Abstract
Inherited pyrimidine 5′-nucleotidase type I (P5′N-1) deficiency is the third most common erythrocyte enzymopathy that causes hemolysis. Fourteen different mutations have been identified to date. We have investigated the molecular bases of the disease by studying the biochemical properties of the recombinant wild-type human enzyme and 4 variant proteins (D87V, L131P, N179S, and G230R) bearing missense mutations found in patients affected by nonspherocytic hemolytic anemia. P5′N-1 is a relatively stable protein and has essentially identical catalytic efficiency toward cytidine monophosphate (CMP) and uridine monophosphate (UMP). All investigated mutant proteins display impaired catalytic properties and/or reduced thermostability, providing a rationale for the pathological effects of the mutations. Despite the substantial changes in the kinetic and thermostability parameters, the enzyme activity detected in the red blood cells of patients homozygous for mutations L131P and G230R exhibits moderate alterations. This suggests that P5′N-1 deficiency is compensated, possibly by other nucleotidases or alternative pathways in nucleotide metabolism. Therefore, nucleotidase activity may not be considered a prognostic indicator in patients affected by the enzymopathy. (Blood. 2005;105:3340-3345)
Introduction
Pyrimidine 5′-nucleotidase type I (P5′N-1), also known as uridine monophosphate hydrolase-1 (UMPH-1), is a member of a large functional group of enzymes (EC 3.1.3.5 and 3.1.3.6) that catalyze the dephosphorylation of various nucleoside 5′-monophosphates to their respective nucleosides. To date, at least 7 different human 5′-nucleotidases have been identified with distinctive kinetic properties and subcellular localization.1 P5′N-1 (called also cN-III) is a cytosolic enzyme that is highly expressed in red blood cells (RBCs).1 Its major role is in the catabolism of the pyrimidine nucleotides, uridine monophosphate (UMP) and cytidine monophosphate (CMP), mainly resulting from RNA degradation during erythrocyte maturation.2 P5′N-1 also possesses phosphotranspherase activities specific for pyrimidine nucleotides, suggesting an additional role in nucleotide metabolism.3 The molecular and enzymatic properties determined for the protein purified from RBCs show a 34-kDa monomeric protein with no disulfide bridges and no phosphate content.4,5 The activity is dependent on the presence of magnesium ions, and it is readily inhibited by heavy metals (Cd2+, Cu2+, Hg2+, Ni2+, Pb2+) and thiol-reactive reagents such as p-chloromercuribenzoate.4 The P5′N-1 gene consists of 10 exons and codes for 2 alternatively spliced mRNAs.2 The shorter mRNA corresponds to a 286 amino acid-long protein that has characteristics of P5′N-1 4,6 ; the longer mRNA should code for a polypeptide of 297 amino acids, but the significance of the 11 amino acid addition at the N terminus is unclear and the enzyme is yet to be characterized.5 An additional alternatively spliced form of P5′N-1 mRNA has been recently identified in reticulocytes, raising to 11 the total number of exons.7
P5′N-1 deficiency is an autosomal recessive disorder characterized by hemolytic nonspherocytic anemia, heavy basophilic stippling in RBC peripheral blood smears, and accumulation of pyrimidine nucleotides within erythrocytes.5,8,9 It has been suggested that P5′N-1 deficiency is the third most common RBC enzymopathy—after glucose-6-phosphate dehydrogenase and pyruvate kinase deficiency10 —that causes hemolysis. P5′N-1 deficiency can also be acquired as a result of lead poisoning or oxidative threat.2,9
Fourteen different mutations have been identified at the DNA level to date7 ; 4 of them are missense mutations and do not affect the protein length, and the remaining 10 (nonsense mutations, deletions, insertions, and alterations of the splicing sites) are expected to generate shorter or aberrant forms of the polypeptide. P5′N-1 deficiency is generally associated with mild to moderate hemolytic anemia,7,11 although at present no correlation between the residual P5′N-1 activity and the severity of clinical manifestations has been documented.
The purpose of this study was to elucidate the molecular defects responsible for the hemolytic anemia observed in patients with P5′N-1 deficiency due to missense mutations.
The paper presents the first in-depth biochemical characterization of the wild-type P5′N-1 erythrocyte enzyme and 4 mutants (D87V, L131P, N179S, and G230R) obtained from Escherichia coli cells as recombinant proteins. The comparison of the mutants with the wild-type enzyme allowed us to define the effects of amino acid replacements on stability and kinetic properties of P5′N-1 and to correlate the genotype to phenotype.
Materials and methods
Materials
Avian myeloblastosis virus (AMV) reverse transcriptase was purchased from Roche Diagnostics (Monza, Italy). Restriction enzymes and Taq polymerase were purchased from New England Biolabs (Beverly, MA). Oligonucleotides were synthesized by Invitrogen (Paisley, United Kingdom). Quick Change XL Site-Directed Mutagenesis Kit was from Stratagene (La Jolla, CA). CMP, UMP, isopropyl-β-d-thiogalactopyranoside, and phenylmethylsulfonyl fluoride were from Sigma-Aldrich (St Louis, MO). Other chemicals were reagent grade.
RNA extraction, cDNA synthesis, and subcloning of PCR product
Peripheral blood was collected from a healthy control after obtaining informed consent. Total RNA was isolated from leukocytes using TRIzol (Life Technologies, Paisley, United Kingdom) and reverse transcribed to cDNA using random hexamer primers and AMV reverse transcriptase. The entire P5′N-1 cDNA obtained was amplified by polymerase chain reaction (PCR) (94°C, 40 seconds; 58°C, 40 seconds; 72°C, 90 seconds; 30 cycles) using primers designed according to the cDNA sequence reported by Marinaki et al.5
The forward primer was 5′-GGCGGGGGTGGTGCTGGCTC; the reverse primer was 5′-GGTGGAGAAAAGGAGCTTCCAG. The PCR product was digested with PvuII to abolish interferring pseudogenes,5 purified with GFX PCR DNA and gel Band Purification Kit (Amersham Pharmacia, Piscataway, NJ), and the undigested product was cloned into pCR II-TOPO vector (TA cloning kit; Invitrogen, Leek, The Netherlands). The recombinant plasmid obtained was designated pMI 2. The insert was sequenced in an ABI PRISM 310 genetic analyzer (Applied Biosystems, Foster City, CA) by the Big Dye termination method. Sequencing showed the presence of a silent mutation at codon 81 (TAC > TAT) corresponding to silent polymorphism codon 92 of the 297-amino acid form described by Marinaki et al.5
Construction of expression vectors
The vectors used to express P5′N-1 and its mutant forms were derivatives of pET-23b vector (Novagen, Nottingham, United Kingdom). The recombinant vector coding for the wild-type form was obtained amplifying by PCR (94°C, 15 seconds; 62°C, 20 seconds; 72°C, 45 seconds; 30 cycles) the coding insert present in pMI 2 (see above paragraph, “RNA extraction, cDNA synthesis, and subcloning of PCR product”), restricting, and inserting the PCR product into NdeI/XhoI sites of pET-23b vector. The oligonucleotides used for PCR were 5′-AGTCAAAGGGACTTCATATGATGCCAGAATTCCA (forward primer, with the included NdeI site underlined) and 5′-AGTCAAAGGGACTTCTCGAGTTATAGAATCTTCTGTAAAATAGA (reverse primer, with the included XhoI site underlined). The insert was assessed by DNA sequencing; the recombinant plasmid obtained was designated pEA-1. To obtain the desired P5′N-1 mutants, the pEA-1 was subjected to site-directed mutagenesis with Quick Change XL Site-Directed Mutagenesis Kit. Both sense and antisense mutagenic oligonucleotides were used. The specific primers were 5′-AAATACTACGCTATTGAAGTTGTTCCTGTTCTTACTGTAGAAGAG (forward) and 5′-CTCTTCTACAGTAAGAACAGGAACAACTTCAATAGCGTAGTATTT (reverse) for D87V; 5′-GTGGCAGAATCTGACGTTATGCCCAAAGAAGGATATGAGAATTTC (forward) and 5′-GAAATTCTCATATCCTTCTTTGGGCATAACGTCAGATTCTGCCAC (reverse) for L131P; 5′-CCCAATGTCAAAGTTGTGTCCAGTTTTATGGATTTTGATGAAAC (forward) and 5′-GTTTCATCAAAATCCATAAAACTGGACACAACTTTGACATTGGG (reverse) for N179S; and 5′-CTGGGAGACTCCCAACGAGACTTAAGAATGGC (forward) and 5′-GCCATTCTTAAGTCTCGTTGGGAGTCTCCCAG (reverse) for G230R. The underlined letters indicate the mutated bases. PCR was performed as following: 95°C, 50 seconds; 60°C, 50 seconds; 68°C, 9 minutes; 18 cycles. The appropriate substitutions and the absence of unwanted mutations were confirmed by sequencing the inserts.
Expression and purification
E coli BL21(DE3) pLysS transformed with the desired expression vectors were grown at 37°C in Luria-Bertani (LB) medium containing 100 μg/mL ampicillin and 50 μg/mL chloramphenicol. When the culture optical density at 600 nm reached a value of 0.4 to 1.0, the expression was induced by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM. Expression of soluble protein was obtained by incubating the induced culture at 30°C for 4 to 5 hours. Mutant enzymes D87V and L131P were obtained after an induction of 12 hours at 21°C.
The enzyme was purified by a simple procedure that included an ammonium sulfate fractionation and 3 chromatographic steps. Typically, E coli BL21 (DE3)pLysS-transformed cells obtained from 3 L of culture were collected by centrifugation and suspended in 200 mL buffer A (50 mM sodium phosphate [pH 7.0], 50 mM KCl, 2 mM MgCl2, 1 mM ethylenediaminetetraacetic acid [EDTA], 2 mM β-mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride). The cell suspension was sonicated at 800 W for 6 minutes, centrifuged, and the supernatant subjected to ammonium sulfate fractionation (45% to 70% saturation). The pellet collected by centrifugation was suspended in buffer A, dialyzed, and applied to a 45 × 5 cm diethylaminoethyl (DEAE)-Sepharose (Amersham Biosciences, Uppsala, Sweden) column equilibrated in buffer A. The recombinant protein was eluted by buffer A, concentrated, dialyzed against buffer B (10 mM sodium phosphate [pH 6.5], 50 mM KCl, 2 mM MgCl2, 2 mM β-mercaptoethanol), and applied to a 19 × 2 cm CHT ceramic hydroxyapatite column (Bio-Rad, Hercules, CA). Protein was eluted with a 300 mL 10 to 500 mM sodium phosphate linear gradient, concentrated, and applied to a Superdex-75 column (Amersham Biosciences) equilibrated in buffer B; P5′N-1 was eluted by buffer B. Typically, the yield was about 5 mg of purified enzyme per liter of culture. Protein concentration was determined according to Lowry et al12 using bovine serum albumin as standard. The homogeneity of the protein was assessed by native gel electrophoresis, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and N-terminal sequencing performed by adsorptive biphasic column technology, using an HPG1000 A protein sequenator (Hewlett-Packard, Palo Alto, CA) with the routine 3.0 chemistry method and PTH4M high-performance liquid chromatography (HPLC) method.
Enzyme activity assay
The enzyme activity of P5′N-1 was determined at 37°C by the HPLC-based method according to Amici et al,13 with minor modifications, using a 250 × 4.6 mm Synergi Polar reverse phase (RP) column. The standard reaction mixture contained 50 mM Tris (tris(hydroxymethyl)aminomethane)-HCl (pH 7.5), 1 mM MgCl2, 1 mM dithiothreitol (DTT), and 1 mM CMP in a final volume of 100 μL. One unit is the amount of enzyme catalyzing the formation of 1 μmol of cytidine per minute under conditions here described. The specific activity was expressed in units per milligram of protein.
Kinetic analyses
Enzymatic activity was assayed at 37°C using various concentrations of CMP or UMP under conditions identical to those described in the above subsection, “Enzyme activity assay,” except for substrates. In all cases the reaction was started by adding CMP or UMP, and the enzyme activity was assayed at least at 10 different concentrations of substrate. All measurements were performed in triplicate. The kinetic parameters were determined with the Enzyme Kinetic Module 1.1 (SPSS Science Software, Chicago, IL). The kcat, or turnover number, is the number of catalytic events per second per active site. Km is the substrate concentration at which the reaction velocity is half maximal. kcat/Km is a measure of how efficiently an enzyme converts substrate to product at low substrate concentrations.
Thermal stability assays
All enzymes were diluted to 100 or to 200 μg/mL in 20 mM potassium phosphate (pH 6.5), 1 mM EDTA, and 2 mM β-mercaptoethanol. Fifty-microliter samples were incubated for an hour to a given temperature. Single samples were removed at intervals, immediately chilled, and assayed by the standard protocol. Relative activity was calculated as the percent of the activity of the enzyme before the incubation. t1/2 is the time required by the enzyme to lose 50% of its activity at a given temperature; T50 is the temperature at which the enzyme loses 50% of its activity in 5 minutes.
Circular dichroism (CD) spectropolarimetry
Far-UV circular dichroism (CD) measurements were performed with a Jasco J-700 spectropolarimeter (Jasco-Europe, Cremella, Italy) using a 1 mm path cell. Scans were conducted between 190 and 250 nm at a speed of 20 nm/min with a spectral band width of 2 nm and a sensitivity of 20 millidegrees. CD spectrum measurements were performed at 25°C and 37°C in 10 mM sodium phosphate (pH 6.5), 50 mM KCl, 2 mM MgCl2, and 2 mM β-mercaptoethanol and represent the average of 10 scans. The protein concentration was 4 to 5 μM. The α-helical and β-sheet content was calculated with K2D software14 (http://www.embl-heidelberg.de/~andrade/k2d/).
Results
The molecular bases of P5′N-1 deficiency have been investigated studying the biochemical properties of the wild-type and 4 mutant forms of the enzyme (D87V, L131P, N179S, and G230R). Table 1 summarizes country of origin, age, and hematologic data concerning the patients affected by P5′N-1 deficiency caused by these missense mutations. The specific activity of purified wild-type P5′N-1 was 48 U/mg. The N-terminal analysis of the protein gave a single sequence (MMPEFQKSSV), which perfectly matches the first amino acids of the 286-form of P5′N-1. For this reason, amino acid numbering adopted in this study is referred to this spliced form (TrEMBL accession no. Q9P0P5; the numbering of nucleotides is referred to cDNA reported in GenBank accession no. AF312735). The protein migrated in 12% SDS-PAGE and eluted from analytical gel filtration as a protein of apparent molecular mass of approximately 34 kDa (data not shown). Thus, the recombinant P5′N-1 is a monomer whose molecular mass value is in agreement with that predicted from the amino acid sequence (32.7 kDa) (TrEMBL accession no. Q9P0P5) and identical to the enzyme obtained from erythrocytes.4
Thermal stability of wild-type and mutant P5′N-1 proteins
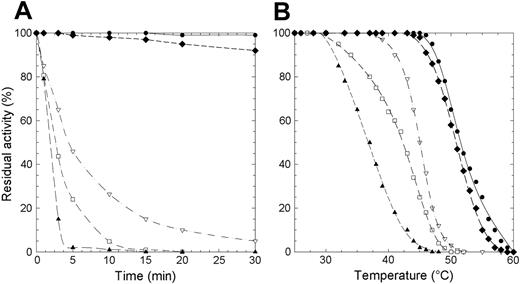
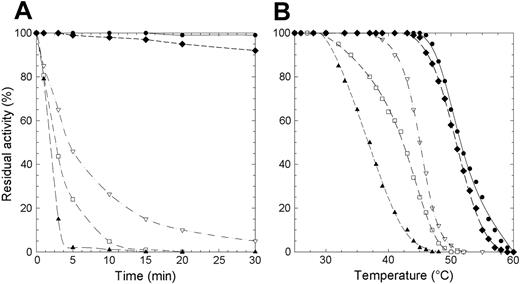
Protein stability is particularly important in mature erythrocytes that lack the machinery to synthesize proteins. To evaluate if the lowered enzyme activity observed in patients homozygous for the mutations here considered was due to a decreased level of P5′N-1 as a consequence of protein instability, we subjected the wild-type and mutant enzymes to thermal stability assays. We assessed that the wild-type enzyme retained full activity after 1 hour of incubation at 46°C. By contrast, at this temperature, 3 of the 4 investigated mutants (D87V, L131P, and G230R) lost more than 50% of their respective activities within the first 5 minutes of incubation (Figure 1A). Incubation of the mutant enzymes at 37°C and subsequent measurement of their residual activity revealed (data not shown) that, even at this temperature, L131P and D87V were very unstable (t1/2 values 5 and 25 minutes, respectively). Finally, a more in-depth thermal analysis performed in a wider range of temperatures (26°C-60°C) (Figure 1B) allowed us to determine the T50 value for all enzymes (T50 is the temperature at which the enzyme loses 50% of its activity in 5 minutes). T50 was 52°C for the wild-type P5′N-1 and N179S mutant, whereas the G230R, D87V, and L131P proteins exhibited significantly lower values (Table 2).
Thermal inactivation of recombinant wild-type and P5′N-1 mutant enzymes. (A) Thermal stability of wild-type and mutant enzymes at 46°C. Aliquots were collected at intervals for measuring residual activity. (B) Plot of the residual activities at 5 minutes versus temperatures. Each enzyme was subjected to heat inactivation in a range of temperature from 26°C to 60°C. After 5 minutes of incubation at a given temperature, the enzyme sample was chilled and the residual activity measured. Residual activity was expressed as percentage of initial activity. • indicates wild type; □, D87V; ▴, L131P; ♦, N179S; ▿, G230R. The solid line differentiates the wild-type from mutants (dashed lines).
Thermal inactivation of recombinant wild-type and P5′N-1 mutant enzymes. (A) Thermal stability of wild-type and mutant enzymes at 46°C. Aliquots were collected at intervals for measuring residual activity. (B) Plot of the residual activities at 5 minutes versus temperatures. Each enzyme was subjected to heat inactivation in a range of temperature from 26°C to 60°C. After 5 minutes of incubation at a given temperature, the enzyme sample was chilled and the residual activity measured. Residual activity was expressed as percentage of initial activity. • indicates wild type; □, D87V; ▴, L131P; ♦, N179S; ▿, G230R. The solid line differentiates the wild-type from mutants (dashed lines).
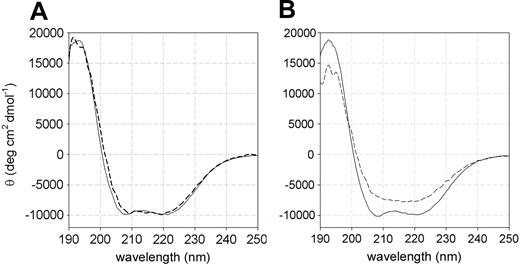
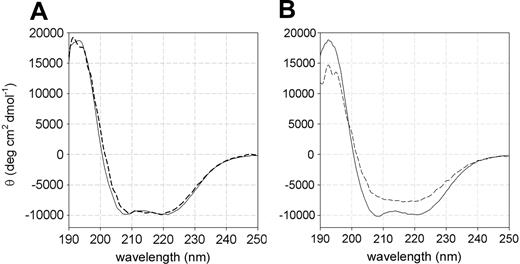
The high instability exhibited by the L131P P5′N-1 with respect to the wild-type enzyme has been further confirmed by far-UV circular dichroism (CD) spectroscopy performed at 25°C and 37°C. At 25°C, both wild-type and L131P mutant forms displayed similar spectra, with an α-helical and β-sheet content of 37% ± 2% and 26% ± 1%, respectively (Figure 2A). Conversely, at 37°C the secondary structure content of the mutant decreased significantly (15% ± 3% for α-helices and 22% ± 2% for β-strands), whereas that of the wild type was unaffected by the temperature increase (Figure 2B).
Far-UV CD spectra of recombinant wild-type and L131P mutant P5′N-1. (A) At 25°C, wild type (solid line) and L131P (dashed line) showed similar spectra with a maximum at 192 nm (18 292 ± 302 deg cm2 dmol-1 and 18 092 ± 212 deg cm2 dmol-1, respectively) and double minima at 208 nm (-9854 ± 128 deg cm2 dmol-1 and -9671 ± 113 deg cm2 dmol-1) and 220 nm (-9793 ± 105 deg cm2 dmol-1 and -9871 ± 101 deg cm2 dmol-1). (B) At 37°C, the wild type (solid line) displayed a spectrum identical to that recorded at 25°C, whereas L131P (dashed line) lost the double minima at 208 and 220 nm and had a reduced maximum at 192 nm (14 252 ± 282 deg cm2 dmol-1).
Far-UV CD spectra of recombinant wild-type and L131P mutant P5′N-1. (A) At 25°C, wild type (solid line) and L131P (dashed line) showed similar spectra with a maximum at 192 nm (18 292 ± 302 deg cm2 dmol-1 and 18 092 ± 212 deg cm2 dmol-1, respectively) and double minima at 208 nm (-9854 ± 128 deg cm2 dmol-1 and -9671 ± 113 deg cm2 dmol-1) and 220 nm (-9793 ± 105 deg cm2 dmol-1 and -9871 ± 101 deg cm2 dmol-1). (B) At 37°C, the wild type (solid line) displayed a spectrum identical to that recorded at 25°C, whereas L131P (dashed line) lost the double minima at 208 and 220 nm and had a reduced maximum at 192 nm (14 252 ± 282 deg cm2 dmol-1).
Catalytic properties of the wild-type P5′N-1 and mutant enzymes
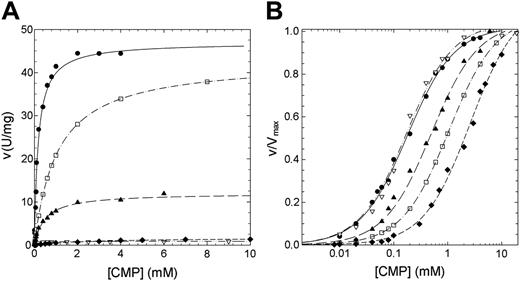
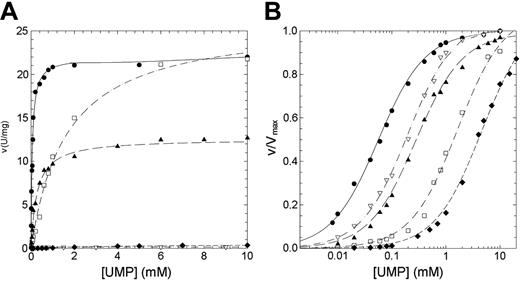
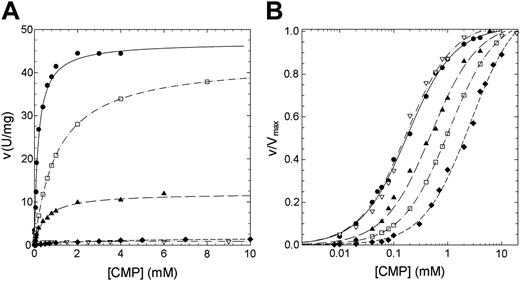
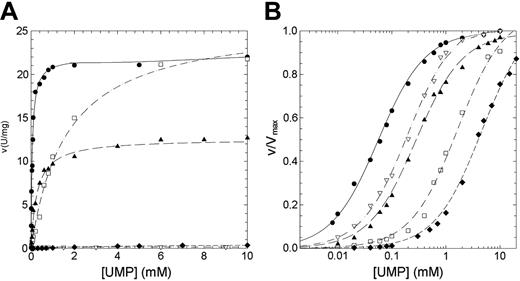
The residual activity values reported for patients affected by nonspherocytic hemolytic anemia due to missense mutations of P5′N-1 range from 2% for N179S up to 65% for G230R mutation (Table 1). Thus, to explain the lowered activity observed in patients homozygous for these mutations, we have undertaken a study of the kinetic properties of the wild-type and mutant P5′N-1 (Figures 3 and 4). Parameters derived from these kinetics are summarized in Table 2. All enzymes exhibited hyperbolic response to both CMP and UMP substrates. For the wild type, the kcat values versus CMP and UMP were 26.14 and 11.98 s-1, respectively; the Km values were 0.18 and 0.056 mM. The catalytic efficiency toward CMP and UMP are therefore very similar (145 and 214 s-1 mM-1, respectively). In all 4 mutants the amino acid replacement causes a reduction of the catalytic efficiency, although to different extent. The kcat values for N179S and G230R enzymes were both heavily altered (more than 1 order of magnitude for N179S and 2 orders for G230R). On the contrary, the kcat value for D87V was unaffected and that for L131P enzyme only slightly reduced. The effect of mutations toward the Kms are highlighted in Figures 3B and 4B. The Km values for G230R and L131P did not change, indicating that either CMP or UMP interacted normally with the active site. On the contrary, the N179S and D87V mutants exhibited large increases in the Km values, particularly versus UMP. The N179S protein also showed a large change in kcat, suggesting that this residue is directly involved in substrate binding and catalysis. Taken all together, these data indicate reductions of the catalytic efficiency of at least a factor of 10 for all investigated mutants.
Steady-state kinetics of recombinant wild-type and mutant P5′N-1 as a function of CMP. (A) Steady-state kinetics of recombinant wild-type and mutant P5′N-1 using CMP as substrate. (B) Semilog plot of the data in panel A to highlight the difference of Km of mutants with respect to the wild-type enzyme. v/Vmax represents the ratio of the initial (v) to the maximal (Vmax) rate of enzyme reaction. Enzyme activity was assayed at 37°C, pH 7.5, as described in “Materials and methods.” • indicates wild type; □, D87V; ▴, L131P; ♦, N179S; ▿, G230R. The solid line differentiates the wild-type from mutants (dashed lines).
Steady-state kinetics of recombinant wild-type and mutant P5′N-1 as a function of CMP. (A) Steady-state kinetics of recombinant wild-type and mutant P5′N-1 using CMP as substrate. (B) Semilog plot of the data in panel A to highlight the difference of Km of mutants with respect to the wild-type enzyme. v/Vmax represents the ratio of the initial (v) to the maximal (Vmax) rate of enzyme reaction. Enzyme activity was assayed at 37°C, pH 7.5, as described in “Materials and methods.” • indicates wild type; □, D87V; ▴, L131P; ♦, N179S; ▿, G230R. The solid line differentiates the wild-type from mutants (dashed lines).
Steady-state kinetics of recombinant wild-type and mutant P5′N-1 as a function of UMP. (A) Steady-state kinetics of recombinant wild-type and mutant P5′N-1 using UMP as substrate. (B) Semilog plot of the data in panel A to highlight the difference of Km of mutants with respect to the wild-type enzyme. Enzyme activity was assayed at 37°C, pH 7.5, as described in “Materials and methods.” • indicates wild type; □, D87V; ▴, L131P; ♦, N179S; ▿, G230R. The solid line differentiates the wild type from mutants (dashed line).
Steady-state kinetics of recombinant wild-type and mutant P5′N-1 as a function of UMP. (A) Steady-state kinetics of recombinant wild-type and mutant P5′N-1 using UMP as substrate. (B) Semilog plot of the data in panel A to highlight the difference of Km of mutants with respect to the wild-type enzyme. Enzyme activity was assayed at 37°C, pH 7.5, as described in “Materials and methods.” • indicates wild type; □, D87V; ▴, L131P; ♦, N179S; ▿, G230R. The solid line differentiates the wild type from mutants (dashed line).
Discussion
Many inherited enzymopathies in red blood cells can result in hemolytic anemia and other abnormalities.15,16 P5′N-1 deficiency is associated to mild to moderate anemia and, in a few cases, learning difficulties.9,17 The diagnosis of P5′N-1 deficiency relies on the reduced specific activity of P5′N-1 and the increased concentrations of pyrimidine nucleotides in the erythrocyte.2 In this study, we have performed a thorough biochemical and molecular analysis of P5′N-1 to provide the basis to establish a correlation between genotype and phenotype. The wild-type erythrocyte P5′N-1 and 4 pathological variants (D87V, L131P, N179S, and G230R) resulting from missense mutations have been purified and characterized. The enzymes have been expressed as recombinant forms to overcome the difficulties of obtaining them in large quantity from blood. By this way, the amount of protein obtained was appropriate for performing an in-depth biochemical characterization of the enzymes. This study is part of a project focused on the molecular biology and biochemistry of inherited hemolytic anemias.18,19
Wild-type P5′N-1
Characterization of the enzyme stability using thermal inactivation assays suggests that P5′N-1 is a relatively stable enzyme with a T50 of 52°C. An important observation is that P5′N-1 can use both CMP and UMP as substrates. The kinetic analysis on the recombinant enzyme shows that catalytic efficiency is essentially the same for the 2 phosphorylated nucleosides. CMP is hydrolyzed with higher turnover number, but the affinity exhibited by the enzyme versus this substrate is lower. Notably, these data partly differ from those obtained using the hemolysate, which indicated a preference for CMP with respect to UMP.3,4 We believe that our kinetic analysis based on highly homogeneous protein provides a more accurate picture of the enzymatic properties of P5′N-1.
D87V mutant form
The mutation D87V (260A > T transversion of P5′N-1 cDNA, GenBank accession no. AF312735) has been identified in a Norwegian family, and it causes a change from a charged to a hydrophobic amino acid.5 The reduced heat stability (t1/2 at 46°C, 3.5 minutes; T50, 10°C lower than the wild type) together with the decreased catalytic efficiency, especially toward UMP (about 25 times), is likely to be the reason for the low activity found in patients. Moreover, with Asp87 being a well-conserved amino acid,5,20 we suggest that this residue might be part of the active site.
L131P mutant form
The mutation L131P (392T > C transition) has been found in a Japanese patient. Recently it has been suggested7 that Leu131 is a functionally important residue for intracellular stability of P5′N-1. By CD analysis and heat inactivation treatments, we proved that the mutant even at physiological temperature showed extreme instability, losing 50% of its initial activity in about 5 minutes. Thus, these findings strongly suggest that the L131P protein is unable to maintain proper folding in vivo, increasing the susceptibility to the degradation by the ubiquitin-proteasome pathway as indicated by Kanno et al.7 It is also notable that the high instability is coupled to a 10-fold reduction of the catalytic efficiency.
N179S mutant form
The mutation N179S (536A > G transition) is a missense mutation that we found in an Italian patient affected by P5′N-1 deficiency.11 The mutation targets an amino acid residue that falls in a region highly conserved among species.5,20 Unlike the other 3 mutants here considered, N179S enzyme turned out to display a stability comparable to the wild-type enzyme. Conversely, it exhibited a greatly impaired kinetic behavior. This suggests that the mutation affects an amino acid residue, which is likely to be directly involved in catalysis and substrate binding.
G230R mutant form
The mutation G230R (688G > C transversion) is the most common missense mutation found in the Japanese population.7 This mutant evidenced a drastic decay in the catalytic activity and a significant reduction in the thermostability. Clearly, position 230 cannot tolerate the large and charged arginine side chain.
Molecular basis of nonspherocytic hemolytic anemia
The general picture emerging from the mutagenesis analyses is that all mutations considered here affect the catalytic properties and/or impair the protein stability. Therefore, these mutations can be expected to produce reductions of the enzyme activity in red blood cells. This is indeed the case for D87V and N179S. Homozygous patients carrying these mutations exhibit nucleotidase residual activities in the range of 2% to 5% (Table 1). Surprisingly, however, patients affected by the mutations L131P and G230S show only moderate alterations in enzyme activity of their red blood cells (Table 1) despite the substantial changes in the kinetic and thermostability parameters. A similar picture has been obtained for other mutations affecting P5′N-1, such as frameshift or nonsense mutations,7,11,21 that are expected to lead to aberrant proteins likely to lack enzymatic activity. Thus, it appears that reductions in P5′N-1 activity caused by mutations can be compensated. This agrees with the observation that P5′N-1-deficient patients exhibit moderate anemia,7,11 except for a few cases.11,17,21
At present we are not able to explain this phenomenon. A possibility is that P5′N-1 deficiency is compensated by other nucleotidases and/or by alternative pathways for nucleotide metabolism. P5′N-2, a deoxyribonucleotidase also expressed in red blood cells that has been shown to be weakly active also toward ribonucleoside 5′-monophosphates,22,23 could partly compensate for the lack of P5′N-1 activity. In addition, the presence in P5′N-1-deficient RBCs of a nonspecific phosphatase having the capability of hydrolyzing nucleoside monophosphates cannot be excluded.24,25 Clinical manifestations are clearly the result of a number of environmental and physiological factors, making it hard to directly correlate the activity of P5′N-1 with the severity of the disease. This suggests that nucleotidase activity may not be considered a prognostic indicator in patients affected by P5′N-1 deficiency.
Prepublished online as Blood First Edition Paper, December 16, 2004; DOI 10.1182/blood-2004-10-3895.
Supported by grants from the University of Pavia Fondi per Ricerca di Ateneo (FAR) and from Ministero dell'Istruzione, dell'Università e della Ricerca Fondo per gli Investimenti della Ricerca di Base (FIRB) and Progetto Genomica Funzionale, legge 449/97).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr R. Tenni for his skillful assistance in the CD analysis and Dr P. Arcidiaco (Centro Grandi Strumenti, Università di Pavia) for performing protein sequencing.