Abstract
The multiplicity of transcription factors involved in hematologic malignancies suggests a complicated scenario in which many different molecular mechanisms lead to malignant transformation. We hypothesized that some of these proteins might physically and functionally interact and thus mechanistically link different diseases. The ETO protein of t(8;21) acute myeloid leukemia (AML) is an excellent candidate as a common factor because it is normally expressed in human hematopoietic cells, it binds to histone deacetylases (HDACs), and it interacts with the PLZF protein of t(11;17) acute promyelocytic leukemia. To determine whether ETO functionally links a broader range of disease entities, we asked whether ETO forms a complex with the Bcl-6 oncoprotein of B-cell lymphomas. We found that ETO and Bcl-6 are coexpressed in normal and malignant lymphoid tissue, where they interact and colocalize in nuclear speckles. ETO binds to the fourth zinc finger of Bcl-6, enhances Bcl-6 repression of artificial and endogenous genes in an HDAC-dependent manner, and forms a complex with Bcl-6 on the promoters of its endogenous target genes in B-cell lymphoma cells. Therefore, ETO is a bona fide corepressor that links the transcriptional pathogenesis of acute leukemias and B-cell lymphomas and offers a compelling target for transcriptional therapy of hematologic malignancies.
Introduction
Hematologic malignancies often manifest with nonrandom translocations or with mutations in genes encoding transcriptional regulatory proteins.1 Such mutations alter the genetic programming of a cell by deregulating the expression of an important transcription regulator or by altering its function.2 This can result in a block of differentiation, altered cell proliferation and self-renewal, or changes in the apoptotic set point of the cell. Unfortunately, the list of such mutations is long and still growing, raising the possibility that dozens of new disease subtypes may have to be classified and characterized. This complexity suggests that it will be difficult to identify a specific molecular target-based therapy that would be robust enough to benefit broad groups of patients. On the other hand, among the complexity, common patterns can be discerned that suggest that common mechanisms are at play in hematologic malignancy and could offer specific remedies for a wide range of tumors. Many fusion proteins generated in leukemia involve transcriptional repressors that recruit histone deacetylases (HDACs).3 It is possible that cofactors (ie, corepressors) that link DNA-binding factors, HDACs, and other components of the chromatin remodeling machinery may play a role in mediating disease as well. One possible mechanism is through aberrant fusion to transcriptional activators or by recruitment of aberrant corepressors. An alternative scenario is that the overexpression or deregulation of an oncogenic DNA-binding transcriptional repressor may yield disease and, hence, that the cofactor is a secondary yet crucial link in repressing target genes and initiating the malignant process.
One such repressor is the fusion partner of AML-1 in t(8;21) acute myeloid leukemia (AML), named ETOMTG8. ETO (eight-twenty-one) is a member of a highly conserved protein family that also includes MTGR1 and ETO-24-9 in mammals. ETO family proteins contain 4 conserved domains (for reviews, see Licht et al10 and Davis JN11 ) that include (1) the TAF110 homology domain; (2) the hydrophobic heptad repeat (HHR domain), which mediates dimerization and interaction with corepressors12,13 ; (3) the nervy domain (NHR3 domain), which interacts with the regulatory subunit of cyclic adenosine monophosphate (cAMP)–dependent kinase PKAII14 ; and (4) a zinc finger (MYND) domain that mediates interaction with the nuclear corepressors N-CoR and SMRT.15-18 In humans, ETO is normally expressed in hematopoietic CD34 cells and peripheral blood lymphocytes.14,19 In t(8;21) AML, AML-1/ETO recruits corepressors and HDACs to AML-1 target genes, leading to aberrant transcriptional repression.15 Furthermore, ETO represses transcription when tethered to DNA by a heterologous DNA-binding motif.12,20 However, ETO itself is not a transcription factor because it does not specifically bind to DNA.21
Given that ETO is a nuclear protein that binds corepressors (such as SMRT and N-CoR) and HDACs but not DNA, we previously hypothesized that ETO itself might normally function as a corepressor for DNA-binding transcriptional repressors. We have shown that ETO could interact with the promyelocytic leukemia zinc finger (PLZF) protein and its malignant counterpart, PLZF/RARα, generated by the t(11;17) retinoid refractory variant of acute promyelocytic leukemia (APL).22,23 This suggests that ETO might function as a corepressor protein involved in the pathogenesis of t(11;17) APL because its binding to PLZF/RARα is through the PLZF motif, and, hence, it is retinoid insensitive.
PLZF is a member of the BTB/POZ (bric-a-brac, tramtrack, broad complex/pox virus zinc finger) family of transcription factors, which also includes Bcl-6, the oncoprotein most frequently involved in human B-cell lymphomas (approximately 40% of patients).24 Overexpression or lack of down-regulation of Bcl-6 in lymphoma can occur by translocation of the gene to a heterologous promoter or through somatic hypermutation of the Bcl-6 locus.24-26 Bcl-6 is normally expressed in germinal-center B cells.27 It mediates cell survival and proliferation and possibly the blockade of terminal differentiation28-30 (and A.M., unpublished results, June 2003). In lymphomas, the inappropriately timed expression of Bcl-6 is believed to block lymphocyte differentiation and to immortalize cells, leaving them competent to continue proliferating and vulnerable to mutagenesis by somatic hypermutation.30,31 Cotransfection experiments with artificial reporter constructs showed that Bcl-6 is a potent transcriptional repressor.32-34 Repression by Bcl-6 is dependent mainly on its BTB/POZ domain, which mediates the recruitment of SMRT/N-CoR/BCoR corepressors and HDACs.35-37 In addition, Bcl-6 contains a second repression domain in a central region and an array of 6 C-terminal Krüppel-like C2H2 zinc fingers that specifically bind a consensus DNA sequence in target gene loci.28,32,33,36,38
Because ETO is expressed in peripheral blood lymphocytes and Bcl-6 is a critical repressor in B cells, we hypothesized that ETO is a bona fide corepressor protein of Bcl-6. Thus, akin to SMRT and N-CoR, ETO would enhance transcriptional repression by multiple DNA-binding transcription factors, including Bcl-6, PLZF, and PLZF/RARα. Such interaction would provide a mechanistic link between proteins targeted in different hematologic malignancies. Accordingly, we now show that ETO and Bcl-6 are, in fact, partner proteins that are coexpressed in normal and malignant B cells. ETO enhanced the ability of Bcl-6 to repress the transcription of reporter genes and endogenous target genes, and it forms a complex with Bcl-6 on DNA binding sites on the promoters of Bcl-6 endogenous target genes. These data indicate that ETO is a genuine transcriptional corepressor and that common elements link the transcriptional pathogenesis of acute leukemias and B-cell lymphomas.
Materials and methods
Cells and culture
293T cells were grown in Dulbecco modified Eagle medium (DMEM; Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (Gibco). Daudi and Raji cells were grown in RPMI 1640 (Gibco) containing 10% fetal bovine serum.
Samples
Cryopreserved (–80°C) tissue samples and paraffin tissue sections from 6 patients with lymphoma were studied. Previously, the samples had been extensively immunophenotyped as part of diagnostic evaluation and were classified according to the Revised European American Lymphoma (REAL) or World Health Organization (WHO) system.39 They consisted of 5 diffuse large B-cell lymphomas (DLBCLs) and 1 follicular lymphoma (FL), grade 2 to 3, and all were of B-cell origin.
Immunohistochemistry
Paraffin tissue sections from the 6 patients with lymphoma (5 DLBCL, 1 FL) and 3 tonsil controls were stained with polyclonal rabbit antiserum directed against the N-terminus of ETO. Immunostaining was performed using an automated immunostainer (Ventana Medical Systems, Tucson, AZ) according to a modified MIP protocol (Ventana Medical Systems) using the ChemMate ABC Peroxidase Secondary Detection System (Ventana Medical Systems). Before staining, the paraffin sections were incubated at 95°C for 40 minutes in Antigen Retrieval Citra Plus buffer (BioGenex, San Ramon, CA).
Plasmids
All the ETO plasmids (HA, Gal DBD, GST) used were kindly provided by Scott Hiebert and were described previously.12,17 The Bcl-6 EF expression vector was previously described.36 The Bcl-6 constructs used in the GST pull-down experiments and the Bcl-6 ΔZF construct were polymerase chain reaction (PCR) generated and were cloned into the T7 plink vector or the EF vector, respectively, and sequenced.40
Coimmunoprecipitation and immunoblotting
293T cells were transfected with Fugene (Roche, Indianapolis, IN) with 2.5 μg diverse hemagglutinin (HA)–tag-ETO or Gal-DBD-ETO constructs and 2.5 μg myc-tag-Bcl-6 plasmids. Twenty-four hours after transfection, cells were lysed (1% NP40, 60 mM Tris HCl, pH 8, 150 mM NaCl with protease inhibitors [Roche]). Daudi and Raji cells were lysed for 15 minutes at 4°C in the same buffer. Lysates were immunoprecipitated for 1 hour at 4°C with 5 μL ETO N-terminal rabbit antibody or with 2 μg c-myc 9E10 (Santa Cruz Biotechnology, Santa Cruz, CA) antibody. Preimmune rabbit or mouse immunoglobulin G (IgG) was used as negative control. Immune complexes were collected on protein A/G-agarose (Roche) for 1 hour at 4°C and then washed 3 times with NP40 buffer and separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Immunoblotting was performed with primary antibodies Bcl-6 D8, c-myc 9E10, Gal4-DBD (Santa Cruz), HA (Covance, Princeton, NJ), or ETO (Ab1) (Oncogene Research, San Diego, CA).
Immunostaining
293T cells transfected with ETO and Bcl-6 or Daudi cells were fixed for 10 minutes in frozen methanol/acetone (70/30, vol/vol) and were stained with ETO N-terminal antibody and Bcl-6 monoclonal antibody (DAKO, Carpinteria, CA). The cells were then secondarily stained with Alexa-conjugated goat antimouse 568 and goat antirabbit 488 antibodies (Molecular Probes, Eugene, OR) and were visualized using a Leica-TCS-SP (UV) confocal microscope (Leica, Heidelberg, Germany).
Transcriptional repression assays
293T cells plated at a density of 40 000 cells per 12-well plate well were transfected using Fugene (Roche) with 200 ng 4xB6BS-TK-luciferase, 200 ng pCMV5-HA-tag-ETO, 200 ng pEF-myc-tag-Bcl-6 or the pEF-empty vector, and 10 ng TK-renilla plasmid as an internal control. Cells were harvested 24 hours later and were assayed using the Dual Luciferase kit (Promega, Madison, WI). Raw values performed in triplicate were normalized to the internal control. For HDAC inhibition studies, 2.5 μM suberoylanilide hydroxamic acid (SAHA) (Biomol Research Laboratories, Plymouth Meeting, PA) or the same volume of ethanol was added for 24 hours before dual luciferase assay.
FACS sorting, RNA extraction, reverse transcription–PCR, and quantitative real-time (Q) PCR
Ten-millimeter dishes of 293T cells were transfected with 2 μg pCMV5-HA-tag-ETO or pCMV5-empty vector, 2 μg pEF-myc-tag-Bcl-6 or pEF-empty vector, and 1 μg green fluorescence protein (GFP) plasmid. At 12 or 24 hours after transfection, 2 × 106 GFP-positive cells were sorted by fluorescence-activated cell sorter (FACS) (FACSVantage; BD Biosciences, Palo Alto, CA), lysed in Trizol (Invitrogen, Carlsbad, CA), RNA extracted and DNAse treated (Promega), reverse transcribed with Superscript II (Invitrogen), and amplified with Taq Platinum (Invitrogen). Cyclin D2 and GAPDH real-time PCR were performed with the Quantitect SYBR green PCR kit (Qiagen, Valencia, CA) with an MJR Opticon thermal cycler (MJ Research, Waltham, MA). Sense (S) and antisense (AS) primers were as follows: cyclin D2, S-5′TGGGTCATCCTTGGTCTATGTGCTC-3′, AS-5′-AGGGTTGTCTTCTCCTCTGGCTTTG-3′; actin, S-5′-GCCCAGAGCAAGAGAGGTAT-3′, AS-5′-GGCCATCTCTTGCTCGAAGT-3′; G3PDH, S-5′-CCAAAATCAAGTGGGGCGATG-3′, AS-5′-AAAGGTGGAGGAGTGGGTGTCG-3′; ETO-ORF, S-5′-CTTATCCTACCAGCCCAATGGCCTG-3′, AS-5′-CTAGAGTGGCTGCTGCTACTGC-3′; ETO3′UTR, S-5′-GAGGAAAGACAACACAACCAACG-3′, AS-5′-CCACAATAAGTCATTACGACCCG-3′; Bcl-6, S-5′-CGGAAGTTTTCAATGATGGGC3′, AS-5′-ATGTCGAGAATGGGATGAGAC-3′
Electrophoretic mobility shift assay
The following probes and competitors were used: B6BS-WT, 5′-GAAAATTCCTAGAAAGCATA; B6BS-mut, 5′-GAAAATTTCGAAAAAGCATA; cyclin D2-WT, 5′-GGGCCATTTCCTAGAAAGCTGCA; cyclin D2-mut, 5′-GGGCCATTTTCGAAAAAGCTGCA. Annealing and end labeling were performed as described.41 Three micrograms nuclear extract was incubated for 30 minutes at room temperature (RT) with or without competitor or antibody. For supershift assay, 2 μg Bcl-6 (C19), ETO (Ab1) (Santa Cruz), or rabbit IgG (Zymed, South San Francisco, CA) was used. The probe was added for 1 hour at 4°C in 100 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) (pH 7.5), 250 mM KCl, 25 mM MgCl2, 50 μM ZnCl2, 20% glycerol, 0.5 μg/μL BSA, and 100 μg/mL polydIdC.
Chromatin immunoprecipitation
Cells were fixed in a final concentration of 1% formaldehyde (Sigma, St Louis, MO) and incubated for 30 minutes (Daudi cells) or 10 minutes (293T cells) at 37°C, quenched with glycine 0.125 M, and lysed for 15 minutes in 1% NP40 buffer at 4°C. Chromatin was sonicated 10 times for 30 seconds (Daudi) or 10 times for 20 seconds (293T cells). Chromatin immunoprecipitation (ChIP) was performed according to the Upstate Biotechnology protocol. ChIP-PCR primers were: cyclin D2 promoter, S-5′-CCTAATCCCTCACTCGCCCC-3′, AS-5′-CATCTGCTGACAAGCCGCC-3′; MIP1α, S-5′-TGCTTAGGGTTGGCAAGG-3′, AS-5′-AGGGATAGGGTTGATGGG-3′; CD69, S-5′-CACCAGGGAAGAAGCGAATGAC-3′, AS-5′-GTCTGTGAGGCTCTGAGGCATC-3′; BCL-6, S-5′-CGATGAGGAGTTTCGGGATGTC-3′, AS-5′-TTTCTGGGGGCTCTGTGGACTAAC-3′.
GST affinity chromatography assays
GST-ETO constructs were expressed as described.36 For proteins with 5 or methionines, 35S-methionine (Amersham) was used with the T7 Quick Coupled System (Promega), and when there were fewer than 5 methionines, 35S-cystine (Amersham, Piscataway, NJ) was used with the TNT T7 System (Promega). Two microliters radiolabeled protein was preincubated with 119 μL binding buffer (100 mM KCl, 20 mM HEPES, pH 7.9, 2 mM EDTA [ethylenediaminetetraacetic acid], 0.1% NP40, 10% glycerol, 0.5% nonfat dry milk, 5 mM dithiothreitol [DTT]) for 30 minutes at RT and was centrifuged, and 100 μL supernatant was transferred to a new tube. Thirty microliters GST-ETO-bead slurry was added, and the mixture was incubated for 1 hour at RT. The beads were washed twice and submitted to SDS-PAGE.
Results
Bcl-6 and ETO are coexpressed in normal and malignant B cells
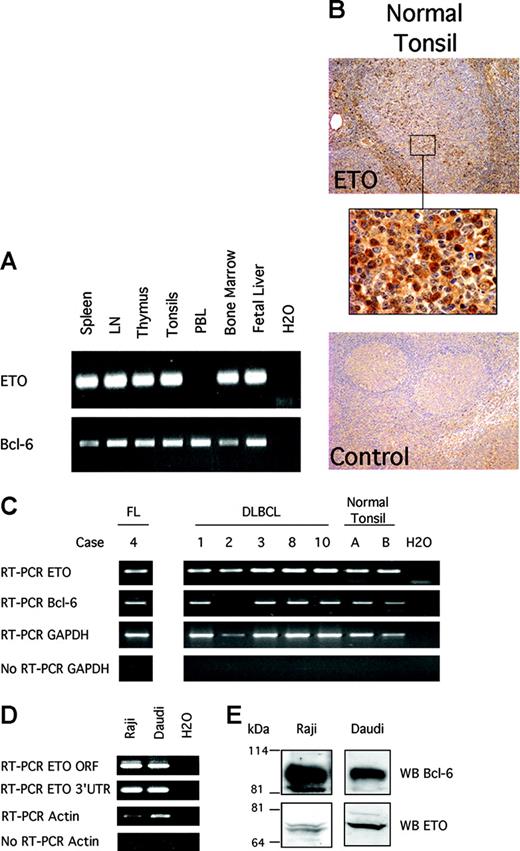
In humans, ETO is expressed in CD34 cells and peripheral blood lymphocytes.14,19 However, it was not known whether ETO was expressed in germinal center B cells, where Bcl-6 is normally expressed. To determine the expression of ETO in hematopoietic and lymphoid tissues, we performed reverse transcription (RT)–PCR on a cDNA panel containing samples from human spleen, lymph node, thymus, tonsils, and bone marrow. Given that ETO family genes are highly similar, we designed 2 sets of specific primers complementary to ETO but not to MTGR1 or ETO-2 to minimize artifacts resulting from cross-hybridization. These corresponded to the coding sequence and the 3′ untranslated region, respectively. We found that ETO and Bcl-6 mRNA were coexpressed in spleen, lymph node, and tonsil (Figure 1A). Accordingly, immunohistochemistry assays performed in normal human lymph node showed ETO expression in germinal center cells (where Bcl-6 is also expressed) and in marginal zone lymphocytes (Figure 1B).
ETO and Bcl-6 are coexpressed in hematopoietic tissues and in normal and malignant B cells. (A) Human tissue cDNA obtained from Clontech (Palo Alto, CA) was used to detect ETO and Bcl-6 by PCR with primers designed to amplify ETO but not ETO-2 or MTGR1. RT-PCR with ETO-specific 3′ UTR primers is shown. (B) Normal human tonsils were submitted to immunohistochemistry for ETO. (Top) Extensive staining for ETO in germinal center and marginal zone lymphocytes (original magnification, × 10). (Middle) × 100 magnification of a section of the germinal center. (Bottom) Rabbit serum control staining (× 10) of tonsil. (C) mRNA was extracted from human FL, human DLBCL, and human normal tonsil and subjected to RT-PCR for ETO or Bcl-6 using primers specific for ETO but not ETO-2 or MTGR1. GAPDH RT-PCR and no-RT-GAPDH PCR reactions were also performed as positive and negative controls, respectively. RT-PCR with ETO-specific 3′ UTR primers is shown. (D) RT-PCR was performed on RNA extract from Raji and Daudi cells. RT-PCR for β actin and no-RT-β actin PCR reactions were also performed as positive and negative controls, respectively. The PCR products were run in 2% agarose gel. RT-PCR with the coding region (ORF) and the 3′UTR ETO-specific primers is shown. (E) Immunoblot of ETO and Bcl-6 was performed on lysates from Daudi and Raji cells with ETO and Bcl-6 polyclonal antibodies.
ETO and Bcl-6 are coexpressed in hematopoietic tissues and in normal and malignant B cells. (A) Human tissue cDNA obtained from Clontech (Palo Alto, CA) was used to detect ETO and Bcl-6 by PCR with primers designed to amplify ETO but not ETO-2 or MTGR1. RT-PCR with ETO-specific 3′ UTR primers is shown. (B) Normal human tonsils were submitted to immunohistochemistry for ETO. (Top) Extensive staining for ETO in germinal center and marginal zone lymphocytes (original magnification, × 10). (Middle) × 100 magnification of a section of the germinal center. (Bottom) Rabbit serum control staining (× 10) of tonsil. (C) mRNA was extracted from human FL, human DLBCL, and human normal tonsil and subjected to RT-PCR for ETO or Bcl-6 using primers specific for ETO but not ETO-2 or MTGR1. GAPDH RT-PCR and no-RT-GAPDH PCR reactions were also performed as positive and negative controls, respectively. RT-PCR with ETO-specific 3′ UTR primers is shown. (D) RT-PCR was performed on RNA extract from Raji and Daudi cells. RT-PCR for β actin and no-RT-β actin PCR reactions were also performed as positive and negative controls, respectively. The PCR products were run in 2% agarose gel. RT-PCR with the coding region (ORF) and the 3′UTR ETO-specific primers is shown. (E) Immunoblot of ETO and Bcl-6 was performed on lysates from Daudi and Raji cells with ETO and Bcl-6 polyclonal antibodies.
Bcl-6 expression is usually deregulated in DLBCL and FL.24,42 To determine whether ETO is expressed in these neoplasms, we performed RT-PCR for both proteins in a panel of tumor tissue samples from patients with DLBCLs and one with FL (Figure 1C). Four of 5 DLBCL patients expressed Bcl-6, whereas 5 of 5 expressed ETO. The FL patient expressed both. We also tested whether 2 Bcl-6–expressing germinal center phenotype B-cell lymphoma lines (Raji and Daudi cells) express ETO mRNA and protein. RT-PCR, Western blot, and immunofluorescence assays confirmed the expression of ETO mRNA and protein in these cells (Figures 1D-E). Therefore, ETO and Bcl-6 are coexpressed in normal and malignant B cells.
Bcl-6 interacts in vivo with ETO
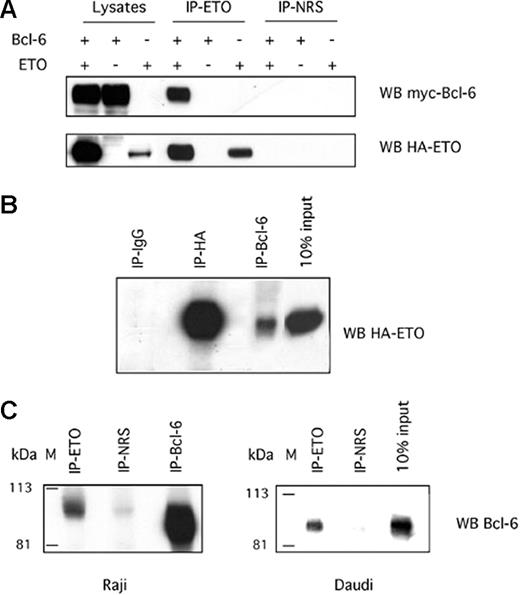
To determine whether Bcl-6 and ETO could interact in mammalian cells, we expressed Bcl-6 and ETO in 293T cells, immunoprecipitated with a polyclonal ETO antibody and immunoblotted for myc-tag Bcl-6 and HA-tag ETO (Figure 2A). Bcl-6 was observed only in immunoprecipitates from lysates containing ETO and Bcl-6, but not in lysates lacking ETO or Bcl-6. No coimmunoprecipitation of Bcl-6 was seen with preimmune serum. In reciprocal experiments, myc-tag Bcl-6 was also able to specifically pull down HA-ETO (Figure 2B). Therefore, ETO and Bcl-6 interact in vivo in mammalian cells. To establish the physiologic relevance of this interaction, lysates from Daudi and Raji B-cell lymphoma cells were immunoprecipitated using a polyclonal ETO antibody and were immunoblotted for Bcl-6 with a monoclonal antibody (Figure 2B). In both cell lines, Bcl-6 was enriched in immunoprecipitates with ETO antibody but not with the preimmune serum. Thus, endogenous ETO interacts with endogenous Bcl-6 in B-cell lymphoma cells.
ETO interacts with Bcl-6 in vivo. (A) 293T cells were transfected with HA-ETO, myc-Bcl-6 plasmid, or the respective empty vectors. Whole-cell lysates were immunoprecipitated with an ETO-N-terminal polyclonal antibody or normal rabbit serum (NRS) as a negative control and immunoblotted for myc to visualize the Bcl-6 ETO expression (upper panel) or for HA to verify ETO expression (lower panel). (B) Reciprocal immunoprecipitation in similarly transfected 293T cells in which control IgG, HA, or myc (to myc-Bcl-6) antibodies were used to pull down HA-ETO. (C) Whole-cell lysates from Raji cells (left) or Daudi cells (right) were subjected to immunoprecipitation of endogenous ETO with an N-terminal polyclonal antibody or rabbit serum control (NRS) and were subsequently immunoblotted for endogenous Bcl-6 with a mouse monoclonal antibody.
ETO interacts with Bcl-6 in vivo. (A) 293T cells were transfected with HA-ETO, myc-Bcl-6 plasmid, or the respective empty vectors. Whole-cell lysates were immunoprecipitated with an ETO-N-terminal polyclonal antibody or normal rabbit serum (NRS) as a negative control and immunoblotted for myc to visualize the Bcl-6 ETO expression (upper panel) or for HA to verify ETO expression (lower panel). (B) Reciprocal immunoprecipitation in similarly transfected 293T cells in which control IgG, HA, or myc (to myc-Bcl-6) antibodies were used to pull down HA-ETO. (C) Whole-cell lysates from Raji cells (left) or Daudi cells (right) were subjected to immunoprecipitation of endogenous ETO with an N-terminal polyclonal antibody or rabbit serum control (NRS) and were subsequently immunoblotted for endogenous Bcl-6 with a mouse monoclonal antibody.
ETO and Bcl-6 colocalize to nuclear speckles
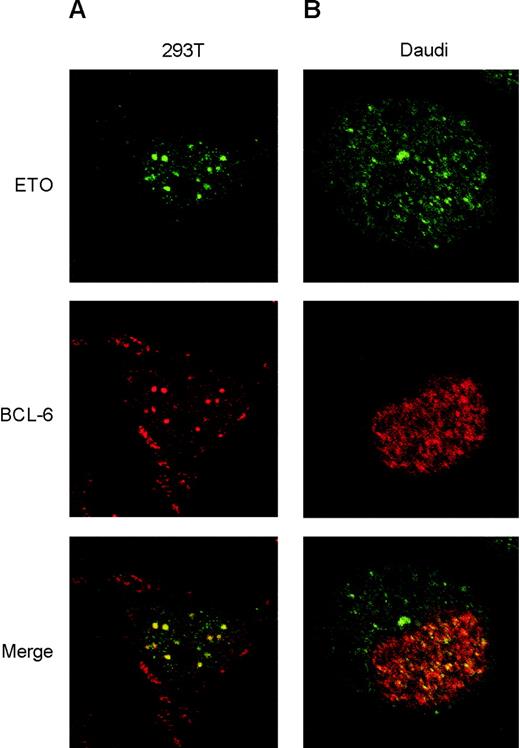
To determine whether ETO localized with Bcl-6, we first established the level of expression of exogenous ETO or Bcl-6 required to reproduce the endogenous nuclear distribution (nuclear diffuse and speckled and nuclear speckled, respectively) in 293T cells and then performed colocalization studies. Imaging of cell sections by confocal laser microscopy showed near total (approximately 80%-90%) overlap of ETO with Bcl-6 nuclear speckles (Figure 3A). The staining pattern was not seen in untransfected cells or when preimmune serum and an irrelevant monoclonal antibody were used (data not shown). Bleed-through across channels was prevented by imaging each channel independently at nonoverlapping wavelengths. The staining pattern of endogenous ETO and Bcl-6 was then determined in Daudi and Raji cells. However, because Raji cell nuclei are lobulated and folded and harder to accurately score, we focused here on Daudi cells. As observed by others in peripheral blood lymphocytes,14 we found abundant ETO in the cytoplasm of these lymphoid cells, though there was nuclear staining as well (Figure 3B). Confocal optical sections of Daudi nuclei demonstrated partial overlap (20%-30%) of ETO and Bcl-6 speckles, consistent with our previous observations of endogenous ETO and PLZF colocalization (A.M., unpublished results, January 2000) and with our results showing approximately 20% coimmunoprecipitation of ETO and Bcl-6. Primary and secondary antibody controls did not manifest any staining (data not shown). These results suggest that endogenous ETO and Bcl-6 interact in the nuclei of lymphoma cells.
ETO colocalizes with Bcl-6 in vivo. (A) 293T cells were cotransfected with levels of ETO and Bcl-6 that reproduce the endogenous nuclear staining pattern of both proteins. Immunofluorescence visualization was achieved by exposing the cells to ETO polyclonal antibody and Bcl-6 monoclonal antibody, followed by Alexa 488 (green)– and Alexa 568 (red)–labeled secondary antibodies, respectively. Cell sections were visualized at × 60 magnification using laser scanning confocal microscopy. Panels show the visualization of ETO or Bcl-6 alone and the overlay of both images in the lower panel. (B) Daudi cells were treated with the same antibodies to detect localization of endogenous ETO and Bcl-6 by immunofluorescence. Cell sections were visualized by confocal microscopy at × 60 magnification, as described for panel A.
ETO colocalizes with Bcl-6 in vivo. (A) 293T cells were cotransfected with levels of ETO and Bcl-6 that reproduce the endogenous nuclear staining pattern of both proteins. Immunofluorescence visualization was achieved by exposing the cells to ETO polyclonal antibody and Bcl-6 monoclonal antibody, followed by Alexa 488 (green)– and Alexa 568 (red)–labeled secondary antibodies, respectively. Cell sections were visualized at × 60 magnification using laser scanning confocal microscopy. Panels show the visualization of ETO or Bcl-6 alone and the overlay of both images in the lower panel. (B) Daudi cells were treated with the same antibodies to detect localization of endogenous ETO and Bcl-6 by immunofluorescence. Cell sections were visualized by confocal microscopy at × 60 magnification, as described for panel A.
Mapping the interaction between ETO and Bcl-6
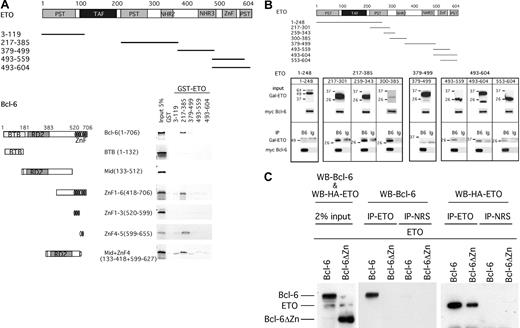
Transcription factors and cofactors interact in specific ways through defined motifs or regions. For example, the N-CoR, BcoR, and SMRT corepressors bind to the BTB/POZ domain of Bcl-6.37 Because ETO and Bcl-6 interact with these corepressors, bridging effects may occur. To determine the nature of the interaction between ETO and Bcl-6, we performed protein-affinity chromatography using GST-ETO fusion proteins. Glutathione-agarose beads were coated with fusion proteins containing the conserved motifs of ETO, and expression was verified by SDS-PAGE–Coomassie blue staining of the purified products (data not shown). Only a fragment containing amino acids 217 to 385 of ETO specifically bound to Bcl-6 (Figure 4A). This region contains the HHR motif of ETO (which mediates ETO homodimerization and heterodimerization) and a more proximal region that mediates the interaction with PLZF. It was not possible to perform this experiment with a shorter fragment lacking the HHR because the resultant fusion protein was highly insoluble.
Mapping the interaction between ETO and Bcl-6 in vitro and in vivo. (A) Protein-affinity chromatography was performed to map the interacting domains of ETO and Bcl-6. Schematic representations of the bacterially expressed GST-ETO and in vitro–transcribed/translated Bcl-6 constructs are shown. Results of the interactions between the different fragments after GST pull-down is shown by fluorography of the 35S-methionine–labeled Bcl-6 constructs. (B) Coimmunoprecipitation was performed in 293T cells cotransfected with myc-tagged Bcl-6 and the GAL4-ETO fusion constructs shown in the schematic representation. The upper panels show 2% input from immunoblots performed with GAL4 polyclonal antibodies and myc-tagged monoclonal antibodies. The bottom panels show immunoblots resulting after immunoprecipitation with myc-tagged mouse monoclonal antibody for Bcl-6 (B6) or mouse control IgG (Ig). The analysis is separated into 4 boxes to show separately the area from the N- to the C-terminus of the ETO protein interacting or not interacting with Bcl-6. (C) Myc-tag Bcl-6 (Bcl-6) or GAL4–Bcl-6 deleted for the zinc finger (Bcl-6ΔZincF) was cotransfected with HA-ETO in 293T cells. Coimmunoprecipitation was performed with the ETO N-terminal polyclonal antibody or its IgG control. Proteins (2% input or immunoprecipitate) were run on 15% SDS-PAGE and immunoblotted with the Bcl-6 mouse monoclonal antibody and the HA antibody.
Mapping the interaction between ETO and Bcl-6 in vitro and in vivo. (A) Protein-affinity chromatography was performed to map the interacting domains of ETO and Bcl-6. Schematic representations of the bacterially expressed GST-ETO and in vitro–transcribed/translated Bcl-6 constructs are shown. Results of the interactions between the different fragments after GST pull-down is shown by fluorography of the 35S-methionine–labeled Bcl-6 constructs. (B) Coimmunoprecipitation was performed in 293T cells cotransfected with myc-tagged Bcl-6 and the GAL4-ETO fusion constructs shown in the schematic representation. The upper panels show 2% input from immunoblots performed with GAL4 polyclonal antibodies and myc-tagged monoclonal antibodies. The bottom panels show immunoblots resulting after immunoprecipitation with myc-tagged mouse monoclonal antibody for Bcl-6 (B6) or mouse control IgG (Ig). The analysis is separated into 4 boxes to show separately the area from the N- to the C-terminus of the ETO protein interacting or not interacting with Bcl-6. (C) Myc-tag Bcl-6 (Bcl-6) or GAL4–Bcl-6 deleted for the zinc finger (Bcl-6ΔZincF) was cotransfected with HA-ETO in 293T cells. Coimmunoprecipitation was performed with the ETO N-terminal polyclonal antibody or its IgG control. Proteins (2% input or immunoprecipitate) were run on 15% SDS-PAGE and immunoblotted with the Bcl-6 mouse monoclonal antibody and the HA antibody.
To test whether this interaction occurs in vivo, we expressed a series of GAL4-DNA binding domain/ETO fusion proteins along with Bcl-6 in 293T cells and performed coimmunoprecipitation experiments. Consistent with our prior results, GAL4-ETO (217-301), GAL4-ETO (259-343), and GAL4-ETO (300-385) interacted with Bcl-6, but the N-terminal portion of ETO did not (Figure 4B), indicating that the interaction surface was long, there were several contacts for Bcl-6 within ETO, or one of the fragments interacted with Bcl-6 through a third bridging factor. For example, low levels of endogenous ETO (or possibly other ETO family members) in 293T cells could interact with Bcl-6 and recruit the ETO (300-385) through the HHR domain. In contrast to in vitro studies, we found that the C-terminal portion of ETO interacts with Bcl-6, though at much lower levels in vivo (Figure 4B). This is likely a result of bridging through proteins such as N-CoR or SMRT, both of which interact with the ETO C-terminal domain15 and are present in 293T cells (A.M., unpublished results, November 2001). Therefore, we conclude that the central region of ETO, possibly including the HHR motif, contacts Bcl-6 and that the C-terminal domain might make secondary contacts with Bcl-6 through bridging by other factors.
Bcl-6 interacts with ETO through the Krüppel-like zinc finger domain
Protein affinity chromatography experiments were performed with [35S]methionine or cysteine-labeled in vitro–translated Bcl-6 deletion mutants, which were allowed to interact with GST-ETO fusion products (Figure 4A). Of note, the BTB/POZ domain, the major repression domain of Bcl-6, does not interact with ETO. This indicates that ETO and the SMRT/N-CoR/BCoR corepressors interact with Bcl-6 through distinct mechanisms. Instead, the interaction was mapped to the Krüppel-like zinc finger domain of Bcl-6. Further mapping of this region indicated that the fourth zinc finger was specifically required for binding the medial region of ETO that binds Bcl-6 (Figure 4A).
To test whether the zinc finger domain of Bcl-6 is required for the interaction to occur in vivo, we transiently transfected the full-length HA-epitope–tagged ETO with full-length myc-Bcl-6 or with GAL4-DNA binding domain Bcl-6 deleted for the zinc finger region (Bcl-6ΔZincF). Consistent with the in vitro data, little or no Bcl-6ΔZincF interacted with ETO, confirming that the zinc finger array of Bcl-6 is the major interaction site for ETO (Figure 4C).
ETO interacts with DNA-bound Bcl-6
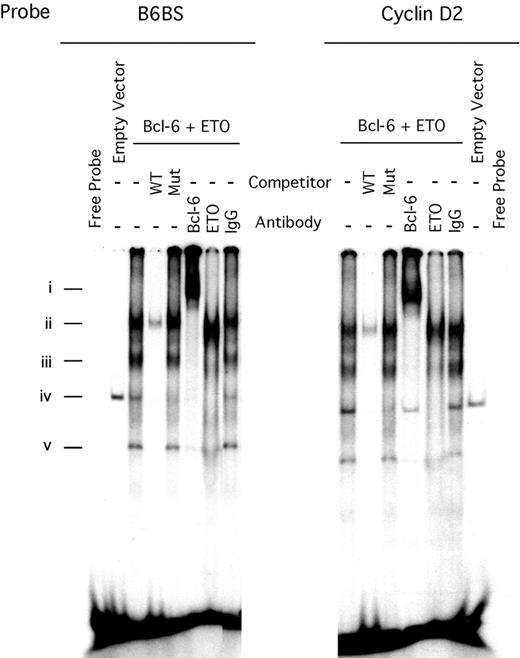
To determine whether the Bcl-6 zinc fingers could interact simultaneously with ETO and DNA, we performed electrophoretic mobility shift assays (EMSAs) using artificial Bcl-6 binding-site (B6BS) or cyclin D2 promoter Bcl-6 binding-site probes (cyclin D2 is a natural target gene of Bcl-628 ). When incubated with nuclear extracts of 293T cells cotransfected with ETO and Bcl-6, the probes yielded 3 prominent DNA-protein complexes (Figure 5Ai, Aiii, Av) that were shifted in the presence of Bcl-6 antibodies. The complexes were competed by a 100-fold excess of unlabeled wild-type probe but not a mutant Bcl-6–binding oligonucleotide. The Bcl-6 complexes were also supershifted or destabilized by the presence of an anti-ETO antibody in the reaction mixture but not by the IgG control. These results show that Bcl-6 can use its zinc fingers to simultaneously bind to ETO and DNA, indicating that ETO could be present at Bcl-6 target genes and influence the transcriptional activity of Bcl-6.
ETO and Bcl-6 form a complex while bound to DNA. (A) EMSA of nuclear extract prepared from 293T cells cotransfected with myc-Bcl-6 and HA-ETO or by the corresponding empty vectors. Three micrograms nuclear extract was incubated with radiolabeled double-stranded Bcl-6 binding-site oligonucleotide (B6BS) or with the cyclin D2 oligonucleotides that contained the Bcl-6 consensus binding site. Lanes 1 and 16, free probe; lanes 2 and 15, empty vector; lanes 3 to 8 and 9 to 14, Bcl-6–ETO-DNA complex; lanes 4 and 10, 100 × wild-type unlabeled cold probe; lanes 5 and 11, 100 × mutated cold probe; lanes 6 and 12, 2 μg rabbit Bcl-6 antibody C19; lanes 7 and 13, 2 μg rabbit ETO antibody Ab1; lanes 8 and 14, 2 μg rabbit IgG.
ETO and Bcl-6 form a complex while bound to DNA. (A) EMSA of nuclear extract prepared from 293T cells cotransfected with myc-Bcl-6 and HA-ETO or by the corresponding empty vectors. Three micrograms nuclear extract was incubated with radiolabeled double-stranded Bcl-6 binding-site oligonucleotide (B6BS) or with the cyclin D2 oligonucleotides that contained the Bcl-6 consensus binding site. Lanes 1 and 16, free probe; lanes 2 and 15, empty vector; lanes 3 to 8 and 9 to 14, Bcl-6–ETO-DNA complex; lanes 4 and 10, 100 × wild-type unlabeled cold probe; lanes 5 and 11, 100 × mutated cold probe; lanes 6 and 12, 2 μg rabbit Bcl-6 antibody C19; lanes 7 and 13, 2 μg rabbit ETO antibody Ab1; lanes 8 and 14, 2 μg rabbit IgG.
ETO enhances Bcl-6 transcriptional repression
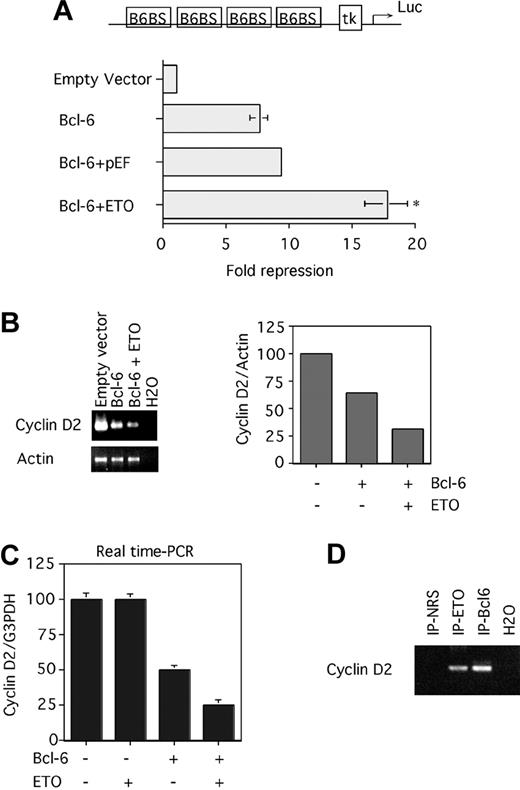
Because ETO and Bcl-6 interact, we next asked whether ETO could modulate transcriptional repression by Bcl-6. 293T cells were cotransfected with expression vectors for both proteins, along with a reporter construct containing 4 Bcl-6 binding sites upstream of a thymidine kinase promoter driving luciferase gene expression (B6BS-tk-luc). Bcl-6 specifically repressed this reporter construct but not a reporter lacking Bcl-6 binding sites (data not shown and37 ). Bcl-6 repression was not altered by cotransfection of the empty vector used to express ETO (Figure 6A). However, the coexpression of ETO increased repression by Bcl-6 from 8- to 18-fold. ETO alone did not affect the transcription of these reporter constructs.
ETO enhances Bcl-6–mediated repression of artificial and endogenous target genes. (A) 293T cells were transfected with a reporter construct containing 4 Bcl-6 binding sites upstream of a thymidine kinase promoter driving luciferase gene expression (B6BS-tk-luc) and with vector control (lane 1), Bcl-6 alone (lane 2), Bcl-6 plus the ETO vector control (lane 3), and Bcl-6 plus ETO (lane 4). Transcriptional output was normalized to an internal control (renilla), and multiple repeats were performed to determine standard deviations. P = .01, representing a significant difference between lanes 3 and 4. This experiment was repeated 6 times. (B) 293T cells were transfected with a GFP-expressing plasmid plus the empty Bcl-6 vector (lane 1), Bcl-6 plus the ETO empty vector (lane 2), and Bcl-6 plus ETO (lane 3). After 24 hours of transfection, the GFP-positive cells were sorted using FACS, and the RNA was submitted to RT-PCR for the cyclin D2 and the actin transcript. After 30 cycles, the PCR products were run on a 2% agarose gel, and the intensity of the bands was analyzed with National Institutes of Health (NIH) image software. The intensities of the respective cyclin D2–actin ratios were plotted on a graph (right panel). (C) 293T cells were transfected with a GFP-expressing plasmid plus the empty vector (lane 1), ETO plus the Bcl-6 empty vector (lane 2), Bcl-6 plus the ETO empty vector (lane 3), and Bcl-6 plus ETO (lane 4). After 12 hours of transfection, the GFP-positive cells were sorted using FACS, and the RNA was submitted to reverse transcription followed by real-time PCR for cyclin D2 and GAPDH transcript. This experiment was performed in triplicate to determine standard deviations. Primers used here were first analyzed for their efficiency, which was almost 100%. (D) 293T cells were cotransfected with ETO and Bcl-6. Chromatin immunoprecipitations were performed after 24 hours with rabbit N-terminal ETO antibody, rabbit polyclonal Bcl-6 antibody, or a rabbit IgG control. ChIP products were PCR amplified using primers surrounding the Bcl-6 binding site on the cyclin D2 promoter. NRS and H2O controls are also shown. Error bars represent SD of 6 experiments.
ETO enhances Bcl-6–mediated repression of artificial and endogenous target genes. (A) 293T cells were transfected with a reporter construct containing 4 Bcl-6 binding sites upstream of a thymidine kinase promoter driving luciferase gene expression (B6BS-tk-luc) and with vector control (lane 1), Bcl-6 alone (lane 2), Bcl-6 plus the ETO vector control (lane 3), and Bcl-6 plus ETO (lane 4). Transcriptional output was normalized to an internal control (renilla), and multiple repeats were performed to determine standard deviations. P = .01, representing a significant difference between lanes 3 and 4. This experiment was repeated 6 times. (B) 293T cells were transfected with a GFP-expressing plasmid plus the empty Bcl-6 vector (lane 1), Bcl-6 plus the ETO empty vector (lane 2), and Bcl-6 plus ETO (lane 3). After 24 hours of transfection, the GFP-positive cells were sorted using FACS, and the RNA was submitted to RT-PCR for the cyclin D2 and the actin transcript. After 30 cycles, the PCR products were run on a 2% agarose gel, and the intensity of the bands was analyzed with National Institutes of Health (NIH) image software. The intensities of the respective cyclin D2–actin ratios were plotted on a graph (right panel). (C) 293T cells were transfected with a GFP-expressing plasmid plus the empty vector (lane 1), ETO plus the Bcl-6 empty vector (lane 2), Bcl-6 plus the ETO empty vector (lane 3), and Bcl-6 plus ETO (lane 4). After 12 hours of transfection, the GFP-positive cells were sorted using FACS, and the RNA was submitted to reverse transcription followed by real-time PCR for cyclin D2 and GAPDH transcript. This experiment was performed in triplicate to determine standard deviations. Primers used here were first analyzed for their efficiency, which was almost 100%. (D) 293T cells were cotransfected with ETO and Bcl-6. Chromatin immunoprecipitations were performed after 24 hours with rabbit N-terminal ETO antibody, rabbit polyclonal Bcl-6 antibody, or a rabbit IgG control. ChIP products were PCR amplified using primers surrounding the Bcl-6 binding site on the cyclin D2 promoter. NRS and H2O controls are also shown. Error bars represent SD of 6 experiments.
Synthetic promoters in transfected episomal plasmids do not necessarily reflect the transcriptional regulatory mechanisms that take place in the genomic chromatin context. Therefore, we tested whether ETO could enhance Bcl-6 repression of one of its natural target genes, in this case the cyclin D2 gene.28 We measured levels of cyclin D2 transcript in mock-transfected 293T cells compared with cells transfected with vectors expressing Bcl-6 or Bcl-6 plus ETO (Figure 6B). A GFP expression vector was added to indicate cells that were successfully transfected 24 hours after transfection. We used FACS to sort GFP-positive cells, and the RNA was extracted. Semiquantitative RT-PCR for cyclin D2 mRNA showed that exogenous Bcl-6 repressed expression of the endogenous cyclin D2 gene by 50%, and cotransfection of ETO doubled Bcl-6 repression of the target gene (Figure 6B). Repetition of these experiments using real-time PCR to quantify cyclin D2 expression more precisely confirmed that ETO doubled repression by Bcl-6, whereas ETO alone did not affect cyclin D2 mRNA (Figure 6C).
To link the interaction between ETO, Bcl-6, and Bcl-6 target genes, we determined whether ETO was present with Bcl-6 at the cyclin D2 promoter by performing ChIPs. 293T cells were cotransfected with ETO and Bcl-6 and were harvested after 24 hours. The cross-linked cell nuclei were sonicated to approximately 500-bp DNA fragments and were immunoprecipitated with Bcl-6 and ETO antibodies. Purified DNA was amplified using specific primers for the cyclin D2 promoter Bcl-6 binding site (Figure 6D). The 122-bp product was seen with ETO and Bcl-6 antibodies but not with the preimmune serum. These results suggest that ETO interacts with Bcl-6 while attached to its binding site on the cyclin D2 promoter in transfected cells.
ETO forms a complex with Bcl-6 on the promoters of endogenous target genes
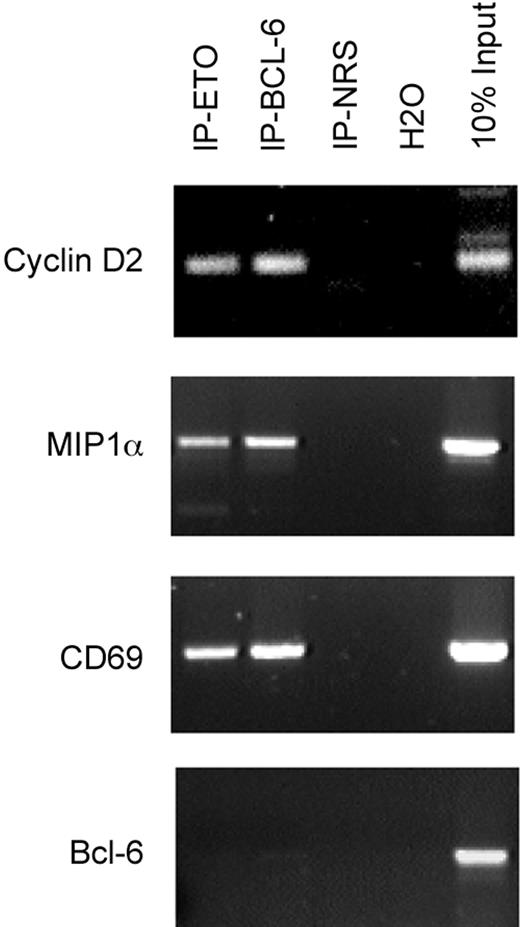
To determine the physiologic relevance of the Bcl-6–ETO interaction, we tested whether ETO is present on Bcl-6 target promoters in B-cell lymphoma cells. ChIP was performed using Bcl-6 and ETO antibodies on chromatin derived from Daudi cells, and PCR was performed on the Bcl-6 binding site of the cyclin D2, MIP1α, and CD69 promoters (previously identified direct Bcl-6 targets28 ). ETO and Bcl-6 antibodies enriched Bcl-6 binding sites of the respective promoters, but preimmune serum reactions did not (Figure 7). As a negative control, a Bcl-6 coding sequence region was not enriched by any of the antibodies. The presence of Bcl-6 and ETO at natural Bcl-6 target genes in vivo indicated that ETO is a bona fide cofactor that interacts with Bcl-6 and Bcl-6 target genes.
ETO and Bcl-6 are present at the promoters of endogenous Bcl-6 target genes in B cells. Chromatin immunoprecipitations were performed on Daudi cells for endogenous proteins with either rabbit N-terminal ETO antibody, rabbit polyclonal Bcl-6 antibody, or rabbit IgG control. ChIP products were PCR amplified using primers surrounding the Bcl-6 binding site on the cyclin D2, CD69, and MIP1α promoters. DNA input as positive control and H2O as negative control are also shown. A control PCR reaction with primers from within ORF of Bcl-6 was also performed as a negative control.
ETO and Bcl-6 are present at the promoters of endogenous Bcl-6 target genes in B cells. Chromatin immunoprecipitations were performed on Daudi cells for endogenous proteins with either rabbit N-terminal ETO antibody, rabbit polyclonal Bcl-6 antibody, or rabbit IgG control. ChIP products were PCR amplified using primers surrounding the Bcl-6 binding site on the cyclin D2, CD69, and MIP1α promoters. DNA input as positive control and H2O as negative control are also shown. A control PCR reaction with primers from within ORF of Bcl-6 was also performed as a negative control.
ETO enhances Bcl-6 transcription repression in an HDAC-dependent manner
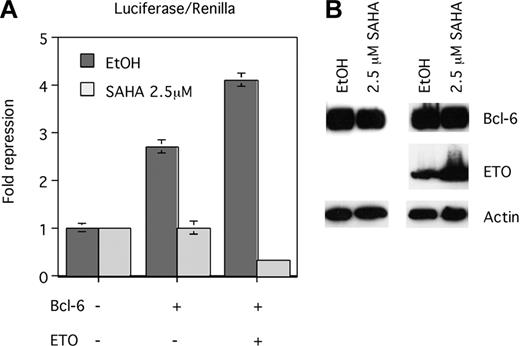
Given the ability of ETO to interact with HDACs12,17 and given that transcriptional repression by a GAL4-ETO fusion protein was impaired by histone deacetylase inhibitor drugs,20 we tested whether ETO enhancement of Bcl-6 repression was dependent on histone deacetylase activity. The B6BS-tk-luc reporter was cotransfected with empty expression vector, with Bcl-6, or with Bcl-6 plus ETO in the presence of SAHA, an HDAC inhibitor active in the micromolar range, compared with vehicle alone.43 At a dose level of 2.5 μM, SAHA abrogated repression by Bcl-6 and the Bcl-6–ETO combination (Figure 8A). Immunoblot analysis verified that these effects were not caused by reductions of Bcl-6 or ETO expression in the presence of SAHA (Figure 8B). These results suggest that enhanced transcriptional repression by ETO and Bcl-6 is mediated through the recruitment of HDAC activity to target promoters.
ETO enhancement of Bcl-6 repression is dependent on HDAC activity. (A) 293T cells were transfected with the Bcl-6 binding site containing reporter construct (B6BS-tk-luc) and with the empty Bcl-6 vector (lane 1), Bcl-6 plus the ETO empty vector (lane 2), and Bcl-6 plus ETO (lane 3). After 4 hours of transfection, 2.5 μM SAHA or the same volume of ethanol was added. After 24 hours, the transcriptional output was determined by luciferase assays normalized to an internal control (renilla). Four repeats were performed to determine standard deviations. (B) Twenty microliters of the same lysates used for luciferase assays were run on 15% SDS-PAGE and were immunoblotted for myc-tag (Bcl-6), HA (ETO), and β-actin. Error bars represent SD of 4 experiments.
ETO enhancement of Bcl-6 repression is dependent on HDAC activity. (A) 293T cells were transfected with the Bcl-6 binding site containing reporter construct (B6BS-tk-luc) and with the empty Bcl-6 vector (lane 1), Bcl-6 plus the ETO empty vector (lane 2), and Bcl-6 plus ETO (lane 3). After 4 hours of transfection, 2.5 μM SAHA or the same volume of ethanol was added. After 24 hours, the transcriptional output was determined by luciferase assays normalized to an internal control (renilla). Four repeats were performed to determine standard deviations. (B) Twenty microliters of the same lysates used for luciferase assays were run on 15% SDS-PAGE and were immunoblotted for myc-tag (Bcl-6), HA (ETO), and β-actin. Error bars represent SD of 4 experiments.
Discussion
Our work stems from the hypothesis that the wild-type counterparts of leukemia oncoproteins might form common regulatory complexes that govern normal hematopoiesis.22 In malignancy, these proteins would still associate, but mutations in one of the members would disrupt normal gene regulation. Accordingly, our previous studies indicated that the ETO protein of t(8;21) AML interacts with and enhances repression by the PLZF protein and PLZF/RARα, its malignant counterpart in t(11;17) APL.22,23 Recently, ETO was also found to interact with the Gfi-1 transcriptional repressor.44 These results suggest that ETO might normally function as a corepressor for transcriptional repressors in normal and malignant cells. However, additional data are required to establish that ETO is a bona fide corepressor. For example, corepressors are likely to have multiple partners so that other ETO-interacting transcription factors must be identified. These interactions should occur between endogenous proteins and in a physiologically relevant context. ETO should enhance repression not only of episomal, artificial reporter constructs but also of endogenous target genes of the partner transcription factors. ETO should form a complex with transcription factors and their DNA target sequences at the promoters of natural, endogenous target genes. Finally, the significance of ETO as a common element in mechanisms of oncogenic transcriptional repression would be greatly enhanced if other partner proteins play key roles in other hematologic malignancies.
Our current studies were designed to address these questions by determining whether ETO is a corepressor for the Bcl-6 B-cell lymphoma oncogenic transcriptional repressor. Accordingly, we found that ETO is endogenously expressed in lymphoid tissues and, most important, in B-cell lymphomas along with Bcl-6. We used specific RT-PCR primers for ETO that do not hybridize to ETO-2 or MTGR1 to establish that the PCR product obtained is specific. This is important because ETO is not expressed in the mouse hematopoietic system.45 Together, our data and those of others indicate that the expression profiles of ETO in mice and humans are different. Our data do not exclude that other ETO family members might directly or indirectly interact with Bcl-6. Future studies of the functions and expression patterns of ETO-2 and MTGR1 are warranted to determine whether they too are corepressors and whether they interact with overlapping or distinct partner proteins.
From the structural standpoint, we were surprised to find that ETO binds to the fourth zinc finger of Bcl-6, the first assignment of a specific zinc finger of Bcl-6 to a protein-protein interaction. The fact that the zinc finger domain of Bcl-6 was required for the recruitment of ETO and that ETO, Bcl-6, and DNA form a complex implies that the zinc fingers of Bcl-6 fold in such a way as to make contact with DNA and partner proteins simultaneously. This is a novel finding. The only other example of a C2H2 zinc finger domain forming a simultaneous complex with DNA and a heterologous protein is that of the YY1 zinc fingers with SP1 and ATF/CREB.46,47 Mascle et al48 found that mutations introduced into the third, fourth, fifth, or sixth zinc finger abrogated DNA binding and transcriptional repression by Bcl-6. This was interpreted as indicating that each of these (including the fourth zinc finger) binds DNA. However, another possibility is that zinc finger domains require a precise folding arrangement, disrupted by mutations. The zinc fingers of Bcl-6 were also shown to associate with class 2 HDACs, though this association could not be narrowed down to a specific finger.48,49
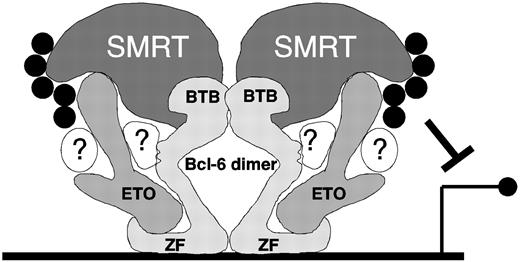
Although these data suggest that the zinc finger domain might function as an autonomous repression motif, there are conflicting reports on this point. In a reporter assay with a Bcl-6 binding site containing reporter, Seyfert et al33 found that the zinc finger domain did not repress transcription. The same group also reported that the zinc finger domain functioned as a weak dominant negative when expressed in Bcl-6–positive Raji cells, indicating that it either does not repress or that it represses less efficiently than full-length Bcl-6.51 In contrast, a GAL4-zinc finger fusion construct inhibited transcriptional activation of a reporter gene containing LexA and GAL4 sites when it was activated by a LexA-VP16 fusion,48 and GAL4 fusions with Bcl-6 deletion mutants lacking the zinc fingers were partially impaired for transcriptional repression compared with full-length Bcl-6 in reporter assays.52 These experiments illustrate the difficulty of determining the functional contribution of the zinc finger domain to transcriptional repression, which is unlikely to be resolved by studying deletion mutants of Bcl-6. Bcl-6 binds directly to N-CoR/SMRT/BCoR corepressors through the BTB/POZ domain, and ETO also binds to N-CoR and SMRT.16-18 We propose that the binding of ETO to the Bcl-6 zinc finger domain could stabilize the interaction of Bcl-6 with SMRT and N-CoR through its C-terminal domain. In this view, the interaction of Bcl-6 with HDACs would be enhanced through the formation of a stable complex in which these corepressors together provide the optimal platform for HDAC action at Bcl-6 target genes (Figure 9). This scenario is also supported by the fact that ETO C-terminal constructs weakly interact with Bcl-6 in vivo but not in vitro, suggesting a bridging interaction with other corepressors.
Model of possible interactions among ETO, Bcl-6, SMRT/N-CoR, and HDACs. Bcl-6 forms an obligate dimer through its BTB domain, which directly contacts the SMRT corepressor.50 The Bcl-6 zinc finger region binds to DNA consensus sites and simultaneously to a central region of ETO. ETO in turn also interacts with SMRT, through its C-terminus (though additional contact sites may exist). SMRT and ETO both recruit HDACs (black circles) to amplify Bcl-6 repression. Undoubtedly, multiple other proteins interact with Bcl-6 and ETO (in white, with question marks) and form a complex repressosome. This model is speculative but is consistent with our data for ETO and SMRT/N-CoR interactions with Bcl-6.
Model of possible interactions among ETO, Bcl-6, SMRT/N-CoR, and HDACs. Bcl-6 forms an obligate dimer through its BTB domain, which directly contacts the SMRT corepressor.50 The Bcl-6 zinc finger region binds to DNA consensus sites and simultaneously to a central region of ETO. ETO in turn also interacts with SMRT, through its C-terminus (though additional contact sites may exist). SMRT and ETO both recruit HDACs (black circles) to amplify Bcl-6 repression. Undoubtedly, multiple other proteins interact with Bcl-6 and ETO (in white, with question marks) and form a complex repressosome. This model is speculative but is consistent with our data for ETO and SMRT/N-CoR interactions with Bcl-6.
Bcl-6 and PLZF interact with the region of ETO containing amino acids 217 to 379, which includes the nuclear localization signal and helical heptad repeat domains. Results from the current study and confocal microscopy studies with endogenous Bcl-6 or PLZF and ETO showed that subsets of ETO speckles colocalize with Bcl-6 or PLZF speckles (A.M., unpublished results, 2003). The same central region of ETO was shown to mediate the localization of ETO to a specific population of nuclear speckles.53 These speckles associated with the nuclear matrix, which is the functional compartment for many transcription factors, including PLZF and probably Bcl-6.23 Accordingly, it is likely that by virtue of its binding to transcription factor partners such as Bcl-6 and PLZF, the ETO central domain targets this corepressor to nuclear speckles that are sites of active transcriptional repression.
A major question to be addressed in future studies is the contribution of ETO to transcriptional and biologic effects of Bcl-6 in B cells. Loss of function studies will be difficult because ETO family members might partially or totally compensate for the down-regulation of ETO, ETO may have unexpected functions in B cells related to nuclear cytoplasmic transport and cytoplasmic functions, and ETO is likely to have many other transcription factor partners. Most critically, any assessment of ETO in mediating the role of Bcl-6 in B-cell biology is complicated by the fact that ETO is not expressed in mouse B cells and that there is no in vitro germinal center reconstitution assay in which to detect the loss of Bcl-6 function because of the lack of ETO expression.
In conclusion, our data indicate that ETO is a bona fide corepressor protein for Bcl-6 and that it functions at least in part through the amplification of HDAC activity, consistent with the fact that ETO directly interacts with HDACs.12 This is supported by the endogenous coprecipitation and colocalization of ETO and Bcl-6 and by the ETO enhancement of Bcl-6 repression of a specific reporter and an endogenous target gene. Furthermore, ETO was present in Bcl-6 DNA complexes by EMSA supershift experiments and was present with Bcl-6 on 3 of its natural target genes in B-cell lymphoma cells. Finally, the involvement of ETO in repressing Bcl-6 target genes broadens the scope of ETO actions in hematologic malignancies to include B-cell lymphomas along with the previously described M2 and M3 acute myeloid leukemias. By enhancing Bcl-6 repression in B-cell lymphoma cells, ETO would contribute to oncogenic gene silencing. ETO is thus a common element of transcriptional control proximal to the general activity of HDACs, and it may represent a valuable target for experimental transcription therapy approaches to hematologic malignancies.
Prepublished online as Blood First Edition Paper, October 9, 2003; DOI 10.1182/blood-2003-06-2081.
Supported by National Cancer Institute grant R21 CA 95487 (A.M.), National Institutes of Health grants R01 CA 59936 (J.D.L.) and R01 CA 71540 (V.J.B.), the American Society of Hematology (A.M.), the Kimmel Foundation (A.M.), the Chemotherapy Foundation (A.M., J.D.L.), and the Sol Sloan Memorial Fund (A.M.). J.D.L. is a recipient of the Burroughs-Wellcome Fund Clinical Scientist Award in Translational Research.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Scott Hiebert and Joe Amman for sharing important reagents and advice. We also thank Graeme Carlisle for performing preliminary experiments leading to the manuscript.