, the relevance of these interactions to differentiation is less well studied. Several other types of transcription factors have been shown to interact with Rb.12 For example; Rb binds members of the AP-1 family, including c-Jun, and enhances transcriptional activity of c-Jun during terminal keratinocyte differentiation.31 C/EBPϵ joins this growing list of transcription factors that interact with and are activated by Rb in differentiating cells.
C/EBPα knockout mice lack neutrophils and eosinophils and C/EBPϵ knockout mice lack secondary granules in their neutrophils. In a previous study we showed that C/EBPϵ and C/EBPα induced the same secondary granule target genes when overexpressed in NIH 3T3.24 Furthermore, the change in differentiation and gene expression induced in 32Dcl3 cells by C/EBPϵ and C/EBPα are indistinguishable,29,32 suggesting that overexpressing cell lines may not always reflect the physiologic effects of C/EBPs. Similarly, in the current study we showed that Rb enhanced the transcriptional activity of C/EBPϵ and C/EBPα in NIH 3T3 cells, suggesting that both proteins can act as Rb partners. This raises the question as to what target genes are endogenously activated by C/EBPϵ and C/EBPα. Additional experiments, including dose-response induction of C/EBPϵ and C/EBPα may help answer this question.
C/EBPϵ is rapidly induced, and Rb is quickly phosphorylated between days 0 and 2 in differentiating NB4 and U937 cells. Yet, a strong interaction between C/EBPϵ and Rb occurs only by day 4 and is no longer detected by day 6. Although we have shown in the in vitro studies that C/EBPϵ can directly interact with Rb, it is possible that in the differentiating cells, a stable complex between C/EBPϵ and Rb requires additional factors that may be transiently expressed. Alternatively, after transcriptional modifications such as phosphorylation of C/EBPϵ may be necessary for a stable C/EBPϵ-Rb interaction.
We have shown that Rb interacts with C/EBPϵ in cells that are differentiating along the granulocytic lineage. This interaction is functionally probably important because in U937 cells, Rb was found in the protein complex that binds the C/EBP DNA binding site. Furthermore, Rb enhanced the C/EBPϵ-mediated transcription of myeloid specific genes, both in reporter assays and endogenously. While Rb has been shown to be absolutely required for terminal differentiation in some cell types, it has not been shown to be critical for granulopoiesis. In fact, a previous study suggested that hypophosphorylated Rb is elevated and essential for monocytic but not neutrophil differentiation.33 This is in contrast to our findings, showing not only accumulation of hypophosphorylated Rb in NB4 and U937 cells during granulocytic differentiation, but also showing that Rb may play an important role in this process by activating C/EBPϵ. Further studies involving inactivation of Rb and disruption of the Rb-C/EBPϵ interaction will provide a better understanding of the significance of Rb in granulopoiesis.
Recent studies demonstrated that C/EBPα binds and represses the second major partner of the Rb/E2F pathway, E2F1.21-23 Along with Rb, the function of E2F1 has been traditionally tightly associated to the regulation of the cell cycle. Recent data suggest that E2F may have roles beyond the cell cycle and E2F targets include genes involved in apoptosis, differentiation, and development.13,34 The repression of E2F1 by C/EBPα was shown to be critical for the terminal differentiation of adipocytes and myeloid cells as well as arrest growth. Unlike the Rb-C/EBP interaction, which involves the different members of the C/EBP family, the E2F1-C/EBP interaction seems to be more restricted, since C/EBPβ was unable to inhibit E2F1 transcription. In the present study, we demonstrated that interaction between C/EBPϵ and E2F1 could be detected in the human myeloid cell lines NB4 and U937 as well as the murine myeloid precursor cell line 32Dc13. In reporter assays, C/EBPϵ repressed transcription from an E2F site, in a manner similar to C/EBPα. This is in agreement with previous results showing that C/EBPϵ, even if to a lesser extent than C/EBPα, inhibited cell proliferation (Park et al8 and data not shown). On the other hand C/EBPβ, which dose not bind E2F1, has been linked to tumorigenesis and cell survival.35
Using a stable inducible U937 cell line, we demonstrated that induction of C/EBPϵ expression resulted in a significant decrease in the levels of endogenous c-Myc mRNA and protein. Moreover, upon induction of C/EBPϵ, this protein could be detected in E2F1 immunocomplexes in U937 cells. Our data support a model in which C/EBPϵ, like C/EBPα, can interfere with E2F1 transcriptional activation, and this may result in repression of c-Myc expression, which is important during terminal granulocytic differentiation.30,36 Since the effect of C/EBPϵ on U937 differentiation is the same as that seen with C/EBPα, it is not clear what role each C/EBP plays in down-regulating c-Myc in vivo. In 32Dcl3 cells, C/EBPα was up-regulated after one day of G-CSF treatment, while high levels of C/EBPϵ were detected by the third day (Schuster et al37 and E. Williamson and H.P.K., unpublished data, June 2003). Interestingly, the kinetics of c-Myc down-regulation correlated with C/EBPϵ induction because strong inhibition of c-Myc was observed only at day 3 of G-CSF treatment. In the present study, we show that C/EBPϵ can be detected in immunocomplexes with E2F1 in 32Dcl3 cells treated with G-CSF. Taken together, these data suggest that either C/EBPϵ is more critical than C/EBPα for the down-regulation of c-Myc or complete inhibition of c-Myc requires both C/EBPϵ and C/EBPα to be present. Additional studies need to determine if C/EBPϵ can directly repress the c-Myc promoter by disrupting E2F1 as well as to correlate the significance of this repression to the ability of C/EBPϵ to induce differentiation.
Since C/EBPϵ interacts both with Rb and E2F1 and since Rb forms a repressive complex with E2F1, we conjecture that C/EBPϵ repression of E2F1 transcription is Rb dependent, perhaps by serving as an adaptor protein between Rb and E2F1. However, C/EBPϵ inhibited E2F1-mediated transcription in a cell line expressing a mutant Rb, indicating that Rb is not necessary for this repression. Again, this finding is similar to earlier results observed with C/EBPα.22 Consequently, Rb and C/EBPs (ϵ and α) act independently to repress E2F. The observation that 2 parallel pathways target E2F1 is not surprising, considering its important role as a cell cycle regulator and its possible role in enhancing apoptosis and differentiation. The C/EBPϵ-mediated repression of E2F1 transcription does not require Rb. However, we have shown that in 32Dcl3 cells, the C/EBPϵ-Rb and C/EBPϵ-E2F1 interactions did not interfere with the binding of Rb to E2F1 (similar results were seen in U937 cells, data not shown). Therefore, all 3 proteins may form a complex at some stage in the differentiation process. We also did not address the question of whether C/EBPϵ binds to the 2 other pocket proteins (p107 and p130) or other E2F family members. Potentially, a complex including one or more of the pocket proteins, along with one or more of the E2F members, might exist with C/EBPϵ.
In this study, we demonstrated that Rb binds and activates C/EBPϵ. We also showed that C/EBPϵ interacts with E2F1 and represses E2F1-mediated transcription. In light of previous and present studies, which demonstrate the importance of these interactions in regulating differentiation, we propose (as shown in Figure 9) that the interactions of Rb-C/EBPϵ and E2F1-C/EBPϵ are important in the terminal granulocytic differentiation induced by C/EBPϵ.
Prepublished online as Blood First Edition Paper, August 28, 2003; DOI 10.1182/blood-2003-01-0159.
Supported in part by National Institutes of Health grants as well as the Parker Hughes and the Horn funds.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
H.P.K. is a member of the Jonsson Comprehensive Cancer Center and the Molecular Biology Institute (UCLA) and holds the endowed Mark Goodson Chair of Oncology Research at Cedars-Sinai Medical Center/UCLA School of Medicine.