Abstract
Aneuploidy, especially monosomy 7 and trisomy 8, is a frequent cytogenetic abnormality in the myelodysplastic syndromes (MDSs). Patients with monosomy 7 and trisomy 8 have distinctly different clinical courses, responses to therapy, and survival probabilities. To determine disease-specific molecular characteristics, we analyzed the gene expression pattern in purified CD34 hematopoietic progenitor cells obtained from MDS patients with monosomy 7 and trisomy 8 using Affymetrix GeneChips. Two methods were employed: standard hybridization and a small-sample RNA amplification protocol for the limited amounts of RNA available from individual cases; results were comparable between these 2 techniques. Microarray data were confirmed by gene amplification and flow cytometry using individual patient samples. Genes related to hematopoietic progenitor cell proliferation and blood cell function were dysregulated in CD34 cells of both monosomy 7 and trisomy 8 MDS. In trisomy 8, up-regulated genes were primarily involved in immune and inflammatory responses, and down-regulated genes have been implicated in apoptosis inhibition. CD34 cells in monosomy 7 showed up-regulation of genes inducing leukemia transformation and tumorigenesis and apoptosis and down-regulation of genes controlling cell growth and differentiation. These results imply distinct molecular mechanisms for monosomy 7 and trisomy 8 MDS and implicate specific pathogenic pathways.
Introduction
The myelodysplastic syndromes (MDSs) are a heterogeneous group of bone marrow (BM) diseases characterized by low blood counts and a proclivity to leukemic transformation.1 The variable manifestations and clinical courses are reflected in evolving classification schemes based largely on the morphology of BM cells on aspirate smears and in biopsies.2 Prognosis is most closely related to the percentages of marrow myeloblasts and also to the presence and character of specific cytogenetic abnormalities.3 Chromosome changes are present by conventional analysis in about 50% of primary and 80% of secondary MDS patients. Monosomy 7 and trisomy 8 are 2 of the most common such findings.4,5 Although pancytopenic complications are the usual cause of death in MDS, the clinical behavior of these 2 syndromes is different.6-9 Monosomy 7 is associated with refractory and often severe pancytopenia and frequent progression to acute myeloid leukemia.9-11 In one large retrospective analysis, patients with chromosome 7 abnormalities had a uniformly fatal course and all evolved to acute leukemia6 ; chromosome 7 abnormalities therefore are an indicator of a poor prognosis in the International Scoring System.6 Monosomy 7 commonly occurs as a late evolution event in aplastic anemia and, again, most patients succumb to refractory cytopenias or to leukemic transformation.6,12 In contrast, MDS with trisomy 8 can have a more moderate course6 ; peripheral blood cytopenias can respond to immunosuppressive treatment,12 and survival with stable hematologic values for years is not unusual.
The molecular mechanisms responsible for monosomy 7 and trisomy 8, as for MDS in general, are poorly understood. Apoptosis of marrow cells is believed to be increased in MDSs. For trisomy 8 patients, we observed overexpression of cell surface Fas and cytoplasmic caspase 3, especially in the cytogenetically abnormal cell population. In more recent studies, we determined highly prevalent CDR3 skewing of clonally expanded cytotoxic lymphocytes, suggestive of an antigen-driven T-cell pathophysiology, and some patients' T-cell clones are specifically cytotoxic for trisomy 8 cells compared with cytogenetically normal cells from the same individual.13-15 These results are consistent with an immune system role in the development of pancytopenia in MDSs, as has also been inferred from abnormalities in MDSs of tumor necrosis factor (TNF), TNF-related apoptosis-inducing ligand receptors (TRAILs), and functional and genetic studies of T cells performed in other laboratories.16-20
Monosomy 7 is even less well understood. Partial or complete absence of chromosome 7 is relatively common in a variety of myeloid malignant diseases. In MDS, monosomy 7 and 7q deletions are frequent among patients with abnormal cytogenetics and may be associated with poor prognosis.6 Chromosome 7 abnormalities are characteristic of therapy-related MDS that follows exposure to alkylator drugs.21 Monosomy 7 occurs in marrow failure states, including not only acquired AA after immunosuppressive therapy but also Kostmann syndrome with granulocyte colony-stimulating factor (G-CSF) treatment,22,23 Fanconi anemia, and Schwachman-Diamond syndrome. The prevalence of chromosome 7 abnormalities may be underestimated by current methodologies, as submicroscopic deletions have been strongly associated with other chromosomal alterations in acute leukemia.24 Nonetheless, the pursuit of a molecular basis for the obvious leukemogenic properties of monosomy 7 has been frustrating. As with other leukemias and cancers, activating ras mutations may be associated with this particular aneuploidy.25 Despite the clinical linkage with G-CSF administration, abnormalities of the G-CSF receptor or its pathway have not been described in monosomy 7. Loss of a tumor suppressor gene would appear probable and multiple loci have been proposed based on mapping studies using artificial chromosome contigs,26,27 but efforts toward identification of a specific tumor suppressor gene or product have been disappointing.28 Although unusual, transiency of chromosome 7 abnormalities, most spectacularly in Down syndrome,29 and spontaneous reversion to normal cytogenetics in marrow failure patients30 are suggestive of environmental pressure for selective clonal expansion in monosomy 7, as appears to also occur in trisomy 8 disease.
That MDS arises in the hematopoietic stem cell has been demonstrated by a variety of cell culture methodologies, including studies of patients with aneuploidy.31 The close correlation of cytogenetic abnormalities with patterns of dysplastic marrow morphology, ineffective hematopoiesis, and leukemic transformation strongly suggests their pathophysiologic roles in marrow failure and leukemogenesis.11,32,33 As early hematopoietic cells are readily identified and isolated based on the presence of the CD34 antigen, we examined purified populations of hematopoietic stem and progenitor cells using DNA microarrays, which allow assessment in minimally manipulated cells of the expression levels of thousands of genes simultaneously. Because CD34 cells are infrequent even in normal BM and particularly so in many MDS patients, we applied the accepted modifications of sample pooling and RNA amplification; these methods allow comparison of gene expression patterns between standard protocols with single-donor and pooled RNA samples and a new T7-linear amplification protocol when RNA quantities are insufficient.34-40 Using this strategy for CD34 cells from MDS characterized by specific chromosomal abnormalities, we found distinctive differences in the patterns of gene expression between monosomy 7 and trisomy 8 patients, with clear implications for diverse pathophysiologies and identification of potentially important molecular disease pathways.
Patients, materials, and methods
Patient samples
BM samples were collected from 17 patients in the current study, each with a diagnosis of primary MDS based on clinical features, morphologic studies, and cytogenetic analyses according to the new World Health Organization (WHO) classification2 and which had not evolved from aplastic anemia. Four patients had monosomy 7 and 13 showed trisomy 8 chromosomal abnormalities (Table 1). An average of 40% monosomy 7 and 50% trisomy 8 cells were identified by fluorescence in situ hybridization (FISH) performed on peripheral blood mononuclear cells depleted of lymphocytes (Table 1), consistent with chromosomal abnormalities previously identified in BM-derived CD34 cells (data not shown). Eight samples from age-matched healthy volunteers were used as controls. BM was obtained by aspiration from the posterior iliac crest into syringes containing media supplemented 1:10 with heparin (O'Neill and Feldman, St Louis, MO). Informed consent was obtained according to protocols approved by the Institutional Review Board of the National Heart, Lung, and Blood Institute.
Enrichment of CD34 cells
BM mononuclear cells (BMMNCs) were isolated by density gradient centrifugation using lymphocyte separation medium (Organon, Durham, NC). After washing in Hanks balanced salt solution (HBSS; Life Technologies, Gaithersburg, MD), cells were resuspended in magnetic-activated cell separation (MACS) buffer (phosphate-buffered saline [PBS] supplemented with 0.5% bovine serum and 2 mM EDTA [ethylenediaminetetraacetic acid]). After incubation with human immunoglobulin (Ig; Fc receptor [FcR] blocking reagent) and monoclonal hapten-conjugated anti-CD34 antibody for 15 minutes at 6°C to 12°C, cells were washed and resuspended in 500 μL of MACS buffer. Magnetically labeled cells were enriched by passing samples through the positive selection column and magnetic field; the column was washed twice with MACS buffer and then transferred to a collection tube. Retained cells were eluted using MACS buffer followed by plunging. The purity of CD34 cells was 88% to 94% as determined by flow cytometry (EPICS Altra; Beckman Coulter, Miami, FL).
DNA microarray analysis
Microarray analysis was performed for individual and pooled samples using the manufacturer's standard protocol (Affymetrix, Santa Clara, CA; www.affymetrix.com) of RNA extraction with Trizol reagent (Life Technologies).41 RNA was extracted separately but then pooled for 8 trisomy 8 patients (nos. 5-12; Table 1), due to the limited number of CD34 cells; for the pool, each patient contributed 600 ng total RNA. Two pools of normal CD34 RNA were created from 6 healthy donors (3 donors/pool; 1.6 μg total RNA/donor). Sufficient RNA could be obtained from monosomy 7 patients to allow 2 individuals (nos. 1-2; Table 1) to be analyzed separately; 4.8 μg total RNA from each patient donor was assessed. Approximately 5 μg of total RNA from each test sample was used to synthesize single-stranded cDNA by reverse transcription with T7 (dT)24 primer in the SuperScript Choice System (Life Technologies). Second-strand cDNA (ds-cDNA) was transcribed in vitro into cRNA and labeled with biotin using the BioArray High Yield RNA Transcript Labeling Kit (Enzo Diagnostics, Farmingdale, NY). Microarray chips (HG-U95Av2 Array; Affymetrix) were hybridized, washed, and stained, followed by scanning with the GeneArray scanner (Affymetrix).
For CD34 cells of 3 individual trisomy 8 patients (nos. 13-15; Table 1) and 2 individual healthy donors, we also employed a 2-round small-sample RNA amplification protocol in preparation for microarray analysis using the RiboAmp OA Kit (Arcturus, Carlsbad, CA; www.arctur.com). Total RNA was isolated using PicoPure RNA isolation Kit (Arcturus). The first round of cDNA synthesis was performed using 300 ng of total RNA from each sample as a starting material. The ds-cDNA from the second-round amplification was used to generate biotin-labeled antisense cRNA. Fragmentation, hybridization, staining, and scanning were performed as described in the preceding paragraph.
Image data were quantified with Microarray Suit version 5.0 (Affymetrix) and further analyzed using GeneSpring version 6.0 software (Sinica Genetics, Redwood City, CA). The samples derived from standard and 2-round amplification protocols were analyzed independently with comparison to appropriate healthy controls. Samples were normalized for expression levels in each chip to the 50th percentile and each gene to the median in order to obtain reference values. We called “present” 5495 of 12 600 genes based on values of 0.01 to 20 determined by filter on the expression level. Differences in expression level using the filter on fold changes and statistical analysis (analysis of variance [ANOVA]) were then calculated for all individual genes in the test samples from the MDS patients, based on comparison with healthy controls. We arbitrarily set a threshold of a 2.5-fold difference to declare whether a gene was considered differentially expressed. Patterns of gene expression were identified using a hierarchical-clustering algorithm as a gene tree or a condition tree to indicate related expression. Nomenclature and functional descriptions were derived from public databases (CMAP42 and DAVID43 ).
Quantitative real-time RT-PCR
Relative expression levels and differences in CD34 cells of individual patients compared with controls were validated using the TaqMan 5′ nuclease real-time reverse transcriptase–polymerase chain reaction (RT-PCR) assay. Samples from 8 patients (4 with monosomy 7 [nos. 1-4; Table 1] and 4 with trisomy 8 [nos. 14-17]) were subjected to real-time RT-PCR using appropriate primers for 10 genes of interest (Table 2) and β-actin as an internal control. Primers were designed using Clonal Manager Suite software except for TRAIL (R&D Systems, Minneapolis, MN). Using reverse transcriptase, cDNA was synthesized from 500 ng of total RNA extracted from CD34 cells; aliquots of cDNA were used as templates for real-time PCR reactions containing primers of either target genes or β-actin. Each amplification was performed in duplicate for each sample in a 25-μL reaction mixture using the components of the TaqMan PCR Core Reagent Kit (Perkin Elmer, les Ulis, France) and 300 nM of forward and reverse primers. The PCR reactions, TaqMan analyses, and subsequent calculations were performed with the ABI Prism 7700 sequence detection system (Perkin Elmer Biosystems, Foster City, CA) and SYBR Green reagents. The threshold cycle (CT) value was defined as the fraction cycle number and set at 10 times the standard deviation above the mean baseline fluorescence calculated from cycles 3 to 15. The fold changes in the target genes normalized to β-actin and relative expression of healthy control were calculated for each sample using the 2-ΔΔCT method, where -ΔΔCT = (CT, Target - CT, Actin) Patient's sample - (CT, Target - CT, Actin) Normal control.
Flow cytometry
Fresh BMMNCs were stained with an antibody cocktail containing fluorescein isothiocyanate (FITC)–conjugated anti-CD34 (clone HPCA-2; BD PharMingen, San Diego, CA) and phycoerythrin (PE)–conjugated anti–c-Mpl (BD PharMingen). Isotype-matched FITC- or PE-conjugated mouse immunoglobulins were used as negative controls. All samples were analyzed using an EPICS Altra flow cytometer (Beckman Coulter).
Results
Global gene expression in cytogenetically defined MDS
We first assessed the reliability of our 2 methods of preparation of RNA for microarray analysis. Starting with 300 ng of total RNA, an average of 29 μg (21.4, 36.4, and 29.3 μg) of biotin-labeled cRNA was generated from individual patient samples using the small-sample protocol compared with 28.6 μg from the pooled patient sample generated using the standard protocol. The hybridization signal intensity of probe sets interrogating the 3′ and 5′ ends of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was only slightly greater in individual samples (7.49, 5.79, and 2.25) than in the pooled sample (3.65). The scale factor (SF) and “present” call, also used to assess technical sensitivity in microarray assays, were comparable: the SF of individual samples after 2-round amplification was 5.38, 7.61, and 4.0, slightly higher but not statistically different than for the pooled sample using the standard protocol (3.81); the “present” call obtained from individual samples (44%, 42%, and 45%) and pooled sample (48%) were also very similar. We further compared data sets for pooled trisomy 8 CD34 cells, extracted and hybridized according to the standard protocol, to each of 3 samples of CD34 cells from individual patients for which RNA had been amplified using the small-sample protocol: the correlation coefficients for these individual samples to the pooled data set were 0.87, 0.75, and 0.74. From these results we inferred sufficient reproducibility for both methods of 2-round amplification and standard protocol and that transcription data obtained would be comparable for individual and pooled patient samples.
Both protocols were used to assess global gene expression in pooled and individual trisomy 8 patient samples compared with respective healthy controls. The expression levels of 63 genes were up-regulated and of another 71 genes down-regulated in pooled samples; in the individual CD34 samples, 78 genes were up-regulated and 89 genes were down-regulated compared with healthy controls. Among the pooled and individual trisomy 8 patient samples, there were 115 commonly dysregulated genes, 54 overexpressed and 61 underexpressed. These results suggested high reproducibility and fidelity of gene expression between the 2 protocols. The commonly dysregulated genes were further analyzed based on functional category.
Overall global gene expression profiling identified dysregulated expression in 5.1% (177 genes) of genes in monosomy 7 patients, more than was observed in trisomy 8 (3.2% or 115 genes), compared with healthy controls. Based on expression analysis systematic explorer (EASE) score calculated, which represents the lower bound of all possible jackknife probabilities and has advantages in terms of penalizing the significance of categories supported by few genes,45 these genes were assigned to categories of signal transduction, oncogene and tumor suppressor, cell cycle regulator, cell growth and maintenance, immune and inflammatory response, apoptosis, metabolism, transport, enzyme, and unknown function. A small number of genes were identified as commonly dysregulated in both cytogenetically abnormal types of MDS. Genes involved in immune and inflammatory roles were frequently found to be overexpressed in trisomy 8 patients, whereas cell growth and maintenance genes were underexpressed in monosomy 7, indicating obvious differences in the gene profiles between these 2 diagnoses (Table 3).
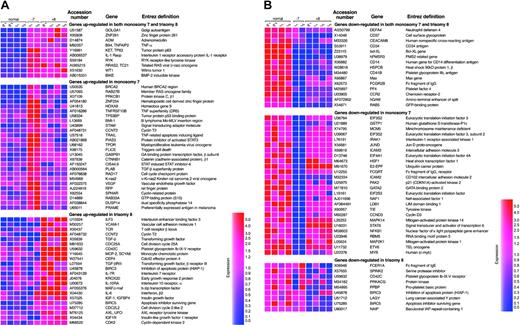
A number of oncogene and tumor suppressor genes were up- or down-regulated in CD34 cells from MDS patients in comparison with controls (Figure 1). A small number of genes were commonly dysregulated in both monosomy 7 and trisomy 8. Of these genes, ZNF26 and RYK receptor-like tyrosine kinase were overexpressed, whereas RAB5 and hPMSR3 showed decreased expression. Furthermore, genes grouped as oncogene and tumor suppressor, many of which have been implicated in the development of solid tumors as well as in hematologic malignancies, were overexpressed in CD34 cells of monosomy 7 and trisomy 8, but each disorder showed a distinct expression profile; expressions of BMI-1, HoxA9, BRCA2, PRAME, and PLAB were high in monosomy 7, and v-abl, MYD18, AXL, and Rad51C were up-regulated in trisomy 8.
Up- or down-regulated gene expression in MDS. Expression levels are shown on the color scale: red indicates overexpression; blue, underexpression. Vertical axes indicate sample resources. Normal indicates healthy control; -7, monosomy 7; +8, trisomy 8; P, pooled sample; and I, individual sample.
Up- or down-regulated gene expression in MDS. Expression levels are shown on the color scale: red indicates overexpression; blue, underexpression. Vertical axes indicate sample resources. Normal indicates healthy control; -7, monosomy 7; +8, trisomy 8; P, pooled sample; and I, individual sample.
Multiple genes (28% and 26% in monosomy 7 and trisomy 8, respectively; EASE score < 10-6 in both monosomy 7 and trisomy 8) involved in signal transduction were up- or down-regulated in MDS. Nine genes were up-regulated and 10 genes down-regulated in both cytogenetic abnormalities including the IL-1 receptor, B-cell growth factor adrenomedullin (ADM), BMP-2–inducible kinase, platelet factor 4 (PF4), platelet glycoprotein IIb (CD41b), cell surface glycoprotein (CD14), neutrophil defensin 4 (DEFA4), chemokine receptor-2 (CCR2), and the IgE receptor; their protein products relate to platelet adhesion and aggregation as well as to neutrophil/monocyte migration and homing. In the up-regulated gene group, ZNF254 (hematopoietic cell–derived zinc finger protein), HOXA9, c-Mpl, and VEGF were abnormal in monosomy 7, as were IGF and IGFR in trisomy 8. Many signal transduction genes were reduced in expression including IRAK1 (IL-1RI–associated kinase-1), MCM5, GATA2, genes of the MAP and EIF families in monosomy 7 and CD42C (platelet glycoprotein Ib-IX-V receptor), PRKACG (protein kinase), and PPBP (proplatelet basic protein) in trisomy 8. These dysregulated genes have been implicated in proliferation and differentiation of hematopoietic cells.
Multiple immune and inflammatory response genes (18%; EASE score < 10-5) were dysregulated in trisomy 8 CD34 cells, whereas only a small number (7.0%) were abnormally expressed in monosomy 7. Most of these genes in trisomy 8 patients were overexpressed including TGF-β, TGF-β receptor III, interferon beta-II, IL-10 RA, IL-6, IL-7 R, MCP-1, ICAM-1, and c-Maf.
Genes (13%) related to apoptosis also demonstrated aberrant expression in MDS patients. TC21(RRAS2), TNF α–induced gene (B94), and a 63-kDa tumor protein (KET) gene that induces apoptosis were up-regulated in both cytogenetic varieties of MDS patients. In contrast, antiapoptotic genes bcl-XL and Max, which block apoptosis by the p53 pathway, were down-regulated. In addition, TRAIL, TRAIL-receptor DR5, and FLICE, which induce cell apoptosis via the Fas/FasL pathway, were overexpressed in CD34 cells of monosomy 7 patients. The inhibitors of apoptosis (IAP) genes BIRC5 and NAIP were down-regulated in trisomy 8.
Genes (20%; EASE score < 10-5) involved in cell cycle regulation and cell proliferation were also dysregulated in MDS patients. Most of these genes showed elevated expression including cyclin-related protein (SPHAR) and cell cycle checkpoint protein (RAD17) in monosomy 7 and CCNT2, CDC25A, CDC45L, CDC2L2, CDK2, CCNE1, and CEP4 in trisomy 8.
Also, 20% (EASE score < 10-5) of genes related to cell growth and maintenance were mainly underexpressed in monosomy 7 patients, including platelet-derived growth factor (PDGF) and inhibin alpha and beta, which control cell survival and growth.
Confirmation of differential gene expression
Because of the limited amount of RNA available from each patient's bone marrow and the limited numbers of patients with cytogenetically defined MDS, demonstration of reproducibility by repetitive experimentation, or even direct comparison of processing and hybridization using scarce patient material, was not feasible. Instead, we employed the microarray data to generate testable hypotheses concerning individual gene expression, which could be addressed by other assays. To validate the microarray data, we determined RNA expression levels for 10 genes of interest in individual MDS patients using real-time RT-PCR. Relative expression levels initially determined by global analysis were correlated with TaqMan results. RNA from CD34 cells of 8 MDS patients (4 monosomy 7 and 4 trisomy 8; of the 4 monosomy 7 patients, 2 had contributed to the previous GeneChip experiments; Table 1) was used for validation studies. We chose genes up-regulated in both monosomy 7 and trisomy 8 (B94), in monosomy 7 only (TRAIL, HOXA9, BRCA2, and TPOR), in trisomy 8 only (TGF-β, IL-6, and VCAM1), as well as PF4 and MCM5, which were down-regulated in MDS CD34 cells. The high correlation between the 2 assays for all 10 genes among all MDS patients validated the identification of dysregulated genes (Figure 2A-B). Similar expression patterns of test genes in CD34 cells were found in individual monosomy 7 or trisomy 8 patients (Figure 2C). There were no systematic differences in the real-time PCR data among trisomy 8 samples containing 2% to 10% blasts. The consistency of these data are a strong argument that the pooling strategy was indeed valid as a predictor of gene activity in these syndromes.
Comparison of gene expression levels by real-time RT-PCR and GeneChip. The fold difference expression of each target gene relative to an internal control gene (β-actin) was studied using the 2-ΔΔCT method. The fold changes of gene expression in 10 genes were assessed in CD34 cells obtained from 4 monosomy 7 and 4 trisomy 8 MDS patients. The average fold change of gene expression level from those monosomy 7 (□) and trisomy 8 (▪) patients was calculated as fold change compared with healthy control by both real-time RT-PCR (A) and microarray assay (B). Real-time PCR data were highly correlated to the GeneChip data by linear regression analysis (r = 0.67; P < .016). Similar gene expression patterns of CD34 cells were found in individual with monosomy 7 (○) or trisomy 8 (•) patients (C).
Comparison of gene expression levels by real-time RT-PCR and GeneChip. The fold difference expression of each target gene relative to an internal control gene (β-actin) was studied using the 2-ΔΔCT method. The fold changes of gene expression in 10 genes were assessed in CD34 cells obtained from 4 monosomy 7 and 4 trisomy 8 MDS patients. The average fold change of gene expression level from those monosomy 7 (□) and trisomy 8 (▪) patients was calculated as fold change compared with healthy control by both real-time RT-PCR (A) and microarray assay (B). Real-time PCR data were highly correlated to the GeneChip data by linear regression analysis (r = 0.67; P < .016). Similar gene expression patterns of CD34 cells were found in individual with monosomy 7 (○) or trisomy 8 (•) patients (C).
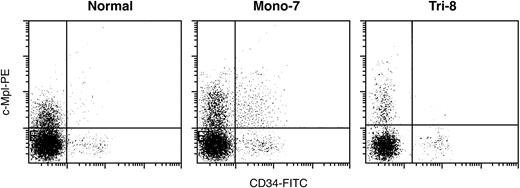
We further verified c-Mpl expression by flow cytometry using fresh BM CD34 cells from MDS patients. Expression of c-Mpl was much higher in CD34 cells from monosomy 7 MDS patients (37% ± 6%; n = 2) than in cells from trisomy 8 MDS patients (14% ± 2%; n = 2; neither had contributed BM cells for the GeneChip experiment) or healthy controls (16% ± 3%; n = 3; Figure 3), again, independently confirming microarray predictions.
Quantitation of c-Mpl expression in CD34 cells by flow cytometry. Expression of c-Mpl was much higher in fresh BM CD34 cells from monosomy 7 MDS patients than in those from trisomy 8 MDS and healthy controls (Normal). Mono-7 indicates MDS patients with monosomy 7; Tri-8, MDS patients with trisomy 8.
Quantitation of c-Mpl expression in CD34 cells by flow cytometry. Expression of c-Mpl was much higher in fresh BM CD34 cells from monosomy 7 MDS patients than in those from trisomy 8 MDS and healthy controls (Normal). Mono-7 indicates MDS patients with monosomy 7; Tri-8, MDS patients with trisomy 8.
Discussion
Oligonucleotide microarrays and gene chips offer powerful tools to measure global gene expression profiles and to describe the “transcriptome” of minimally manipulated healthy and abnormal cells.37,44,46 For human cells, such analyses have been applied to describe differences between normal and malignant cells,35 to delineate stages of malignant evolution,47 to discriminate subsets of disease by genetic rather than morphologic criteria,48 and to develop prognostic “cassettes” of gene activities in order to predict outcomes and drug toxicities.48-50 MDS is an attractive candidate for a genomics strategy for several reasons. First, MDS pathogenesis and pathophysiology are uncertain. Second, repeated attempts at classification reflect considerable nosologic confusion, and even the most current scheme2 depends largely on morphologic criteria. Third, MDS in many patients represents a preleukemic state, affording the possibility of defining early genetic factors of susceptibility for malignant transformation.
Because MDS is not marked by a proliferative marrow process and BM cell populations are heterogeneous, and also due to the considerable diagnostic heterogeneity in MDS case series, an array approach has been limited. To overcome these obstacles, we restricted our study to MDS defined by clear cytogenetic criteria. Because the National Institutes of Health (NIH) is essentially a quaternary referral center, our patient population may not be entirely representative (as selected for features of hematopoietic failure, age and willingness to travel, appropriateness for clinical research protocols, and other factors); confirmation of our results by other investigators with marrow from other MDSs, similarly characterized by their particular aneuploidy, will be necessary, using global array or specific RNA or protein assays.
The cells of interest in bone marrow failure syndromes are available for laboratory study in very limited numbers. In the current experiments, CD34 cells containing the hematopoietic progenitor and stem cell populations were collected and isolated, and, when necessary, pooled samples were employed. Such an approach has been used for CD34 cells from normal BM and blood,38 for scant cell populations in AA,41 and in paroxysmal nocturnal hemoglobinuria,51 and, in our current work, we further sought to confirm the validity of results with comparison to samples from individual patients assessed using the small-sample microarray protocol and for individual genes by gene amplification and flow cytometry. The reliability of the small-sample protocol has been extensively tested by Affymetrix in our experiments41,51 (for example, the correlation coefficient for K562 cells processed by these 2 methods in our laboratory was 0.87 in unpublished experiments preliminary to the present work, A.M., August 2001) and in other laboratories.52-54 In the current experiments, approximately the same proportion of genes were found to be up- or down-regulated in pooled and individual samples and there was good if not perfect identification of the same dysregulated genes in the pooled and individual samples. Nevertheless, the necessary use of pooled samples may conveniently reduce interindividual variation but also risk distortion of the data by an extreme outlying result from one sample. This appeared to be unlikely given the good agreement of the microarray data, intended for purposes of hypothesis generation, with confirmatory testing of individual samples by alternative assays such as PCR (for mRNA expression) and flow cytometry (for protein expression; however, more gene products will need to be analyzed). Additionally, a number of dysregulated genes in our microarray results were previously identified as being abnormally regulated in other experiments and using alternative laboratory methods. A second caution applies to the still tentative nature of functional annotation in drawing conclusions as to pathophysiologic relevance. Like others, we relied on software to assign gene functions to known broad categories. Of course, many genes will have multiple roles in cellular function; many gene functions have been assigned only due to the convenience of current experimental assays and may not reflect a physiologic role, and many, perhaps most, gene functions are unknown or incompletely understood.
The broad pattern of altered gene behavior we observed in the 2 cytogenetically distinct dysplasia syndromes correlated well with the marrow failure nature of trisomy 8 and the preleukemia of monosomy 7. Blast percentages were on average higher in monosomy 7 marrows compared with trisomy BM, and some differences between the 2 syndromes might be related to gene expression patterns uniquely present in myeloblasts. A rigorous fine analysis of such a relationship was not feasible, given the small numbers of patients and the limited variability in blast percentage; for trisomy 8, at least, we saw no obvious differences in the real-time PCR data among samples containing 2% to 10% blasts. Whether certain gene expression patterns correlate with blasts deserves a separate study in MDS, leukemia, or both; regardless, microarrays likely provide more information relevant to the pathophysiology of MDS than does notation of blast numbers, even if such a relationship existed.
By global gene expression analysis, some common genetic characteristics were present for both our monosomy 7 and trisomy 8 MDS patients. Our results can be compared with limited relevant published data relating to gene expression in MDS. In the work of Miyazato et al,55 the goal was to distinguish MDS, especially in older patients, from acute leukemia (such an effort at discrimination of these 2 syndromes in the most recent classification, in which the limit of blast cells in the BM has been reduced to 20%, eliminates a large category of refractory anemia with excess blasts in favor of “lumping” with acute myelogenous leukemia2 ). Fluorescent-labeled cDNA was generated from the RNA of patients and healthy controls and hybridized to custom cDNA microarrays designed for the analysis of cancer-related genes. AC133 (prominin 1), a marker of primitive normal hematopoietic progenitors also frequently expressed on leukemic blasts, was used to isolate cell populations. On comparison of 5 patients with MDS with excess blasts and acute leukemia derived from MDS to 5 patients with new onset acute leukemia, a few genes were identified as putatively “MDS-specific,” including those encoding for mitogen-activated protein kinase kinase (Dlk) (also known as fetal antigen-1 and stromal cell protein-1), Tec protein tyrosine kinase (Tec), and inositol 1,4,5-triphosphate receptor type 1; the MDS patient samples showed considerable heterogeneity in their hybridization patterns, limiting the analysis. In our study, we did not appreciate abnormal expression of these 3 genes in monosomy 7 and trisomy 8 cases, but our patients suffered pancytopenia rather than leukemic transformation. A different strategy was employed by Hofmann et al,56 who compared MDS patients categorized as low risk with those of high risk, according to the International Prognostic Scoring System; RNA was extracted from purified CD34 cells of individual patients' BM and Affymetrix HG-U95Av2 microarrays were hybridized. Even using a stringent 5-fold difference to arbitrarily delineate up- and down-regulated genes compared with normal CD34 cells, a large number of genes were identified as differentially regulated in the MDS patient populations. In comparison of our global results, only a limited number of genes appear to be differentially regulated in our trisomy 8 patients in comparison to the low-risk category patients in Hofmann et al56 and similarly for our monosomy 7 cases compared with their high-risk cases. For example, both our groups found SCYA18 up-regulated in trisomy 8 and low-risk MDS and P63, PF4, and ZNF261 dysregulated in monosomy 7 and trisomy 8 and high-risk MDS. However, the explanation for failure to detect most genes similarly dysregulated almost certainly is secondary to the very heterogeneous patient population in the California series: most of the patients had normal cytogenetics and deletions of chromosomes 5 and 11 were the most common abnormalities; only a single each of their patients had monosomy or trisomy 8, in both as part of a complex aberrant karyotype. In the work of Ueda et al,57 AC133 was again used for cell isolation, and patterns obtained from blast cells of 30 patients with MDS were compared with 2 healthy controls; the chips were designed to assess mainly transcription factors and membrane proteins or proteins that mediate cell signaling or redox potential. They identified 11 “poor prognosis” and 6 “good prognosis” genes that appeared to relate to progression of MDS from a primarily marrow failure state to frank leukemia through the stage of refractory anemia with excess blasts. Remarkably, none of the prognostically important genes implicated by either Hofmann et al56 or Ueda et al57 appeared in both analyses, and we only observed one gene, IL1-R, implicated in progression by the latter laboratory to be up-regulated in monosomy 7 patients. Innovative as were these early attempts at gene profiling in MDS, failure of agreement among them suggests that the usefulness of their results may be limited by the criteria employed for selection of patients and that the anticipated heterogeneity of pathogenetic processes in MDS is reflected in the detailed genetic analysis provided by array approaches.
As have others, we observed several genes involved in normal early hematopoietic cell proliferation to be up-regulated in CD34 cells of MDS patients, including zinc finger protein 261 (ZNF261) and B94, which is also in blast cells from acute myeloid leukemia.58 Conversely, we observed down-regulation of several genes annotated as antiapoptotic genes, such as Bcl-XL and Max, in both monosomy 7 and trisomy 8 MDS patients, consistent with resistance to apoptosis among some CD34 cells in MDS. Aberrant activity of these genes, important for cell proliferation and survival, may be functionally linked to the obvious abnormalities in cell proliferation, differentiation arrest, trilineage dysplasia, and leukemic transformation typical of MDS. Other genes that we observed to be dysregulated in both monosomy 7 and trisomy 8 may relate to abnormal function of mature progeny. PF4, which regulates the proliferation of megakaryocytes and the activity of platelets, was down-regulated (consistent with Hofmann et al's results in high-risk MDS56 ). Several other down-regulated genes encode proteins that protect cells from damage or regulate functions of platelets, neutrophils, monocytes, and lymphocytes including CD41b, DEFA4, CCR2, and CD14. If extrapolation of genetic dysregulation in progenitor cells can also be made to mature progeny, alterations in the differentiation programs may underlie the neutrophil and platelet qualitative dysfunctions that exaggerate the clinical consequences of otherwise moderate degrees of neutropenia and thrombocytopenia in MDS.
The gene expression patterns for MDS defined by trisomy 8 or monosomy 7 were very different from each other. Trisomy 8 MDS was notable for overexpression of immune and inflammatory genes such as TGF-β, TGF-β receptor, IL-10, IL-7R, VCAM-1, and c-Maf; some of these gene products also have roles in cellular proliferation and differentiation and in angiogenesis.59-63 Important cell cycle control and signal transduction genes and oncogenes were also up-regulated, including CCNE1, CDK2, IGF, IGFR1, v-abl, Rad51, and AXL, whereas apoptotic inhibitors (BIRC5 and NAIP) were down-regulated. Abnormal expression of these genes has been observed in many cancers and their functions associated with resulting chromosome instability and increased cell survival in the tumor microenvironment of hypoxia, low pH, and low glucose.64-67 We also compared these patterns to relevant published microarray data. For CD34 cells from patients with MDS and trisomy 8, there were multiple genes similarly dysregulated in comparison to CD34 cells in aplastic anemia.41 For example, in the functional category of immune/inflammatory genes, 6 of 9 genes up-regulated in MDS with trisomy 8 were similarly changed in AA (TGF-β, TGF-βRIII, IL-6, IL-10RA, IL-7R, MCP-2, and VCAM-1); these results are consistent with the similar clinical behavior of these patients, whose pancytopenia frequently responds to immunosuppression and in whom trisomy 8 is associated with a favorable prognosis. In contrast, we found very little in common in the global gene expression patterns between trisomy 8 MDS and acute myeloid leukemia in which trisomy 8 was the unique cytogenetic abnormality.68 Trisomy 8 is a relatively frequent type of aneuploidy but carries a poor prognosis in acute myeloid leukemia; in comparison to leukemia with normal cytogenetics and to normal CD34 bone marrow cells, myeloblasts from trisomy 8 patients showed increased expression of genes located on chromosome 8, indicative of a gene-dosage effect.68 We did not observe such a pattern in MDS with trisomy 8, however. Clinical studies also have suggested differences between trisomy 8 MDS and trisomy 8 acute myeloid leukemia.69 In combination with functional studies from our laboratory,13 the microarray results provide genetic mechanisms for proliferation and survival of cytogenetically abnormal cells in this subtype of MDS, despite evidence for targeting of trisomy 8 cells by T-cell clones, and a partial explanation for pancytopenia and responsiveness to treatment directed at the immune system. Importantly, specific genes dysregulated in trisomy 8 CD34 cells by array have been shown to be functionally active in cell culture and histochemical analyses: CDK2 and c-myc activity would provide a molecular explanation for trisomy 8 cell survival despite triggering of early apoptotic events, leading to peripheral pancytopenia in the setting of bone marrow cellularity, morphologic evidence of dysplasia, and ineffective hematopoiesis.
For monosomy 7 MDS, we were especially interested in dysregulated expression of genes that might figure in the marked predisposition to leukemic transformation conferred by this cytogenetic abnormality. Monosomy 7 CD34 cells overexpressed several known oncogenes, including HOX9A, BRCA2, PRAME, BMI-1, and PLAB, aberrancies of which have been implicated in cell transformation in leukemias and solid tissue cancers,35,37,70-77 as well as poor responses to chemotherapy in hematologic malignancies.78,79 Abnormal expression of 2 oncogenes, HOX9A and PLAB, has been useful in the classification of acute myeloid versus lymphoid leukemias and in monitoring minimal residual disease.37,75 Other genes prominently dysregulated in monosomy 7 cells promote cell proliferation and differentiation, including the cell cycle regulator SPHAR, the DNA replication checkpoint gene Rad17, and the signal transduction gene TPO; in contrast, genes with broadly antagonistic functions were down-regulated, including the cyclin-dependent kinase inhibitor p21, several signal transducers such as GATA2, and members of the mammalian mitogen-activated protein (MAP) family.80-82 In general, the gene expression pattern in monosomy 7 progenitor cells was consistent with functional characteristics of high proliferation and malignant potential.
Comparison of our results to microarray data for acute myeloid leukemia is of interest for the possibility of identifying genes dysregulated early in leukemogenesis, but such an analysis is constrained by the limited data available in published studies. In some reports, the emphasis has been on defining prognostic categories rather than comparison to normal cells,83 and in other papers the focus is on only a few dysregulated genes.68 In one recent global expression analysis of hematopoietic stem cells from Japanese patients, M2 leukemia was compared with leukemia associated with MDS (none of the latter were monosomy 7)84 : of the 52 genes apparently down-regulated in leukemia, only 8 of these genes (EIF-3, EF2, eIF-4, CCNI, H-SP1, PAB1, ORF1, and CCND3) were down-regulated in our study of monosomy 7 in MDS; of the 5 genes up-regulated in acute myeloid leukemia, only one (BAX) was similarly changed in our patients. As did Virtaneva et al68 in acute myeloid leukemia, we also saw low expression of the gene MCM5. These comparisons are informal and limited, however, as our studies of monosomy 7 CD34 cells were performed with normal CD34 cells as controls for gene expression rather than to examine patterns of gene expression in leukemic blasts in comparisons of histologic subtypes or for purposes of gauging survival, responsiveness to chemotherapy, and risk of relapse. The observation of a subset of oncogenes and cell proliferation genes, as well as a number of apoptosis genes (TRAIL, TRAIL-DR, and FLICE; previously recognized to be up-regulated in certain MDS subtypes14,15,18,85-87 ) as dysregulated in monosomy 7, is suggestive of a limited subset of genes altered early in leukemogenesis. Confirmation and extension of this hypothesis will come from both functional studies of novel genes identified in this preleukemic state and in serial microarray experiments performed in individual patients and appropriate cohorts from the time of appearance of the chromosome abnormality through to the development of frank leukemia, studies now feasible for limited tissue samples and underway in our laboratory.
Prepublished online as Blood First Edition Paper, August 17, 2004; DOI 10.1182/blood-2004-01-0103.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.