Abstract
The zebrafish cloche (clo) mutation affects the earliest known step in differentiation of blood and endothelial cells in vertebrates. We established clo/gata1-GFP transgenic line with erythroid-specific green fluorescent protein (GFP) expression, which allowed differentiation of clo and wild-type siblings at the midsomitogenesis stages before morphologically visible phenotypes appeared. To discover novel genes potentially involved in hematopoietic and vascular development, we performed microarray analysis of more than 15 000 zebrafish genes or expressed sequence tags (ESTs) in clo mutant embryos. We isolated the full-length sequences and determined the expression patterns for 8 novel cDNAs that were significantly down-regulated in clo–/– embryos. Dual specificity phosphatase 5 (dusp5), cadherin 5 (cdh5; VE-cadherin), aquaporin 8 (aqp8), adrenomedullin receptor (admr), complement receptor C1qR-like (crl), scavenger receptor class F, member 1 (scarf1), and ETS1-like protein (etsrp) were specifically expressed in the vascular endothelial cells, while retinol binding protein 4 (rbp4) was expressed in the yolk syncytial layer and the hypochord. Further functional studies of these novel genes should help to elucidate critical early steps leading to the formation of vertebrate blood vessels.
Introduction
During embryogenesis, blood and endothelial cell development are closely associated with each other. In mammalian embryos, the blood islands on the yolk sac consist of both endothelial and blood cells.1 Both cell types are thought to emerge from a common precursor, the hemangioblast.2 The zebrafish has recently emerged as an advantageous model organism to study how the evolutionary conserved network of vertebrate blood vessels arises during development. In zebrafish embryos, both endothelial and blood cell precursors originate in the lateral plate mesoderm and during the middle and late somitogenesis stages reside in the region known as the intermediate cell mass (ICM).3 During vasculogenesis, the endothelial cell progenitors (angioblasts) give rise to the major blood vessels of the trunk, the dorsal aorta, and axial vein, as well as the endocardium of the heart.4,5 During subsequent angiogenesis, the axial vessels sprout to form secondary vessels in the trunk region of a zebrafish embryo.6
During the last decade, the molecular mechanisms of blood and blood vessel formation started to be elucidated. Scl/tal1 transcription factor functions at the level of hemangioblast, affecting both hematopoietic and vascular development in both mouse and zebrafish.7-9 Transcription factors including gata2, gata1, cmyb, and transcription factor–interacting proteins such as lmo2 gradually restrict the subpopulation of blood-cell precursors to adopt the erythroid fate (for a review, see Perry and Soreq10 ). Vascular endothelial growth factor A (vegf-a) is required for vascular endothelial development in both mouse and zebrafish and functions through tyrosine kinase receptors vegfr1/flt1 and vegfr2/flk1.11-13 Angiopoietin receptor tyrosine kinase tie2 is important in angiogenesis, while tie1 is critical for endothelial cell integrity and survival.14-16 Zebrafish homologs of tie1 and tie2 are also expressed in vasculature.17 However, complete signal transduction pathways involved in hematopoiesis, vasculogenesis, and angiogenesis are still unknown.
Zebrafish mutagenesis studies resulted in the discovery of multiple mutants affecting different stages of blood or blood vessel formation.18-21 The cloche (clo) mutation affects a very early step in hematopoiesis and vasculogenesis.22-25 cloche mutants lack nearly all blood cells, head and trunk endothelial cells, as well as endocardium. Expression of all known blood or endothelial cell–specific genes, including scl, lmo2, gata1, gata2, flt1, and flt4, is almost completely lost in cloche mutants.7,22-25 The molecular nature of the clo mutation has yet to be identified.
In the current study, we aimed to discover novel genes that could be involved in hematopoiesis, vasculogenesis, and/or angiogenesis. We performed global gene expression analysis of the zebrafish cloche mutant embryos using microarrays. To find novel genes potentially involved in early hematopoiesis and vasculogenesis/angiogenesis, we chose to analyze cloche mutants at the earliest developmental stages possible. We established the clo/gata1-GFP zebrafish line in which green fluorescent protein (GFP) reporter was expressed in hematopoietic cells under control of the gata1 promoter.26 Expression of gata1-GFP in zebrafish carrying the clo mutation allowed detection of homozygous mutant embryos much earlier than when using just morphologic criteria. RNA from cloche mutant embryos and their wild-type siblings was analyzed by oligonucleotide-based microarrays corresponding to more than 15 000 zebrafish genes or expressed sequence tags (ESTs). Expression of 23 genes, 13 of them potentially novel, was down-regulated more than 2-fold in cloche mutant embryos. We isolated cDNA sequences and determined developmental expression patterns for 8 genes, all novel or previously unassociated with blood or blood vessel formation. Of the genes, 7 were expressed specifically in vascular endothelial cells, suggesting their possible involvement in vasculogenesis and/or angiogenesis. The current study describing the detailed sequence and expression analysis of a number of novel vasculature-specific zebrafish genes is well complemented by a parallel study by Weber et al,27 which reports global expression analysis and comparison of several hematopoietic and vascular zebrafish mutants.
Materials and methods
Microarray analysis
cloche homozygous embryos and their siblings were segregated at the 12- to 14-somite stages based on their gata1-GFP expression and frozen on dry ice. As a quality control, a subset of the segregated embryos was kept until later stages, when the morphologic phenotype was apparent. Total RNA from batches of approximately 50 embryos was purified using the RNAquous-4PCR kit (Ambion, Austin, TX). Hybridization and data acquisition were performed at the Microarray Core Facility at Kimmel Cancer Center, Thomas Jefferson University, Philadelphia, PA. The zebrafish gene expression microarrays were built from Compugen (San Jose, CA)/Sigma-Genosys (The Woodlands, TX) oligonucleotide sets covering more than 15 000 unique genes. The 65-mer 5′ amine-modified oligos were chemically immobilized on the 3D hydro gel matrix at the surface of glass slides. The RNA was labeled with biotin by in vitro transcription and hybridized to oligo probes built on the chip. Signals of the probe/target complex were detected by streptavidin–Alexa 647 conjugates. Hybridization was repeated 4 times with the RNA from the mutant embryos and their siblings using HS 4800 hybridization station (Tecan, Maennedorf, Switzerland). The acquired fluorescence intensity values were normalized for each experiment, background was subtracted, and median values from the 4 experiments were used to calculate the fluorescence intensity ratio in mutant embryos versus their siblings. Genes with very low expression levels (< 20% of the medium gene expression value in wild-type embryos) were not analyzed further. We analyzed the data for all other genes that showed more than 2-fold difference in expression between the mutant embryos and their siblings. The normalized fluorescence intensity data from individual hybridization experiments for these genes were screened to remove the outlying data points that showed more than 2-fold deviation from the median value. Expression of all of the remaining genes previously unassociated with blood or blood vessel formation was verified by real-time reverse-transcriptase–polymerase chain reaction (RT-PCR).
Image acquisition and processing
To acquire in-situ hybridization images, embryos younger than 24 hpf were positioned in 1X phosphate-buffered saline (PBS) solution on an agarose-coated Petri dish. An Axiocam digital camera (Zeiss, Oberkochen, Germany) mounted on a Stemi SV11 dissecting microscope (Zeiss) was used to capture images of these embryos. Live embryos and in-situ–stained embryos 24 hpf and older were mounted on a slide in 2% methylcellulose solution (1500 centipoises; Sigma), covered with a coverslip, and imaged with an Axiocam digital camera (Zeiss) mounted on an Axioplan 2 compound microscope (Zeiss) equipped with 5×/0.15 and 10×/0.30 Plan-Neofluar objective lenses (Zeiss). OpenLab 4.0.2 software (Improvision, Lexington, MA) was used to capture all images; Photoshop 7.0 software (Adobe Systems, San Jose, CA) was used for further image processing.
Real-time RT-PCR
Total RNA purified from clo–/– and wild-type embryos as described in “Microarray analysis” was used to generate cDNA using a mix of random hexamers (Roche Applied Science, Indianapolis, IN) and the Powerscript reverse transcriptase (BD Biosciences, San Jose, CA). Real-time PCR was performed using the iCycler iQ Real-Time PCR Detection System (BioRad, Hercules, CA) and the iQ SYBR Green Supermix (Biorad). Relative cDNA amounts were calculated using the iCycler program (BioRad) and normalized to the expression of elongation factor 1a (EF1a). PCR profiles and primer sequences are listed in the Supplemental Tables, available on the Blood website (see the Supplemental Tables link at the top of the online article).
Clone isolation
EST clone MPMGp609A1434Q8 (GenBank sequence ID AI793509, contains complete open reading frame [ORF] and 3′ untranslated region [UTR] of etsrp), clone MPMGp609M1343Q8 (GenBank no. AI965222, contains 3′UTR of cadherin 5), clone MPMGp609B1324Q8 (GenBank no. BI896312, matches to the previously published fli1 mRNA sequence), clone MPMGp609I2222Q8 (GenBank no. AI601685, contains complete ORF and 3′UTR of dusp5), clone MPMGp609J0253Q8 (GenBank no. AJ236884, matches to rbp4 mRNA), clone CHBOp575D0817Q3 (GenBank no. AW077654, matches to spi1 mRNA), and clone WUSMp624M242Q2 (GenBank no. AI384298, contains 3′UTR of an mRNA encoding for a protein similar to scavenger receptor class F, member 1 [SCARF1]) were obtained from RZPD (Berlin, Germany) and sequenced. As the sequence of the clone MPMGp609M1343Q8 did not contain an ORF, 2 kb of additional 5′ sequence was isolated by 2 subsequent rounds of PCR from a post-somitogenesis stage cDNA library (kindly donated by S. C. Ekker). An EST clone MDR1743-7601147 (GenBank no. AW115759, contains complete ORF and 3′UTR of C1qR-like mRNA) was obtained from Open Biosystems (Huntsville, AL). cDNAs encoding admr and aqp8 were isolated by PCR from the postsomitogenesis stage cDNA library using AI626636 and AI105766 sequence information, respectively. The cDNA sequences of aqp8, admr, cdh5, dusp5, crl, and scarf1 have been deposited into the GenBank under the corresponding accession numbers AY732215 to AY732220.
In situ hybridization
In situ hybridization was performed as previously described.28 To synthesize dioxigenin (DIG)–labeled probes, etsrp, cdh5, and rbp4-pSPORT1 constructs were linearized with SalI and transcribed with SP6 RNA polymerase (Promega, Madison, WI). scarf1-pBK-CMV construct was digested with EcoRI and transcribed with T7 RNA polymerase. crl insert was amplified from pME18S-FL3 vector using flanking vector primers with T7 promoter sequence attached, and transcribed with T7 RNA polymerase. admr-pCR4-TOPO and aqp8-pCR4-TOPO constructs were digested with NotI and transcribed with T3 RNA polymerase. scarf1-pBK-CMV construct, digested with XhoI, transcribed with T3 RNA polymerase yielded sense RNA, which was used as a negative control during in situ hybridization.
Results
Microarray analysis
The goal of this study was to identify novel genes that could function in early blood and blood vessel development by performing microarray analysis of the zebrafish clo mutant embryos. In order to identify clo–/– embryos at early stages of development, we established a homozygous gata1-GFP clom39+/– line by crossing clom39 carriers to the hematopoietic cell–specific gata1-GFP line.26 clo–/– embryos could be readily recognized at the 12- to 14-somite stage by the complete absence of fluorescence in the hematopoietic precursor cells (Figure 1). Nonspecific expression of gata1-GFP in the anterior tissues served as an internal control for the expression of the transgene. Segregated clo–/– embryos and their siblings were frozen at the 15-somite stage, followed by RNA purification and global gene expression analysis using the Compugen-based oligonucleotide microarrays (“Materials and methods”). The ratio of the relative gene expression level in the clo–/– embryos versus their siblings was calculated. We further analyzed sequences of 35 genes that differed by 2-fold or more in their expression levels between clo–/– embryos and their siblings. Of the genes, 10 have been previously known to be expressed in hematopoietic or endothelial cells in zebrafish. These include draculin (drl), scl, lmo2, gata1, spi1, klf4, and 3 embryonic globin genes (Table 1, indicated by double-dagger symbol [‡]). The expression of all other genes that were either unknown, novel, or previously unassociated with hematopoiesis or vascular formation was verified using real-time RT-PCR. Of the genes, 13 were confirmed to have significantly reduced expression in clo–/– embryos, and none of the genes could be confirmed to have a significantly increased expression in clo–/– embryos. We obtained gene sequences for 8 selected genes (Table 1, indicated by dagger symbol [†]) and determined their developmental expression patterns.
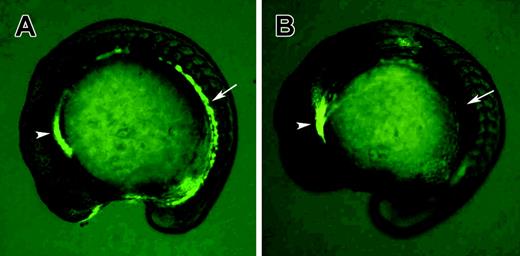
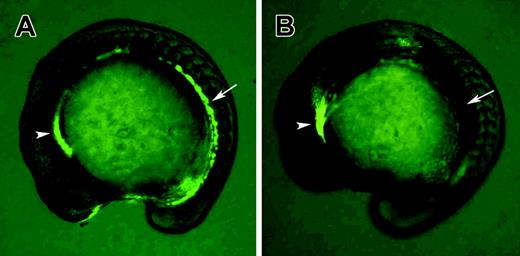
Gata1-GFP homozygous transgenic fish were used to identify clo–/– mutants at the 15-somite stage. (A) Wild-type embryo; (B) clo–/– embryo. Note that GFP fluorescence in hematopoietic precursor cells (arrows) is absent in the clo–/– embryo, while nonspecific GFP expression in the anterior region is not affected (arrowheads).
Gata1-GFP homozygous transgenic fish were used to identify clo–/– mutants at the 15-somite stage. (A) Wild-type embryo; (B) clo–/– embryo. Note that GFP fluorescence in hematopoietic precursor cells (arrows) is absent in the clo–/– embryo, while nonspecific GFP expression in the anterior region is not affected (arrowheads).
Dusp5 encodes a vasculature-specific MAP kinase phosphatase
A cDNA clone corresponding to the EST sequence AI601685 was obtained and fully sequenced. This cDNA contained a complete ORF encoding a zebrafish homolog of the dual specificity phosphatase 5 (DUSP5). Zebrafish DUSP5 protein contains 368 amino acids (AAs) and shows the highest homology (66% similarity, 52% identity) to the rat DUSP5. It contains the Rhodanese homology (RHOD) and the dual specificity phosphatase domains (Figure 2). RHOD domain includes an alpha beta fold, which is found duplicated in Rhodanese protein; it is likely to play a role in protein interactions.29 The dual specificity phosphatases inactivate their target kinases by dephosphorylating both the phosphoserine/threonine and phosphotyrosine residues. DUSP5 is known to inactivate a mitogen-activated protein (MAP) kinase extracellular signal-related kinase 1 (ERK1).30,31 However, DUSP5 has not been directly associated with hematopoietic or vascular development so far.
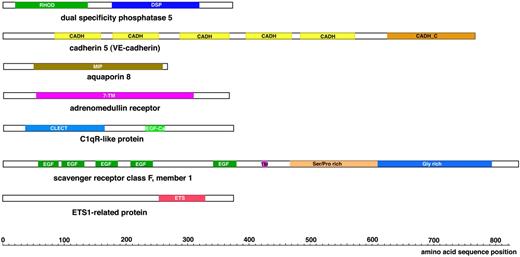
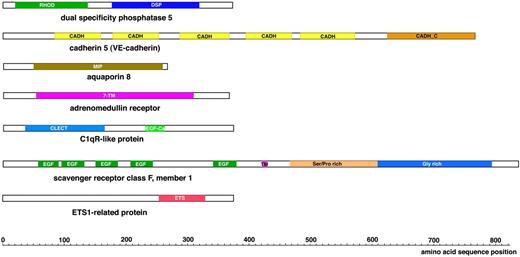
Schematic view of the protein domains present in the novel vasculature-specific zebrafish proteins. RHOD denotes a Rhodanese homology domain; DSP, dual specificity phosphatase; CADH, cadherin repeat; CADH_C, cadherin cytoplasmic domain; 7-TM, 7 transmembrane domains; MIP, major intrinsic protein family domain; CLECT, C-type lectin; EGF-Ca, calcium binding EGF-like homology domain; EGF, EGF-like homology domain; TM, transmembrane domain; and ETS, ETS family DNA binding domain.
Schematic view of the protein domains present in the novel vasculature-specific zebrafish proteins. RHOD denotes a Rhodanese homology domain; DSP, dual specificity phosphatase; CADH, cadherin repeat; CADH_C, cadherin cytoplasmic domain; 7-TM, 7 transmembrane domains; MIP, major intrinsic protein family domain; CLECT, C-type lectin; EGF-Ca, calcium binding EGF-like homology domain; EGF, EGF-like homology domain; TM, transmembrane domain; and ETS, ETS family DNA binding domain.
As determined by in situ hybridization, zebrafish dusp5 is expressed in all vascular endothelial cells and their presumptive precursors at all stages analyzed up to 26 hours after fertilization (hpf) (Figure 3). Vasculature-specific dusp5 expression is down-regulated shortly afterward. At 48 hpf, dusp5 is found in a group of nonvascular cells within the forebrain (Figure 3E-F). Vascular expression of dusp5 was also reported in the parallel microarray study.27
Cadherin 5/vascular endothelial (VE) cadherin
A cDNA clone of 1.27 kb size corresponding to the EST sequence AI965222 was obtained and fully sequenced. No ORF was detected, however. We isolated additional gene fragments by performing 2 rounds of PCR from a postsomitogenesis stage cDNA library (kindly donated by S. C. Ekker), resulting in the isolation of 3.5 kb of contiguous gene sequence. However, no ORF longer than 200 bp was present. The isolated sequence completely matched the genomic sequence located adjacent to cadherin 5 (cdh5; VE cadherin); there was a 70-bp gap between the published cdh5 3′UTR and our sequence.32 It is highly likely that the sequence isolated by us represents the 3′UTR of cdh5.
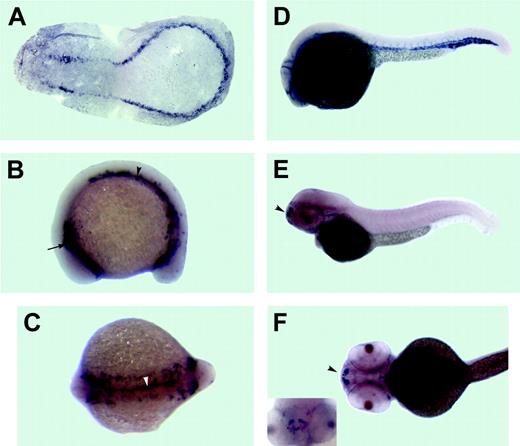
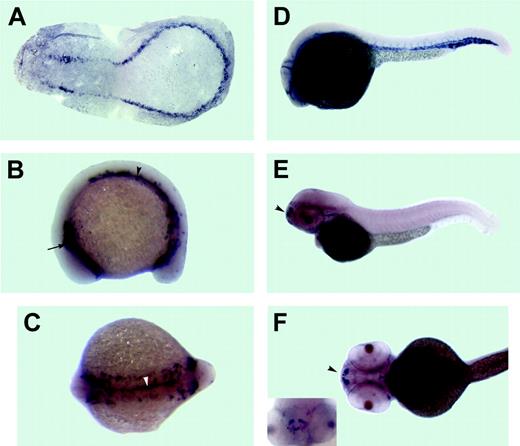
Expression pattern of the dual specificity phosphatase 5 (dusp5) as analyzed by in situ hybridization. Anterior is to the left in all panels. (A)The 3-somite stage. A flat mount of the deyolked embryo. dusp5 is expressed in presumptive endothelial precursor cells, which form 2 bilateral stripes of cells that merge in the posterior. The 15-somite stage: (B) lateral view and (C) dorsal view. dusp5 is expressed in 2 bilateral stripes of cells within the lateral mesoderm in the anterior (arrow in B) and middle/posterior (arrowhead in B) parts of an embryo. In addition, a stripe of dusp5-expressing cells is apparent at the midline and extends through the middle and posterior parts of an embryo (arrowhead in C). (D) The 26-hpf stage. dusp5 expressed in all endothelial cells of the axial, head, and intersomitic vessels. The 52-hpf stage: (E) lateral view, (F) ventral view, and (inset) rostral view. dusp5 is expressed in a subset of nonvascular cells in the forebrain (arrowhead).
Expression pattern of the dual specificity phosphatase 5 (dusp5) as analyzed by in situ hybridization. Anterior is to the left in all panels. (A)The 3-somite stage. A flat mount of the deyolked embryo. dusp5 is expressed in presumptive endothelial precursor cells, which form 2 bilateral stripes of cells that merge in the posterior. The 15-somite stage: (B) lateral view and (C) dorsal view. dusp5 is expressed in 2 bilateral stripes of cells within the lateral mesoderm in the anterior (arrow in B) and middle/posterior (arrowhead in B) parts of an embryo. In addition, a stripe of dusp5-expressing cells is apparent at the midline and extends through the middle and posterior parts of an embryo (arrowhead in C). (D) The 26-hpf stage. dusp5 expressed in all endothelial cells of the axial, head, and intersomitic vessels. The 52-hpf stage: (E) lateral view, (F) ventral view, and (inset) rostral view. dusp5 is expressed in a subset of nonvascular cells in the forebrain (arrowhead).
We determined the expression pattern of the cadherin 5. The RNA was localized to all vascular endothelial cells or their precursors at the 15-somite, 26-hpf, and 52-hpf stages (Figure 4A-C). Strong expression was also observed at 52 hpf in a group of cells within the diencephalon (Figure 4D).
Aquaporin 8
Gene- and vector sequence–specific primers were used to amplify the cDNA corresponding to the EST sequence AI105766 by PCR from the postsomitogenesis stage cDNA library. The isolated fragment contained a complete ORF encoding a 260-AA zebrafish homolog of aquaporin 8 (Aqp8) (54% identity, 74% similarity to rat Aqp8). Aquaporins belong to the major intrinsic protein superfamily and function as membrane channels that selectively transport water. Mammalian aquaporins 8 were found to be expressed in the liver, kidney, pancreas, testis, and gastrointestinal tract.36 However, Aqp8 has not been associated with hematopoietic or vascular development so far.
As analyzed by in situ hybridization, at the 15-somite stage aqp8 was localized to the endothelial cell precursors exclusively in the posterior part of the embryo (Figure 4E). At 26 hpf, aqp8 was abundantly expressed in the dorsal aorta, intersegmental vessels, and caudal vein plexus region (Figure 4F). There was very little expression in the cardinal vein. At 36 hpf, aqp8 expression was observed in the dorsal aorta, intersegmental vessels, dorsal longitudinal anastomotic vessel, and heart endocardium (Figure 4G). At 72 hpf, aqp8 was localized to the dorsal vessel and the pectoral fin bud vessel (Figure 4H).
Adrenomedullin receptor
To isolate cDNA corresponding to the EST sequence AI626636, we used gene- and vector-specific primers for PCR amplification from the postsomitogenesis stage cDNA library. The isolated fragment contained the complete ORF encoding a zebrafish protein of 358 AA that was 33% identical and 45% similar to the human adrenomedullin receptor (ADMR) throughout the entire length of the protein. ADMR contains 7 transmembrane domains and belongs to the family of G-coupled receptors. The ADMR protein may function as a receptor for adrenomedullin, a potent vasodilator peptide.37
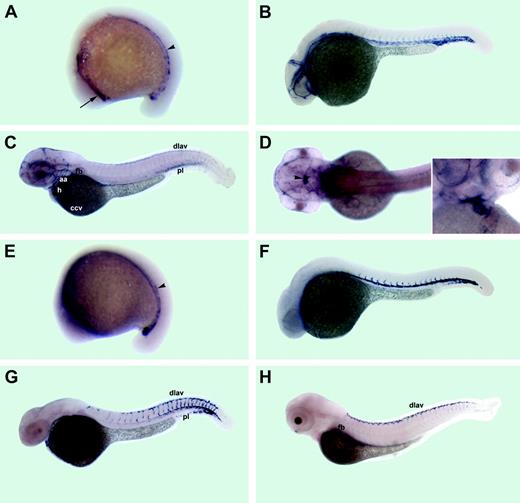
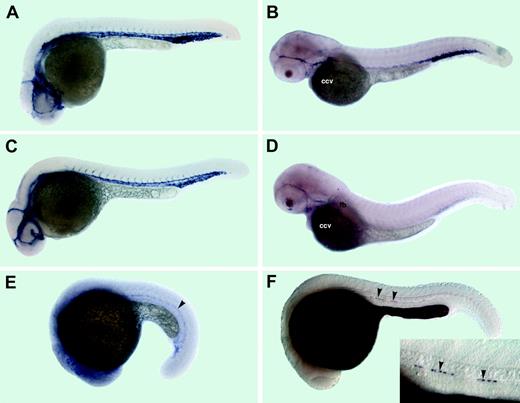
Expression patterns of the cadherin 5 and aquaporin 8. Anterior is to the left in all panels. (A-D) cdh5 expression; (E-H) aqp8 expression. (A) The 15-somite stage. cdh5 mRNA is expressed in 2 distinct anterior (arrow) and middle/posterior (arrowhead) domains within lateral mesoderm. (B) The 26-hpf stage. cdh5 mRNA is expressed in all vascular endothelial cells of axial, intersegmental, and head vessels. (C) The 52-hpf stage, lateral view. cdh5 mRNA is localized to multiple head vessels, aortic arches (aa), pectoral fin bud (fb) vessel, heart (h) endocardium, the common cardinal vein (ccv), the cardinal vein plexus region at the tail (pl), posterior intersegmental vessels, and the dorsal longitudinal anastomotic vessel (dlav). (D) The 52-hpf stage, dorsal view. cdh5 mRNA is expressed in a group of cells within the diencephalon (arrowhead). Inset, ventral view of heart area. cdh5 is expressed in heart endocardium. (E) The 15-somite stage. aqp8 mRNA is expressed within lateral mesoderm in the posterior half on embryo (arrowhead). (F) The 26-hpf stage. aqp8 mRNA is localized to the dorsal aorta, cardinal vein plexus region, and intersegmental vessels. (G) The 36-hpf stage. aqp8 is expressed in the posterior part of the dorsal aorta, intersegmental vessels, the dorsal longitudinal anastomotic vessel (dlav), the cardinal vein plexus region (pl), and heart endocardium. (H) The 72-hpf stage. aqp8 is expressed in the dorsal longitudinal anastomotic vessel (dlav), the pectoral finbud vessel (fb), and faintly in the posterior intersegmental vessels.
Expression patterns of the cadherin 5 and aquaporin 8. Anterior is to the left in all panels. (A-D) cdh5 expression; (E-H) aqp8 expression. (A) The 15-somite stage. cdh5 mRNA is expressed in 2 distinct anterior (arrow) and middle/posterior (arrowhead) domains within lateral mesoderm. (B) The 26-hpf stage. cdh5 mRNA is expressed in all vascular endothelial cells of axial, intersegmental, and head vessels. (C) The 52-hpf stage, lateral view. cdh5 mRNA is localized to multiple head vessels, aortic arches (aa), pectoral fin bud (fb) vessel, heart (h) endocardium, the common cardinal vein (ccv), the cardinal vein plexus region at the tail (pl), posterior intersegmental vessels, and the dorsal longitudinal anastomotic vessel (dlav). (D) The 52-hpf stage, dorsal view. cdh5 mRNA is expressed in a group of cells within the diencephalon (arrowhead). Inset, ventral view of heart area. cdh5 is expressed in heart endocardium. (E) The 15-somite stage. aqp8 mRNA is expressed within lateral mesoderm in the posterior half on embryo (arrowhead). (F) The 26-hpf stage. aqp8 mRNA is localized to the dorsal aorta, cardinal vein plexus region, and intersegmental vessels. (G) The 36-hpf stage. aqp8 is expressed in the posterior part of the dorsal aorta, intersegmental vessels, the dorsal longitudinal anastomotic vessel (dlav), the cardinal vein plexus region (pl), and heart endocardium. (H) The 72-hpf stage. aqp8 is expressed in the dorsal longitudinal anastomotic vessel (dlav), the pectoral finbud vessel (fb), and faintly in the posterior intersegmental vessels.
At the 4-somite stage, admr expression was observed in widely scattered cells above the yolk, possibly yolk syncytial layer (data not shown). At the 15-somite stage, weak staining of 2 stripes in the lateral mesoderm at the anterior part of an embryo was observed (data not shown). At 26 hpf, admr was expressed in all vascular endothelial cells (Figure 5A). At 52 hpf, admr expression was restricted to the main axial vessels, a subset of head vessels, and the common cardinal vein (Figure 5B).
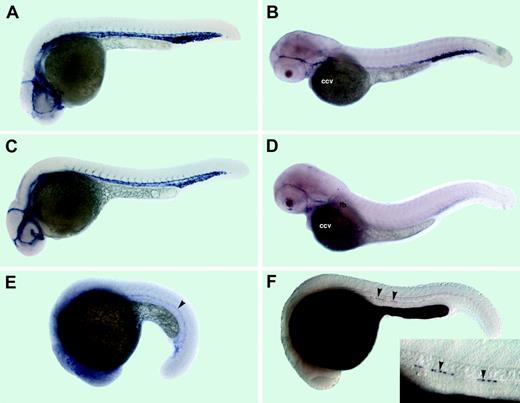
Expression patterns of the adrenomedullin receptor (admr), C1qR-like (crl), scavenger receptor class F (scarf1), and retinol binding protein 4 (rbp4) mRNAs. (A-B) admr expression, (C-D) crl expression, (E) scarf1 expression, and (F) rbp4 expression. (A) The 26-hpf stage. admr is localized to endothelial cells of axial, head, and intersegmental vessels. (B) The 52-hpf stage. admr expression is restricted to the main axial vessels, a subset of head vessels, and the common cardinal vein (ccv). (C) The 26-hpf stage. crl is expressed in endothelial cells of axial, head, and intersegmental vessels. (D) The 52-hpf stage. crl is expressed in the common cardinal vein (ccv), the pectoral fin bud vessel (fb), and a subset of head vessels. (E) The 22-somite stage. scarf1 is localized within lateral mesoderm to the presumptive endothelial cell precursors (arrowhead). (F) The 24-hpf stage. rbp4 mRNA is localized to the yolk syncytial layer and several hypochordal cells (arrowheads); higher magnification in the inset.
Expression patterns of the adrenomedullin receptor (admr), C1qR-like (crl), scavenger receptor class F (scarf1), and retinol binding protein 4 (rbp4) mRNAs. (A-B) admr expression, (C-D) crl expression, (E) scarf1 expression, and (F) rbp4 expression. (A) The 26-hpf stage. admr is localized to endothelial cells of axial, head, and intersegmental vessels. (B) The 52-hpf stage. admr expression is restricted to the main axial vessels, a subset of head vessels, and the common cardinal vein (ccv). (C) The 26-hpf stage. crl is expressed in endothelial cells of axial, head, and intersegmental vessels. (D) The 52-hpf stage. crl is expressed in the common cardinal vein (ccv), the pectoral fin bud vessel (fb), and a subset of head vessels. (E) The 22-somite stage. scarf1 is localized within lateral mesoderm to the presumptive endothelial cell precursors (arrowhead). (F) The 24-hpf stage. rbp4 mRNA is localized to the yolk syncytial layer and several hypochordal cells (arrowheads); higher magnification in the inset.
Complement receptor C1qR-like
We obtained and fully sequenced an EST corresponding to the AW115759 sequence. The cDNA contained a complete ORF encoding a novel zebrafish protein of 368 AA. This protein contained C-type lectin (CTL) and calcium binding epidermal growth factor (EGF)–like homology domains (Figure 2). CTL domains are known to bind carbohydrates, protein ligands, or inorganic surfaces.38 The zebrafish protein displayed low but significant homology (17% identity, 27% similarity to human C1qRp) to the complement component receptor precursor C1qRp family (Figure 6), therefore we named this protein C1qR-like (Crl). A different gene encoding a zebrafish protein highly homologous to C1qRp was found in the zebrafish genome (data not shown), therefore Crl is unlikely to be an ortholog of C1qRp.
No localized expression of crl was observed at the 4-somite stage. At the 18-somite stage, crl was expressed in 2 stripes of the presumptive endothelial cell precursors (data not shown). At 26 hpf, crl was expressed in all vascular endothelial cells (Figure 5C). At 52 hpf, expression of crl was restricted to some head vessels, the common cardinal vein, and the pectoral fin vessel (Figure 5D).
Scavenger receptor class F, member 1
We obtained and fully sequenced an EST corresponding to the AI384298 sequence. The cDNA clone contained only the 3′UTR sequence with no apparent match in the database. We performed a genomic walk using the whole shotgun genomic fragments from the zebrafish genome sequencing project. This way we obtained a partial protein sequence with high homology to the human scavenger receptor class F, member 1 protein (Scarf1), known to be expressed in endothelial cells (Figure 2).39 Indeed, weak expression of the zebrafish scavenger receptor in endothelial cells was observed at the 22-somite and 24-hpf stages (Figure 5E; data not shown).
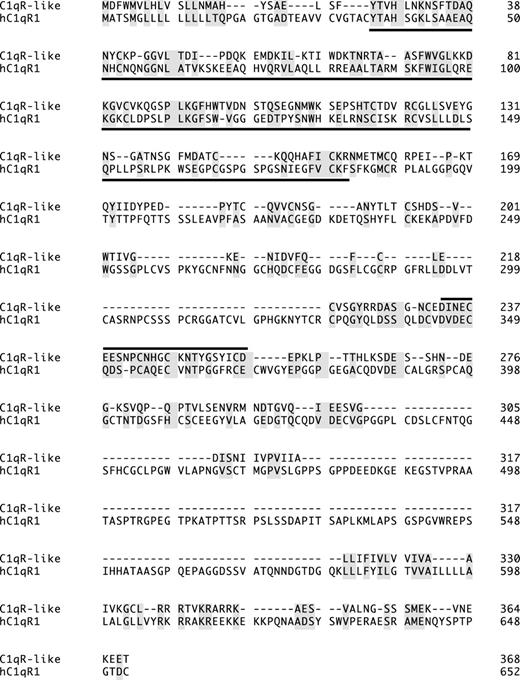
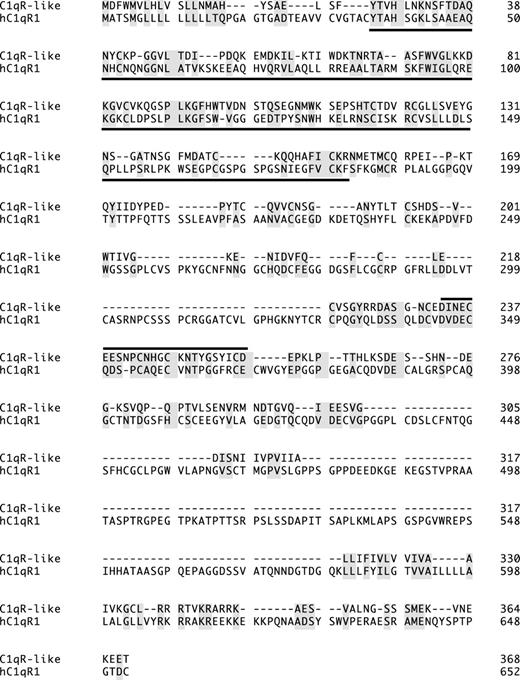
Zebrafish C1qR-like protein is distantly related to the human complement receptor C1qR precursor protein (hC1qR1). Lectin homology domain is underlined; a line above a section indicates EGF-Ca domain; identical and similar amino acids are shaded. GeneWorks 2.5 software (Oxford Molecular Group, Campbell, CA) was used for the alignment.
Zebrafish C1qR-like protein is distantly related to the human complement receptor C1qR precursor protein (hC1qR1). Lectin homology domain is underlined; a line above a section indicates EGF-Ca domain; identical and similar amino acids are shaded. GeneWorks 2.5 software (Oxford Molecular Group, Campbell, CA) was used for the alignment.
Retinol binding protein 4
Sequence AJ236884 corresponded to the zebrafish retinol binding protein 4 (rbp4) mRNA. rbp4 mRNA was localized to the yolk syncytial layer at all stages analyzed (Figure 5F; data not shown). In addition, rbp4 was expressed in several cells within the hypochord at the 22-somite/26-hpf stages (Figure 5G; data not shown). Only several rbp4-expressing hypochordal cells were observed in each embryo; some of the cells were clustered together. Although the hypochord has been known to be involved in vasculogenesis in other vertebrates,40 reduction of rbp4 expression may be a secondary phenotype due to the loss of hematopoietic and endothelial cells in clo–/– mutant embryos.
A novel endothelial cell–specific ETS-domain protein
Expression in clo–/– mutants
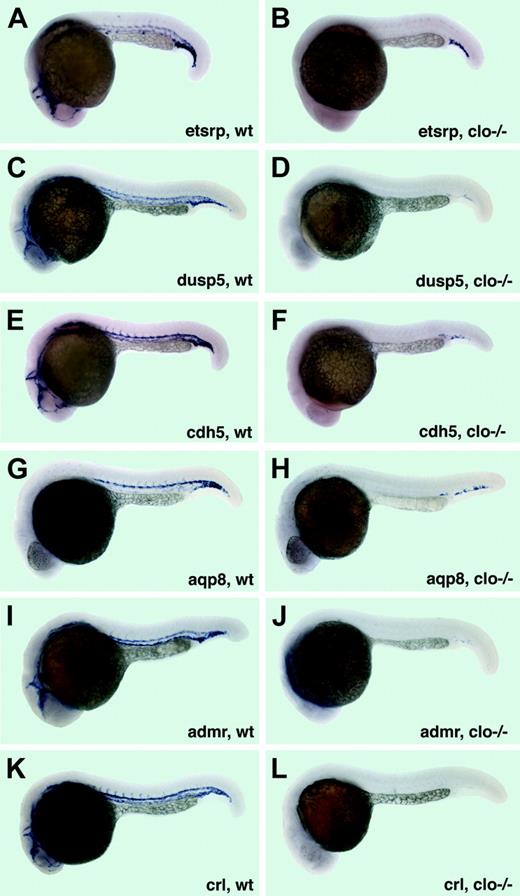
Expression of the 6 novel endothelial-specific genes was analyzed in clo–/– embryos and their siblings and 22- to 24-hpf stages. In all cases, no expression was detected in the clo–/– embryos except for a few cells in the cardinal vein plexus region (Figure 7). A few remaining endothelial cells have been previously observed in this region in clo–/– mutants.23,24 Even in the cardinal vein plexus region, expression of dusp5 and admr was very weak, while crl expression was completely absent in clo–/– embryos (Figure 7D,J,L). In addition, approximately one fourth of the progeny from clo+/– carriers showed no detectable expression of etsrp, dusp5, admr, and crl genes at the 15- to 18-somite stages as analyzed by in situ hybridization (data not shown).
We attempted to determine the chromosomal locations of all novel genes using the zebrafish genome sequencing assembly Zv4. The chromosomal location for all genes that matched to assembled regions is shown in Table 1. Expression of the gene responsible for the cloche phenotype may be down-regulated in clo mutants by a nonsense-mediated RNA decay pathway (if the mutation is a premature STOP codon) or by a negative feedback loop. Previously, cloche mutation has been mapped to the chromosome 13, simple sequence repeats (SSR) marker z1496, close to the telomeric region, which has not been assembled into the chromosomal contig yet.8 Among the sequences analyzed, BI842407 is the only sequence located on chromosome 13; it maps to the 15.3-Mb region, far from either end of the 37.7 Mb chromosome 13. Therefore none of the novel genes that we mapped is likely to be responsible for the cloche phenotype.
Discussion
In this manuscript, we report microarray analysis of the zebrafish cloche mutant embryos that lack hematopoietic and vascular endothelial cells. We report sequences and expression patterns for 8 novel genes that function downstream of cloche. We demonstrate that 7 of them are specifically expressed in vascular endothelial cells at early stages of development. Together with Weber et al,27 this is one of the first reports of global gene expression analysis in an early vertebrate embryos deficient in hematopoietic and vascular endothelial cells. Information revealed by this study will be useful for designing functional studies aimed at elucidating molecular mechanisms underlying the formation of blood and vascular cells during vertebrate embryogenesis.
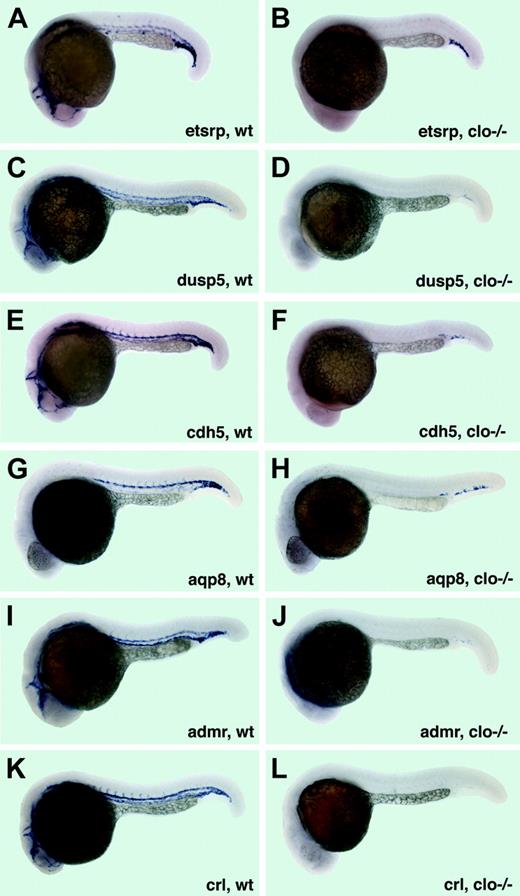
Expression of vascular-specific genes in cloche mutants at 22 to 24 hpf. (A-B) etsrp expression in the vasculature of wild-type siblings (A) and clo–/– embryos (B). Note that only a few etsrp-expressing cells in the cardinal vein plexus region are remaining in clo mutant embryos. (C-D) dusp5 expression in wild-type siblings (C) and clo–/– embryos (D). Note that only a few cells with faint dusp5 expression in the cardinal vein plexus region are remaining in clo mutant embryos. (E-F) cdh5 mRNA expression in wild-type siblings (E) and clo–/– embryos (F). Note that only a few cdh5-expressing cells in the cardinal vein plexus region are remaining in clo mutant embryos. (G-H) aqp8 expression in wild-type siblings (G) and clo–/– embryos (H). Note that only a few aqp8-expressing cells in the cardinal vein plexus region are remaining in clo mutant embryos. (I-J) admr expression in wild-type siblings (I) and clo–/– embryos (J). Note that only a few cells with a very faint admr expression in the cardinal vein plexus region are remaining in clo mutant embryos. (K-L) crl expression in wild-type siblings (K) and clo–/– embryos (L). Note that no crl-expressing cells are remaining in clo mutant embryos.
Expression of vascular-specific genes in cloche mutants at 22 to 24 hpf. (A-B) etsrp expression in the vasculature of wild-type siblings (A) and clo–/– embryos (B). Note that only a few etsrp-expressing cells in the cardinal vein plexus region are remaining in clo mutant embryos. (C-D) dusp5 expression in wild-type siblings (C) and clo–/– embryos (D). Note that only a few cells with faint dusp5 expression in the cardinal vein plexus region are remaining in clo mutant embryos. (E-F) cdh5 mRNA expression in wild-type siblings (E) and clo–/– embryos (F). Note that only a few cdh5-expressing cells in the cardinal vein plexus region are remaining in clo mutant embryos. (G-H) aqp8 expression in wild-type siblings (G) and clo–/– embryos (H). Note that only a few aqp8-expressing cells in the cardinal vein plexus region are remaining in clo mutant embryos. (I-J) admr expression in wild-type siblings (I) and clo–/– embryos (J). Note that only a few cells with a very faint admr expression in the cardinal vein plexus region are remaining in clo mutant embryos. (K-L) crl expression in wild-type siblings (K) and clo–/– embryos (L). Note that no crl-expressing cells are remaining in clo mutant embryos.
In this study, we used one of the first commercially available zebrafish oligonucleotide-based microarrays designed by Sigma-Genosys. The oligos correspond to more than 1300 published mRNA sequences and more than 14 000 unique cDNA sequences. The actual number of unique genes represented in the microarray is probably lower. Among the EST sequences that we obtained and sequenced, 2 of them corresponded to the previously published genes fli1 and spi1 (pu.1). However, they were initially annotated as unique “no match” sequences because of the lack of overlap between the sequenced stretches of ESTs and the published mRNA sequences. Although estimates for a number of transcribed zebrafish genes vary, only a subset of all zebrafish genes is represented in this microarray. A different microarray (Affymetrix, Santa Clara, CA), which contains a partially overlapping set of zebrafish genes, has been used in the parallel study by Weber et al,27 thus making both studies complementary to each other.
By using this microarray, we confirmed the down-regulation of 10 previously known zebrafish hematopoietic and endothelial cell–specific genes in clo mutants. These genes serve as a positive control for the quality of the microarray data. Some of the previously known hematopoietic-specific genes that we did not find include αe2-globin and αe3-globin sequences, which were absent from the microarray. A number of hematopoietic genes are expressed elsewhere in the embryo (eg, gata2 displays blood and neuronal specific expression),3,41 therefore their down-regulation in cloche mutants is not as significant. Yet some genes such as endothelial cell–specific vegfr2/flk124 were missed because the variation between the experiments was too large when compared with a relatively low expression of flk1. We confirmed significant down-regulation in clo mutants for 13 of 24 novel genes by real-time RT-PCR analysis. For 11 genes, RT-PCR analysis showed no significant difference between the clo mutant embryos and their siblings. The confirmation success rate may seem low due to the fact that we chose not to limit ourselves to the microarray sequences that showed highly statistically significant changes in expression. Instead, we verified all of the sequences that showed the largest changes in expression by an independent method of real-time RT-PCR, thus providing independent and definite confirmation of the microarray results. Surprisingly, we did not find a single gene up-regulated more than 2-fold in clo mutant embryos. This is consistent with the idea that the cloche gene regulates hematopoiesis and vasculogenesis mainly by transcriptional activation and not repression.
We found 7 of 8 novel genes analyzed to be expressed in the vasculature system in a developing embryo. Moreover, their expression was transient in most cases, suggesting that these genes may have a regulatory and not a housekeeping function. A little surprisingly, none of the novel genes was expressed in the blood cells. However, among all of the genes that we found down-regulated in clo mutants, 9 were expressed in blood cells and 9 in vasculature (scl was counted for both). It appears that zebrafish blood cells may have been studied more extensively in the past and more genes have been already discovered.
The genes down-regulated in clo mutants include almost exclusively transcription factors and receptors. Transcriptional regulation is one of the major ways of cell fate specification, therefore it is not so surprising to find a large number of transcription factors and cofactors. The small number of intracellular signal transduction molecules (other than transcription factors) that we found may reflect the fact that these signal transducers are not restricted to hematopoietic or vascular endothelial cells and may be present in multiple cell types. Interestingly, we found multiple transmembrane receptors and no secreted signaling molecules down-regulated in clo mutants. It is known that environment plays critical role in specifying and regulating the differentiation and migration of hematopoietic cells and angioblasts. For example, a potent angiogenesis inducer vegf is expressed in the mesoderm close to endothelial cell progenitors,42,43 while its receptor vegfr2/flk1 is restricted to endothelial cells.24 During angiopoietin (Ang), Tie, and semaphorin plexin signaling, tie and plexin receptors are expressed within vasculature,17,44 while ang and semaphorin ligands are expressed in the neighboring mesodermal cells.44,45 Our findings are consistent with hematopoietic cells and angioblasts expressing receptors that transduce signals emanating from the neighboring cells.
The signaling pathways that function during hematopoietic and vascular development are highly conserved. The novel vascular-specific genes described in this study may function in vasculogenesis and/or angiogenesis during vertebrate development. This study will help to decipher multiple signaling pathways that shape the developing hematopoietic and vascular systems.
Prepublished online as Blood First Edition Paper, March 31, 2005; DOI 10.1182/blood-2004-12-4653.
Supported by grants from the National Institutes of Health (grants R01 DK54508 and T32 HL069766).
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.