Abstract
Mutations in the ELA2 gene encoding neutrophil elastase (NE) are present in most patients with severe congenital neutropenia (SCN). However, the mechanisms by which these mutations cause neutropenia remain unknown. To investigate the effects of mutant NE expression on granulopoiesis, we used the HL-60 promyelocytic cell line retrovirally transduced with the G185R NE mutant that is associated with a severe SCN phenotype. We show that the mutant enzyme accelerates apoptosis of differentiating but not of proliferating cells. Using metabolic labeling, confocal immunofluorescence microscopy, and immunoblot analysis of subcellular fractions, we also demonstrate that the G185R mutant is abnormally processed and localizes predominantly to the nuclear and plasma membranes rather than to the cytoplasmic compartment observed with the wild-type (WT) enzyme. Expression of the G185R mutant appeared to alter the subcellular distribution and expression of adaptor protein 3 (AP3), which traffics proteins from the trans-Golgi apparatus to the endosome. These observations provide further insight into potential mechanisms by which NE mutations cause neutropenia and suggest that abnormal protein trafficking and accelerated apoptosis of differentiating myeloid cells contribute to the severe SCN phenotype resulting from the G185R mutation.
Introduction
Severe congenital neutropenia (SCN) is a disorder of ineffective neutrophil production (absolute neutrophil count [ANC], 0.5 × 109/L) associated with recurrent infections and an increased risk for acute myelogenous leukemia.1-4 The observation that patients with SCN experience myeloid maturation arrest at the promyelocyte stage in the bone marrow has led to the suggestion that a defect in maturation or the premature death of developing myeloid cells, or both, contributes to the pathogenesis of the disease. Given that most patients respond to recombinant human granulocyte-colony-stimulating factor (G-CSF) with increased ANC (usually more than 1.0 × 109/L),5-7 myeloid maturation signaling appears intact. Thus, accelerated apoptosis seems more likely in SCN, as recently reported for other congenital disorders associated with neutropenia.8
Although heterozygous mutations in the ELA2 gene encoding neutrophil elastase (NE) have been detected in most patients with SCN and cyclic neutropenia (CyN),9-13 it is not yet known how these mutations lead to neutropenia. Nearly 50 different mutations scattered throughout the promoter and coding regions, resulting in base substitutions, missense, deletion, and truncation mutations, have been identified.9-13 In SCN, most of the mutations cluster around exons 4 and 5, corresponding to the carboxy-terminus of the mature enzyme. A severe phenotype with an ANC typically less than 0.1 × 109/L, also associated with an increased risk for leukemic transformation, was shown to result from the substitution of Gly to Arg at position 185 (G185R).14
In this study, we used HL-60 cells retrovirally transduced with the G185R NE mutant to investigate the mechanisms responsible for ineffective neutrophil production in SCN. We show that the G185R mutant is aberrantly processed and targeted intracellularly and that its expression induces premature activation of “death” pathways. Our data are the first to link a specific NE mutation with a pathogenetic mechanism in SCN and to provide further insight into this rare disorder.
Materials and methods
Cloning of WT NE and generation of WT and G185R NE retroviral constructs
A full-length human cDNA clone for NE was isolated from cDNA prepared from the bone marrow cells of appropriately consenting donors using polymerase chain reaction (PCR) and primers corresponding to the published sequence of human NE (NM 001972). Wild-type (WT) NE cDNA was subcloned into the pCDNA 3.1D-V5-HIS-TOPO vector (Invitrogen, Carlsbad, CA), and the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) was used to generate the G185R mutant. PCR was performed with primers spanning the entire coding region of NE to create a 5′ EcoRI and a 3′ XhoI site (forward primer, 5′-GTAGAATTCACCATGACCCTCGGC-3′; reverse primer, 5′-TGCTCGAGTCAGTGGGTCCTGCTGGC-3′). PCR products were subcloned into the pCR4-TOPO vector (Invitrogen), which was subsequently cut with EcoRI and XhoI to release the WT NE and the G185R mutant constructs for cloning into the MIEG3 retroviral vector. The MIEG3 bicistronic vector contains an internal ribosomal entry site followed by the enhanced green fluorescence protein (EGFP)15 and was generously provided by Dr David Williams (University of Cincinnati, OH). The orientation and sequence of all constructs were confirmed by DNA sequencing on an Applied Biosystems 3100 capillary sequencer (Applied Biosystems, Foster City, CA). MLV-MIEG3-NE and MLV-MIEG3-G185R retroviral vectors were used to generate VSV-G pseudotyped recombinant MLV viral particles by transient transfection of 293T cells (CRL-11268; American Type Culture Collection [ATCC], Manassas, VA) using the pHIT 60 and pMD.G plasmids.16 The recombinant VSV-G-pseudotyped MLV vector was produced by transient 3-plasmid transfection of 293T cells, as described previously.17
Transduction of HL-60 cells
HL-60 cells (2 × 105/mL) in 6-well plates were transduced with replication-defective retroviral particles in the presence of 5 mg/mL polybrene (Aldrich, Milwaukee, WI). The plates were then spun at 1000g for 1 hour, transferred to a humidified incubator (5% CO2), and cultured at 37°C for 17 hours. Cells were washed twice to remove polybrene and were resuspended in StemPro-34 serum-free medium (Gibco-BRL, Grand Island, NY). After culture for 48 to 72 hours, the cells were sorted for EGFP-positivity using a Becton Dickinson FACS Vantage SE (Becton Dickinson Immunocytometry Systems, Manassas, VA). Individual clones expressing vector only, exogenous WT NE, or the G185R mutant were isolated by limiting dilution, cell sorting, or both on a BD CloneCyt Plus (Becton Dickinson Immunocytometry Systems) using a single-cell deposition system. Flow cytometry was used to confirm EGFP expression in all expanded clones (96% EGFP-positive) before their use in subsequent assays.
Confirmation of WT and mutant NE expression in transduced HL-60 cells
Total RNA was extracted with Trizol (Invitrogen), and reverse transcription was carried out using SuperScript II RNase H-Reverse Transcriptase (RT) (Invitrogen) and oligo (dT). Reactions lacking RT were included as controls for contaminating DNA. To distinguish endogenously expressed WT NE from ectopically expressed WT NE and to confirm expression of the G185R mutant, mismatched PCR with the primer pair HNE-FOR S173S GCAGGAACCCTGGGATCGCCAGC (forward) and HNE-REV S173S CCGTTGCAGACCAAGGGGAGGCC (reverse) was used to create a novel StuI site in the ectopically expressed NE mRNAs. PCR reactions were performed in the presence of 5% dimethyl sulfoxide (DMSO) and 1.5 mM MgC2 with denaturation for 30 seconds at 95°C, annealing for 30 seconds at 65°C, and extension for 30 seconds at 72°C for 30 cycles, followed by a final extension for 7 minutes. PCR products were digested with StuI (Roche, Mannheim, Germany), separated on 1% agarose gels, and visualized by ethidium bromide staining. The integrity of the cDNAs was confirmed by analysis of hypoxanthine phosphoribosyl transferase I (HPRT) amplicons.
Analysis of cell growth and differentiation
Transduced and untransduced HL-60 cells (2 × 105 cells/mL) were cultured in StemPro-34 serum-free media supplemented with 2 mM glutamine, penicillin, and streptomycin. Cell proliferation and viability were assessed by counting trypan blue-stained cells. To induce neutrophil differentiation, DMSO (1.25% vol/vol) was added to exponentially growing cells. Differentiation was assessed at various time points by examination of Wright-Giemsa-stained cells under light microscopy and by flow cytometric analysis of CD11b expression. For light microscopy, cells were examined at 100 × magnification (1.25 numerical aperture) using an Olympus CK2 microscope (Olympus, Melville, NY) with MagnaFire 2.0 digital microimaging software with Lucis 4.2 image content analysis software (Image Content Technology, Franklin, MA). For detection of CD11b, cells (1 × 106) were washed with ice-cold Hanks balanced salt solution (HBSS) containing 1.0% bovine serum albumin (BSA) and 0.1% sodium azide (HBSS-BSA-NaAz) to minimize nonspecific binding of labeled antibodies and then were incubated with antihuman CD11b-phycoerythrin (PE) (BD PharMingen, San Diego, CA) in HBSS-BSA-NaAz at 4°C in the dark for 1 hour, washed twice with HBSS-BSA-NaAz, fixed in 1% paraformaldehyde, and analyzed on a BD FACSCalibur using Cell Quest software (Becton Dickinson Immunocytometry Systems). To control for nonspecific antibody binding, the cells were also stained with a PE isotype control antibody (mouse immunoglobulin G-1 [IgG-1] R-PE; Caltag Laboratories, Burlingame, CA).
Apoptosis was serially examined in cells differentiating along the neutrophil pathway in response to DMSO or exponentially growing in enriched media devoid of serum, a rich source of protease inhibitors. Apoptosis was monitored using an Annexin V-Biotin Apoptosis Detection Kit (Oncogene Research Products, San Diego, CA) and flow cytometry. Cells were incubated with biotin-conjugated Annexin, washed, incubated with streptavidin Cy5 (Caltag), and analyzed by 3-color flow cytometry for EGFP, propidium iodide (PI), and Annexin V-Cy5 with a minimum of 10 000 events per sample analyzed.
Subcellular fractionation
Subcellular fractions were prepared from cells treated with DMSO for 5 days. The cells were spun down and resuspended in Krebs ringer phosphate (130 mM NaCl, 5 mM KCl, 1.27 mM MgSO4, 0.95 mM CaCl2, 5 mM glucose, 10 mM Na2HPO4/NaH2PO4, pH 7.4) containing 5 mM phenylmethylsulfonyl fluoride (PMSF) for 5 minutes on ice.18 Samples were centrifuged at 400g for 5 minutes, resuspended (108 cells/mL) in disruption buffer (100 mM KCl, 3 mM NaCl, 1 mM ATPNa2, 3.5 mM MgCl2, 10 mM piperazine N,N′-bis2[ethanesulfonic acid] pH 7.2) containing a protease inhibitor cocktail (Roche), then pressurized under nitrogen for 5 minutes at 380 psi in a nitrogen bomb (Parr Instrument, Moline, IL). Cavitates were collected dropwise in disruption buffer with a final concentration of 0.5 mM EGTA (ethyleneglycotetraacetic acid), then centrifuged at 400g for 15 minutes. Postnuclear supernatants were applied to discontinuous Percoll gradients (1.050/1.065/1.09 g/mL) and centrifuged at 37 000g for 30 minutes at 4°C,19 and 6-mL fractions were collected from each gradient by aspiration from the bottoms of the tubes. After the removal of Percoll by centrifugation at 100 000g for 90 minutes at 4°C, the resultant fractions were resuspended in lysis buffer for immunoblot analysis.
Immunoblot analysis
A BCA Kit (Pierce Biosciences, Rockford, IL) was used to determine protein concentrations in whole-cell lysates (WCLs) and subcellular fractions. Proteins (50 μg/lane) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions, transferred to nitrocellulose membranes (Bio-Rad Laboratories, Hercules, CA), and subjected to immunoblot analysis. Blots were initially blocked with 0.1% Tris-buffered saline (TBS) containing 5% Tween and nonfat milk to reduce nonspecific binding, then were incubated with antibodies to human NE (Calbiochem, San Diego, CA; Cedar Lane, Ontario, Canada), the sigma subunit of human AP3 (BD PharMingen), or myeloperoxidase (MPO) (BD PharMingen). Horseradish peroxidase (HRP)-conjugated antibodies to the relevant species used for generating the primary antibodies were used as secondary antibodies, and immunoreactive proteins were visualized by enhanced chemiluminescence (Amersham Pharmacia Biotech, Piscataway, NJ).
Metabolic labeling
Cells were washed with phosphate-buffered saline (PBS) and were resuspended in methionine- and cysteine-free RPMI media (107 cells/mL) and incubated for 30 minutes at 37°C to deplete intracellular methionine and cysteine pools. Cells were then incubated for 30 minutes with [35S]-methionine and [35S]-cysteine (75 μCi [2.775 MBq]/mL), washed once in PBS, and resuspended (107 cells/mL) in RPMI medium replete with methionine and cysteine. At varying times (0-7 hours), aliquots (2 × 107 cells) were removed, pelleted, washed in PBS, and lysed in ice-cold lysis buffer. To analyze newly synthesized NE, the lysates from each time point were counted on a beta counter, and equal counts were incubated with antihuman NE (Cedar Lane) for 1 hour, then with Protein A-agarose slurry (25 μL) (Invitrogen) for 1 hour. Immunoprecipitates were washed in ice-cold lysis buffer and resolved on 12% polyacrylamide gels, dried gels were subjected to autoradiography, and immunoreactive bands were quantified on a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Confocal immunofluorescence microscopy
Cells (5 × 104) were cytospun onto noncharged slides (Becton Dickinson), fixed for 20 minutes in 4% paraformaldehyde, washed again with PBS, and permeabilized with 1% Triton X-100 for 30 minutes at room temperature and washed with TBS containing 0.1% sodium azide. To reduce nonspecific antibody binding, slides were incubated in 1% BSA in TBS for 1 hour at room temperature before incubation with rabbit polyclonal antibody to human NE (Elastin Products, Owensville, MO) overnight at 4°C. Slides were then washed for 30 minutes in TBS containing 0.1% sodium azide and were incubated for 1 hour with goat antirabbit Alexa633 (Molecular Probes, Eugene, OR). Nuclei were stained with Hoechst stain (Molecular Probes) for 15 minutes at room temperature. Slides were washed and air dried before they were mounted on coverslips with Pro-long Antifade mounting media (Molecular Probes), and then they were examined under a Zeiss LSM 510 multiphoton confocal microscope (Zeiss, Oberkochen, Germany) equipped with a c-Apochromat 63 ×/1.2 corr objective. Zeiss LSM5 Image software was used for image processing.
Results
Detection of ectopically expressed WT and G185R NE forms in HL-60 cells
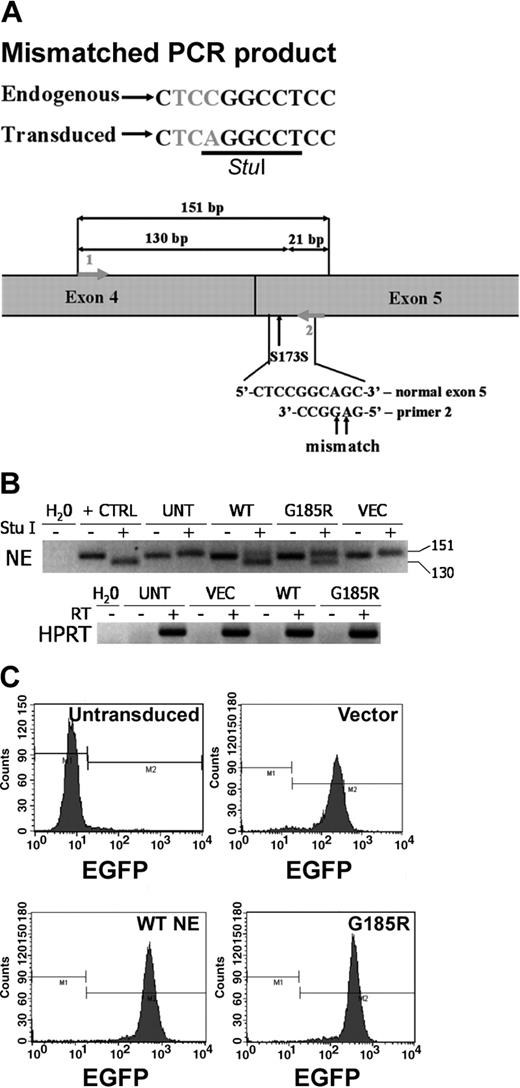
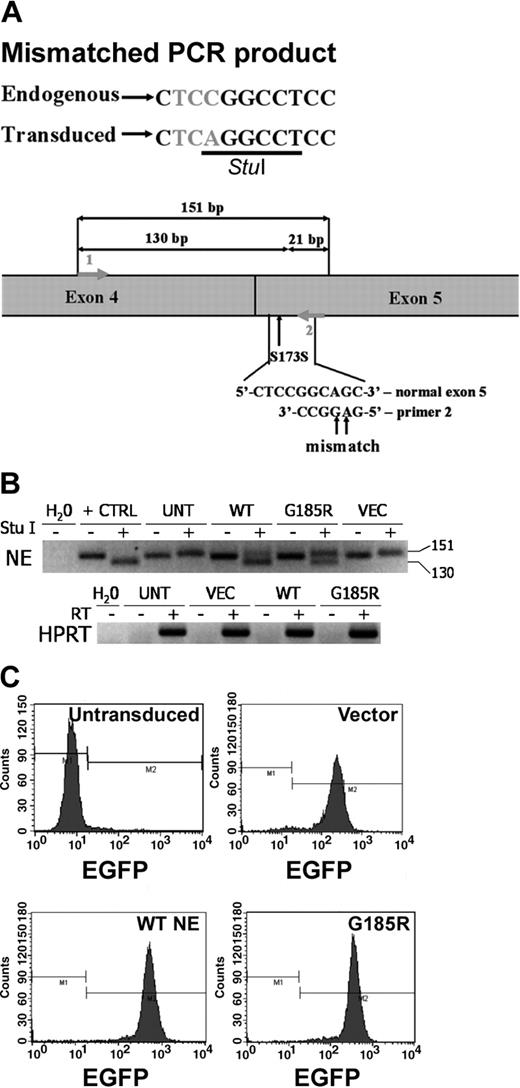
The full-length NE cDNA clone we isolated was found by DNA sequence analysis to contain a C to A substitution in the codon for Ser173. The identical base substitution has previously been identified in healthy persons and has been shown to be a polymorphism (Figure 1A).20 Using this cDNA and site-directed mutagenesis, the G185R NE mutant previously described in patients with SCN was generated.14 The polymorphic WT NE and G185R mutants were cloned into the MIEG3 bicistronic retroviral vector to permit coexpression of the respective NE form and EGFP in transduced HL-60 cells. The polymorphism at Ser173 was used to our advantage to create a StuI restriction site to permit the detection of the transduced WT NE and the G185R NE mutant using mismatched PCR. Because the StuI restriction site is absent in mismatched PCR amplification products arising from endogenously expressed NE mRNA transcripts in HL-60 cells, endogenously expressed WT NE can be distinguished from ectopically expressed WT NE and from ectopically expressed G185R NE. As shown in Figure 1B, StuI digestion generates a 130-base pair (bp) fragment from transduced WT NE transcripts and from G185R transcripts, whereas the endogenously expressed WT NE transcript is detected as a 151-bp fragment. EGFP expression was used for sorting to select positive clones. Clones used in all subsequent assays were more than 96% positive for EGFP expression (Figure 1C).
Expression of WT NE and the G185R mutant in HL-60 cells. (A) Schematic representation of mismatched PCR showing the polymorphism at S173 used to distinguish endogenous NE from untransduced cells (UNT) and ectopically expressed WT NE and the G185R mutant. (B) StuI restriction analysis of reverse transcription-PCR (RT-PCR) products demonstrating the presence of ectopically expressed WT NE or the G185R mutant (130 bp) and endogenously expressed NE (151 bp) in expanded clones. The negative image of an ethidium bromide-stained agarose gel is shown. Water (H2O) indicates negative control; +CTRL, pCDNA3.1 plasmid containing NE as a positive control; VEC, cells transduced with empty MIEG3 vector. (C) Flow cytometric analysis of expanded clones demonstrating more than 96% EGFP-positive cells. M1 and M2 represent negative and positive gates for EGFP fluorescence, respectively.
Expression of WT NE and the G185R mutant in HL-60 cells. (A) Schematic representation of mismatched PCR showing the polymorphism at S173 used to distinguish endogenous NE from untransduced cells (UNT) and ectopically expressed WT NE and the G185R mutant. (B) StuI restriction analysis of reverse transcription-PCR (RT-PCR) products demonstrating the presence of ectopically expressed WT NE or the G185R mutant (130 bp) and endogenously expressed NE (151 bp) in expanded clones. The negative image of an ethidium bromide-stained agarose gel is shown. Water (H2O) indicates negative control; +CTRL, pCDNA3.1 plasmid containing NE as a positive control; VEC, cells transduced with empty MIEG3 vector. (C) Flow cytometric analysis of expanded clones demonstrating more than 96% EGFP-positive cells. M1 and M2 represent negative and positive gates for EGFP fluorescence, respectively.
G185R expression affects differentiating but not proliferating cells
NE is transcribed in developing myeloid cells at the promyelocyte stage of maturation. Given that HL-60 cells exhibit a promyelocyte phenotype and are capable of differentiating into mature neutrophils in response to various stimuli such as DMSO,21 this cell line was used as a model system for investigating the effects of expression of mutant NE forms on the growth and maturation of differentiating myeloid cells.
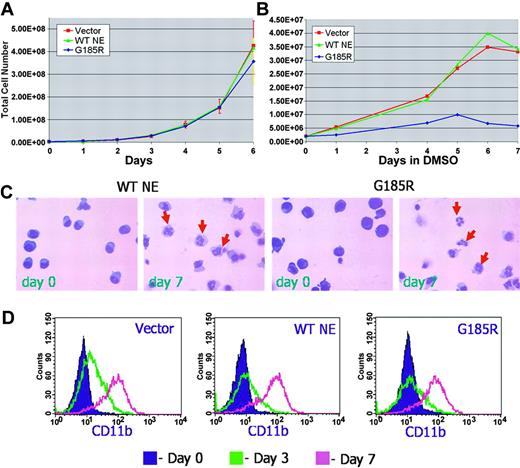
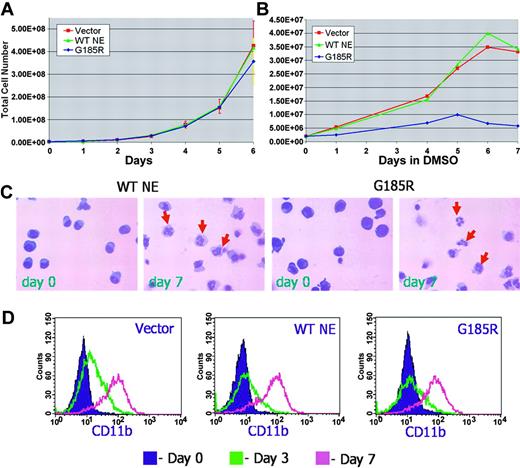
The effect of the G185R mutant on cell proliferation was initially examined in untransduced and transduced HL-60 cells cultured in the absence of differentiation-inducing agents. As shown in Figure 2A, the transduction of HL-60 cells with the empty MIEG3 vector or with WT NE had no effect on their growth. Expression of the G185R mutant also had no effect on HL-60 cell proliferation. Growth curves for cells ectopically expressing WT NE or the G185R mutant appeared virtually superimposable, indicating that expression of the G185R NE mutant did not inhibit the proliferation of cells at the promyelocyte stage of differentiation.
G185R mutant does not inhibit neutrophilic differentiation. Growth curve of (A) proliferating and (B) differentiating HL-60 cells transduced with empty MIEG3 vector (red squares and line), WT NE (green triangles and line), or the G185R mutant (blue diamonds and line). (C) Wright-Giemsa stains of WT NE- and G185R-transduced cells show mature neutrophils (arrows) at day 7 of DMSO treatment. (D) Flow cytometric analysis of CD11b expression in transduced cells after culture in DMSO-containing media (1.25% vol/vol) for day 0 (purple shaded curve), day 3 (green curve), and day 7 (pink curve).
G185R mutant does not inhibit neutrophilic differentiation. Growth curve of (A) proliferating and (B) differentiating HL-60 cells transduced with empty MIEG3 vector (red squares and line), WT NE (green triangles and line), or the G185R mutant (blue diamonds and line). (C) Wright-Giemsa stains of WT NE- and G185R-transduced cells show mature neutrophils (arrows) at day 7 of DMSO treatment. (D) Flow cytometric analysis of CD11b expression in transduced cells after culture in DMSO-containing media (1.25% vol/vol) for day 0 (purple shaded curve), day 3 (green curve), and day 7 (pink curve).
We next examined the effect of the G185R mutant in transduced cells undergoing neutrophilic differentiation in response to DMSO (1.25%). Notably, decreased numbers of terminally differentiated neutrophils consistently arose from cells expressing the G185R mutant (Figure 2B). To confirm that the G185R mutant did not disrupt the program for neutrophilic differentiation and maturation to account for the decreased number of neutrophils observed, transduced cells cultured in DMSO-containing media were serially examined over time. As shown in Figure 2C, typical changes in nuclear morphology indicative of neutrophilic maturation were observed in cells expressing the G185R mutant, similar to the changes observed in cells transduced with WT NE. Similar levels of inducible CD11b expression were also detected in differentiating cells transduced with the G185R mutant, WT NE, and vector only (Figure 2D).
G185R mutant induces accelerated apoptosis of differentiating cells
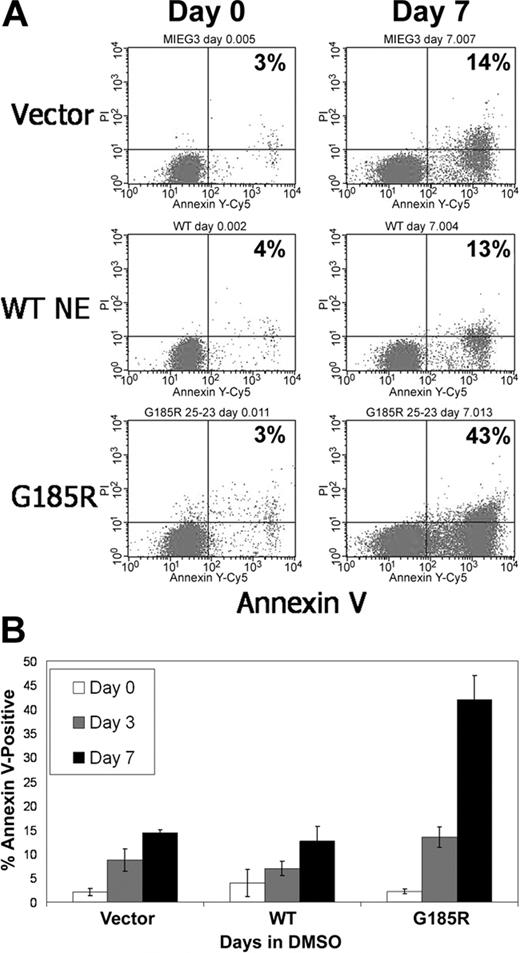
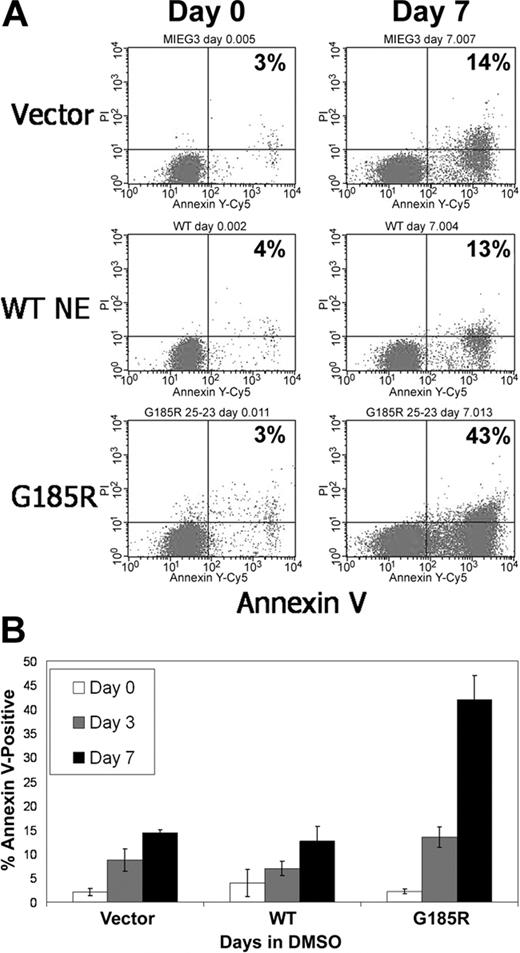
We next investigated whether the reduced numbers of terminally differentiated neutrophils arising from G185R-transduced cells resulted from decreased cell survival. As shown in Figure 3A, exponentially growing G185R-transduced cells showed no evidence of increased apoptosis. Similar numbers of Annexin V-positive cells (3%-4%) were detected when cells transduced with empty vector, WT NE, or the G185R mutant were grown in media devoid of differentiation-inducing agents. However, when cells were cultured in DMSO-containing media to induce neutrophilic differentiation, a significant increase in the number of Annexin V-positive cells was consistently observed in cells expressing the G185R mutant compared with empty vector or with WT-transduced cells. In 15 independent experiments, a greater than 3-fold increase in the number of apoptotic cells was detected in cultures of G185R-transduced cells grown in DMSO-containing media for 7 days compared with vector only- and WT NE-transduced cells (43% vs 13%-14%; Figure 3B).
G185R NE mutant induces accelerated apoptosis of differentiating HL-60 cells. (A) Annexin V staining and flow cytometric analysis showing increased numbers of Annexin V-positive cells in G185R-transduced cells at day 7 of DMSO treatment. Percentages indicate fraction of Annexin V-positive cells. (B) Bar graph depicting the percentage of Annexin V-positive cells in transduced cells cultured in DMSO-containing media for day 0 (□), day 3 (▦), and day 7 (▪). Results shown are representative of 3 different experiments, with standard deviations.
G185R NE mutant induces accelerated apoptosis of differentiating HL-60 cells. (A) Annexin V staining and flow cytometric analysis showing increased numbers of Annexin V-positive cells in G185R-transduced cells at day 7 of DMSO treatment. Percentages indicate fraction of Annexin V-positive cells. (B) Bar graph depicting the percentage of Annexin V-positive cells in transduced cells cultured in DMSO-containing media for day 0 (□), day 3 (▦), and day 7 (▪). Results shown are representative of 3 different experiments, with standard deviations.
Aberrant intracellular processing of the G185R mutant
NE is a highly processed enzyme that is initially synthesized in the ER as pre-pro-neutrophil elastase (PreProNE). A 27-amino acid presequence is cleaved during translation to form pro-neutrophil elastase (ProNE), which is then glycosylated and transported to the Golgi apparatus for trafficking to the plasma membrane or granules. During transit from the Golgi apparatus to granules, the C-terminal extension of glycosylated ProNE and the propeptide are cleaved.22
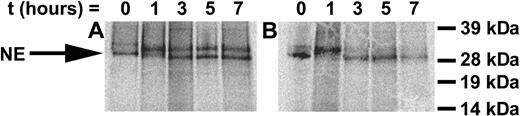
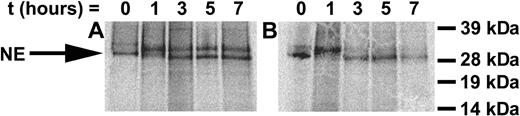
To determine the mechanisms mediating the accelerated apoptosis of differentiating cells expressing the G185R mutant, we investigated whether the mutant enzyme was abnormally processed. Transduced cells were metabolically labeled, and the synthesis and processing of NE were examined in pulse-chase experiments. As shown in Figure 4, at time zero, PreProNE and ProNE are detected in WT NE- and G185R-transduced cells. Subsequent glycosylation of ProNE is observed in WT- and G185R-transduced cells at 1 hour, and by 3 hours the fully processed NE form can be detected in both. In cells ectopically expressing WT NE, the unprocessed glycosylated ProNE form constitutes most of the NE detected throughout the 7-hour time course. In contrast, the fully processed enzyme is the predominant form detected in G185R-expressing cells, which is apparent as early as 3 hours. These results are consistent with aberrant processing of the G185R mutant.
Synthesis and intracellular processing of WT NE and the G185R mutant. Pulse-chase analysis of metabolically labeled cells transduced with (A) WT NE or (B) G185R mutant NE form. NE appears as a doublet of Mr approximately 32 and 29 kDa. Arrow indicates the migration of proenzyme rapidly processed in G185R-transduced cells.
Synthesis and intracellular processing of WT NE and the G185R mutant. Pulse-chase analysis of metabolically labeled cells transduced with (A) WT NE or (B) G185R mutant NE form. NE appears as a doublet of Mr approximately 32 and 29 kDa. Arrow indicates the migration of proenzyme rapidly processed in G185R-transduced cells.
Altered subcellular distribution of the G185R mutant
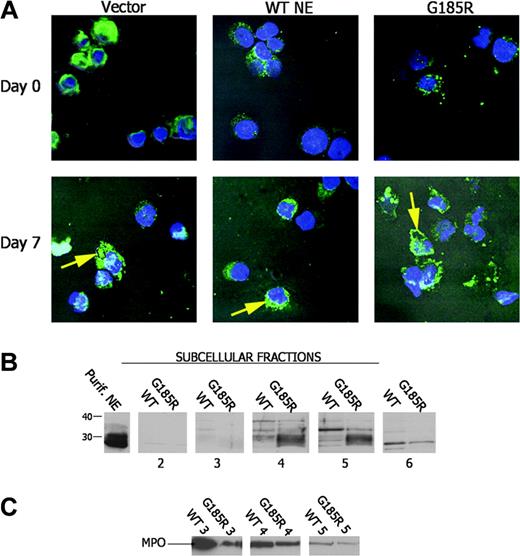
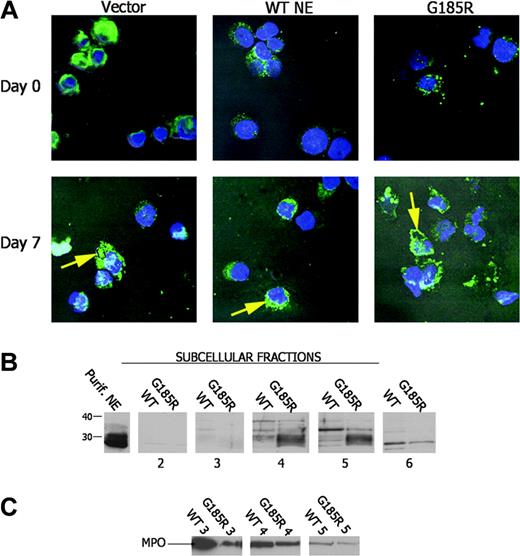
Given that the pulse-chase studies suggested abnormal processing of NE in cells expressing the mutant enzyme, we next examined the subcellular distribution pattern of NE in transduced cells using confocal immunofluorescence microscopy. As shown in Figure 5A, ectopic expression or overexpression of WT NE did not affect subcellular localization of the enzyme. Endogenously and ectopically expressed WT NE were found to be diffusely distributed throughout the cytoplasm. In contrast, in G185R-expressing cells, NE predominantly localized to the nuclear and plasma membranes in a distinct globular pattern consistent with altered subcellular targeting. Targeting to the membrane became increasingly apparent as neutrophilic maturation progressed during DMSO treatment.
Aberrant subcellular localization of NE in G185R cells. (A) Confocal immunofluorescence microscopy shows the subcellular distribution of NE (green) with counterstaining of nuclei (Hoechst, blue). (B) Immunoblot analysis of subcellular fractions from WT NE- and G185R NE-transduced cells blotted with an antibody to NE. NE is seen as a broad low molecular-weight species localizing to fractions 4 to 6 in G185R cells. (C) The blot in panel B was stripped and reblotted with antibody to MPO showing an identical distribution pattern for MPO in WT- and G185R-transduced cells.
Aberrant subcellular localization of NE in G185R cells. (A) Confocal immunofluorescence microscopy shows the subcellular distribution of NE (green) with counterstaining of nuclei (Hoechst, blue). (B) Immunoblot analysis of subcellular fractions from WT NE- and G185R NE-transduced cells blotted with an antibody to NE. NE is seen as a broad low molecular-weight species localizing to fractions 4 to 6 in G185R cells. (C) The blot in panel B was stripped and reblotted with antibody to MPO showing an identical distribution pattern for MPO in WT- and G185R-transduced cells.
To confirm the abnormal distribution pattern of NE observed by confocal microscopy in G185R-transduced cells, subcellular fractions were prepared from cells cultured in DMSO-containing media for 5 days and were analyzed by immunoblot analysis. Subcellular fractions were prepared using nitrogen cavitation and Percoll gradient centrifugation, as previously described by Borregaard.19 As shown in Figure 5B, WT NE predominantly localized to fraction 5, whereas a distinctly different localization pattern was observed in cells transduced with the G185R mutant. In G185R cells, NE localized to fraction 4 and to fraction 5 and appeared as a broader low molecular-weight band compared with the distinct high molecular-weight species observed in cells transduced with the WT form (Figure 5B). The broad low molecular-weight band detected in G185R cells suggests that the fully processed form of NE accumulates in these cells, in agreement with the findings from the pulse-chase analysis.
To confirm that the expression of the G185R mutant did not induce generalized protein mistargeting and to demonstrate that delocalization of NE was specific to the G185R mutant, the subcellular localization of another primary neutrophil granule enzyme was examined. As shown in the immunoblot in Figure 5C, MPO was found to localize to the identical subcellular compartment in cells transduced with either WT NE or the G185R mutant.
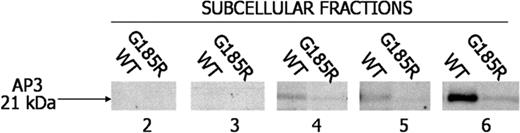
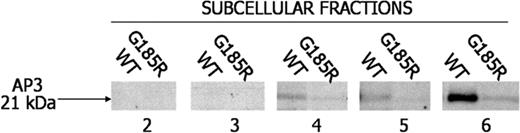
Because it is postulated that AP3 directs intracellular targeting of NE,23 we were interested in examining the interaction of NE with AP3 in the transduced cells. Notably, AP3 was found to localize to fractions 4, 5, and 6 of WT NE-transduced cells but was undetectable or only faintly detectable in the identical fractions from G185R-transduced cells (Figure 6).
Loss of immunologically detectable AP3 in G185R cells. Subcellular fractions from WT NE- and G185R NE-transduced cells were blotted with antibody to AP3. AP3 localizes to fractions 4 to 6 in WT cells but is undetectable in identical fractions from G185R-transduced cells.
Loss of immunologically detectable AP3 in G185R cells. Subcellular fractions from WT NE- and G185R NE-transduced cells were blotted with antibody to AP3. AP3 localizes to fractions 4 to 6 in WT cells but is undetectable in identical fractions from G185R-transduced cells.
Discussion
NE is a neutral serine protease stored in the primary granules of neutrophils and released after their activation.24,25 The expression of NE is transcriptionally regulated and restricted to the promyelocyte stage of granulocyte development.26 NE is initially synthesized as the inactive zymogen PreProNE and is subsequently modified at the amino- and carboxy-termini during granule sorting. The enzyme is activated by proteolysis. The stepwise processing and distinct subcellular localization of NE are postulated to provide a protective mechanism against inappropriate degradation of cellular substrates by the enzyme.24-26
Until recently, a role for NE in hematopoiesis was unknown. Positional cloning to identify the gene(s) involved in the pathogenesis of deficient neutrophil production in SCN and CyN has revealed the unexpected finding of mutations in the ELA2 gene encoding NE in most patients with these disorders. Recent studies by our laboratory27 and El Ouriaghli et al28 demonstrating that NE proteolytically cleaves G-CSF receptor (G-CSFR) and its ligand have provided further evidence for a role of NE in hematopoiesis as a potential negative regulator of granulopoiesis. Despite these important observations, the mechanisms by which mutations in NE lead to neutropenia remain unclear. Initial reports suggested that the mutations in SCN and CyN affected opposite sides of the NE molecule. Using molecular modeling, the mutations in SCN were suggested to cluster on the face of the enzyme opposite its active site.9 However, more recent studies suggest this may not be the case.14 Studies in mice have also not been informative because knock-out mice lacking NE have been found to have normal granulopoiesis, and knock-in mice harboring the V72M mutant NE have normal neutrophil counts with no evidence of maturation arrest in their marrow.29,30
In the current study, we have examined the biologic consequences of expression of the G185R NE mutant on the growth and differentiation of transduced HL-60 cells, a human promyelocytic leukemia cell line. The G185R mutant is of particular interest because it is reported to be associated with a more severe form of SCN (typically ANC less than 0.1 × 109/L) and a high frequency of leukemic transformation.14 Our data provide an explanation for the mechanisms by which a specific mutation in NE in patients with SCN contributes to neutropenia.
We show that heterozygous expression of the G185R mutant in HL-60 cells to reproduce the in vivo situation in patients with SCN has no effect on myeloid cell proliferation but decreases the number of terminally differentiated neutrophils that develop in response to differentiation stimuli. The reduced number of neutrophils arising from G185R-transduced cells was shown to result from an accelerated rate of apoptosis reproducible in more than 15 independent experiments and with multiple clones of transduced cells. However, because Annexin V reportedly binds to activated yet viable B cells31 and to nonapoptotic neutrophils of patients with neutropenia resulting from Barth syndrome,32 results obtained using Annexin staining alone should be interpreted with caution.
Recent observations in patients with mosaicism for normal and mutant NE forms have suggested the cells expressing low levels or no mutant NE protein survive to become mature neutrophils. Notably, we detected no appreciable change in EGFP positivity in G185R-transduced cells during granulocytic differentiation. Because EGFP expression should mirror expression of the transduced NE forms in our model system using the MIEG3 bicistronic vector, we can reasonably infer that the G185R mutant is still expressed in terminally differentiated cells.
We show that the G185R mutant is also aberrantly processed and targeted. Using confocal immunofluorescence microscopy, the mutant was found to predominantly localize to the plasma membrane, with a smaller fraction detected in the region corresponding to the nuclear membrane. In contrast, endogenous and ectopically expressed WT NE localized primarily to the cytoplasmic compartment. Our data also indicate that mislocalization of the G185R mutant does not result from a generalized defect in protein trafficking but is specific to the mutant enzyme. Subcellular analysis of another primary granule enzyme, MPO, in the same cells revealed that only the mutant NE form, not MPO, was mistargeted. These results were independently confirmed by confocal microscopy (MPO data not shown) and by immunoblot analysis.
A mechanism leading to the decreased survival of myeloid cells, as we have observed, would readily explain the maturation arrest observed in the marrow of patients with SCN. Moreover, recent studies of freshly isolated cells from patients with SCN have provided direct in vivo evidence for accelerated apoptosis of myeloid progenitor cells in patients with SCN.33,34 However, in the latter studies, no attempt was made to correlate the presence or absence of an NE mutation with accelerated apoptosis. Heterozygous expression of mutant NE forms from patients with SCN in the rodent RBL-1 basophilic/mast cell line and the murine 32D myeloid cell line was previously reported to produce a modest dominant-negative effect on the protease activity of the WT enzyme, but no effect on cell survival was observed.10 In contrast, transient expression of mutant NE forms in the human U937 monocytic cell line was reported to result in decreased cell survival.11
Collectively, these observations suggest 2 general mechanisms by which mutant forms of NE may affect hematopoiesis. The mutant NE forms may produce a cytotoxic effect when expressed in promyelocytes, resulting in apoptosis of neutrophil precursors at the promyelocyte stage of development. Alternatively, the mutant NE forms may alter the fate of developing myeloid cells to favor monopoiesis over granulopoiesis, possibly through interactions with the Notch receptor pathway.35-37 However, in preliminary experiments, we have demonstrated that G185R cells undergoing monocytic differentiation in response to treatment with phorbol myristate acetate (PMA) also appear to have decreased viability (data not shown). Thus, our data with the G185R mutant support the first hypothesis with the caveat that the mutant is not globally cytotoxic but, rather, induces apoptosis only in differentiating myeloid cells.
Additional insight has recently come from studies by Benson et al23 in which mutations in the beta subunit of the adaptor protein complex 3 (AP3), which traffics proteins from the trans-Golgi apparatus to the endosome, were identified in dogs with CyN. AP3 was shown to interact with NE, and the C-terminal tail of ProNE was shown to inhibit interactions between AP3 and NE. Using computer algorithms, these investigators identified 2 predicted transmembrane domains in NE, suggesting the enzyme exists in soluble and membrane-bound forms. A role for AP3 in directing NE to granules was suggested. Mutations in SCN were postulated to disrupt the AP3 recognition signal, leading to mistrafficking and to excessive routing of the enzyme to the plasma membrane, whereas localization of the mutations to the predicted transmembrane domains in NE was postulated to result in excessive deposition of the enzyme in granules.37
In the current study, we have directly examined the effect of expression of an NE mutant isolated from patients with SCN on subcellular localization of the enzyme. Our data demonstrate increased trafficking of the mutant to the plasma membrane, in agreement with the hypothesis put forth by Benson et al.23 We also find that in addition to the plasma membrane, the G185R mutant localizes to the nuclear membrane. The G185R mutation localizes to one of the predicted transmembrane domains of NE. A Gly to Arg substitution is an unconserved mutation predicted to disrupt the transmembrane domain. The hypothesis put forward by Benson et al23 predicts that this mutation favors processing of the C-terminus of NE, which is precisely what our data demonstrate. Both the pulse-chase and the subcellular fractionation studies demonstrate accumulation of the fully processed form of NE in cells expressing the G185R mutant. However, the consequences of altered processing of the enzyme are not consistent with the prediction of Benson et al,23 who predicted that mutations favoring processing of the C-terminus would result in increased trafficking to granules. Nevertheless, our demonstration of subcellular targeting of the G185R mutant to the plasma membrane is in good agreement with the relationship proposed by Benson et al23 between phenotype and subcellular localization, in which mutations causing SCN target NE to the plasma membrane.
We have also shown that AP3 is undetectable in the same subcellular fractions as NE in G185R-transduced cells. In contrast, we show that NE colocalizes with AP3 in HL-60 cells endogenously expressing the WT enzyme and in HL-60 cells ectopically expressing WT NE. Because AP3 has been shown to interact in vitro with so-called tail-less forms of NE,23 our data demonstrating the G185R mutation favors formation of the fully processed tail-less form of NE, together with the finding that NE does not colocalize with AP3 in G185R-transduced cells, suggest 2 possibilities—either the mutation alters interactions between AP3 and NE, or the G185R mutant itself may, in fact, digest AP3 or one of its subunits, resulting in degradation or down-regulation of the entire AP3 complex. Analysis of the tertiary structure of NE (1HNE) showing that glycine 185 is in proximity to the predicted AP3 (μ3A) recognition sequence (LYPDA)38 supports the first hypothesis because a substitution with arginine at this position could alter the structure of this recognition sequence by steric or electrostatic interactions. Alternatively, expression of the G185R mutant could affect transcription of one of the subunits of AP3, leading to the observed decrease in AP3 protein levels in G185R-transduced cells. Notably, in Hermansky-Pudlak syndrome type 2 (HPS-2)— another human disease associated with neutropenia that results from mutations in the AP3b1 gene encoding the b subunit of AP3—AP3 tetramers are disrupted and the other subunits are degraded.37 As a consequence, NE is directed to the plasma membrane, as in the observations we have made with the G185R mutant.
Although Benson et al23 reported that mutant NE forms were mistargeted in CyN and SCN, no effect of the mutant enzyme forms on survival of differentiating myeloid cells was reported, nor did Benson et al demonstrate an alteration in differentiation favoring monocytopoiesis over neutropoiesis in vitro. Our data, however, not only demonstrate that the G185R mutant is mistargeted to the plasma membrane, they show that expression of this mutant in differentiating HL-60 cells induces premature apoptosis. Although we have not identified the specific cellular targets of the mutant enzyme, one possibility is that the mislocalized NE form aberrantly degrades G-CSF, G-CSFR, or both. We and others have previously shown that both are in vitro substrates for NE and that membrane-targeted NE is known to cleave other membrane proteins.39-42 Alternatively, aberrant interaction of the mutant enzyme with Bcl-2 family members during its processing and subcellular targeting could indirectly contribute to the decreased survival of differentiating cells in patients with SCN or could directly contribute by activation of one or more effecter caspases in a manner similar to the mitochondrial-dependent apoptosis induced by granzyme B in NK cells.43 A mechanism involving decreased Bcl-2 expression was recently reported in patients from the original Kostmann cohort.34 This hypothesis is particularly intriguing because G-CSF has been shown to inhibit cytochrome c release and mitochondrial-dependent apoptosis.44 Such a mechanism would fit well with the observed clinical response of patients with SCN to only supraphysiologic doses of G-CSF. Another possibility is that interactions between the mutant NE form and Notch mediate the apoptosis observed in G185R cells because Notch and NE have been shown to interact, and Notch receptor activation is reported to inhibit apoptosis in erythroleukemia cells.42,45 Perhaps a Notch receptor is important in extending the survival of developing myeloid cells, and the expression of mutant NE forms interferes with Notch-dependent inhibition of apoptosis. Finally, it is also possible that NE interacts with a molecule expressed during differentiation, but not in proliferating cells, such that once cells exit the cell cycle and the differentiation program is initiated, the protein is expressed and available to interact with NE. Future studies should help to clarify the specific substrates of mutant NE forms isolated from patients with SCN and CyN.
Prepublished online as Blood First Edition Paper, January 18, 2005; DOI 10.1182/blood-2004-07-2618.
Supported by National Cancer Institute grants CA75226, CA82859, and CA16058.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.