Abstract
Previously, we demonstrated that enforced activation of signal transducer and activator of transcription 5 (STAT5A) in human cord blood (CB)–derived stem/progenitor cells results in enhanced self-renewal and impaired myelopoiesis. The present study identifies C/EBPα as a critical component that is down-regulated by STAT5. Microarray and reverse transcriptase–polymerase chain reaction (RT-PCR) analysis on STAT5A1*6-transduced CD34+ cells identified C/EBPα as the most prominently down-regulated gene. To determine the cell-biological relevance of these observations, a 4-OHT-inducible C/EBPα-ER protein was co-expressed with the STAT5A1*6 mutant in CB CD34+ cells using a retroviral approach. Re-expression of C/EBPα in STAT5A1*6 cells resulted in a marked restoration of myelopoiesis. The proliferative advantage imposed on CD34+ cells by STAT5A1*6 depended on the down-modulation of C/EBPα, as reintroduction of C/EBPα induced a quick cell-cycle arrest and the onset of myeloid differentiation. Long-term culture–initiating cell (LTC-IC) frequencies were elevated from 0.8% ± 0.6% to 7.8% ± 1.9% by STAT5A1*6 as compared with controls, but these elevated LTC-IC frequencies were strongly reduced upon re-introduction of C/EBPα in STAT5A1*6 cells, and no second cobble-stone area–forming cells (CAFCs) could be generated from double-transduced cells. Enumeration of progenitors revealed that the number of colony-forming cells (CFCs) was reduced more than 20-fold when C/EBPα was co-expressed in STAT5A1*6 cells. Our data indicate that down-modulation of C/EBPα is a prerequisite for STAT5-induced effects on self-renewal and myelopoiesis.
Introduction
Hematopoiesis is initiated in the bone marrow, where a limited number of pluripotent hematopoietic stem cells (HSCs) give rise to lineage-restricted progenitors that ultimately develop into mature erythroid, myeloid, or lymphoid cells. HSCs can both self-renew as well as differentiate, and these processes are in part regulated by the hematopoietic microenvironment (the osteoblastic niche) and hematopoietic growth factors that induce signal transduction.1,2 The initiation of signal transduction results in the activation of transcription factors such as PU.1, C/EBPα, GATA1, and PAX5, which have emerged as key switches of self-renewal, differentiation, and lineage determination during hematopoiesis.3
Recently, we and others have shown that activation of the transcription factor signal transducer and activator of transcription 5 (STAT5) has profound effects on self-renewal and lineage commitment of HSCs.4,5 Introduction of the constitutively activated mutant STAT5A1*6 into cord blood–derived CD34+ cells resulted in enhanced self-renewal and impaired myelopoiesis, while differentiation was diverted toward an erythroid cell fate.5 In murine cells, the absence of STAT5 signaling resulted in a profound defect in competitive repopulation of Stat5a/b knockout cells,6,7 while introduction of activated STAT5A1*6 into murine CD34– c-Kit+Sca-1+Lin– cells led to a drastic expansion of multipotential progenitors and promoted HSC self-renewal ex vivo.4 In addition, enforced activation of STAT5A greatly facilitated the generation of embryonic stem (ES) cell–derived hematopoietic stem cells with long-term self-renewal capacity in vitro that also contributed to hematopoiesis in vivo.8,9 Besides the effects of STAT5A on ES-derived HSC self-renewal, we also observed that myeloid differentiation was impaired.8 Although several lines of evidence indicate that STAT5 signaling has profound effects on hematopoietic lineage-commitment decisions and HSC self-renewal, little is known about the underlying mechanisms.
The transcription factor CCAAT enhancer binding protein-α (C/EBPα) is a key regulator of granulopoiesis.10 Cebpa–/– mice have profound defects in granulocyte differentiation,11 and C/EBPα binding sites have been identified in promoters of various myeloid-restricted genes such as the granulocyte-colony stimulating factor (G-CSF) receptor and neutrophil elastase.12,13 It has been noted that C/EBPα is expressed at low levels in immature HSCs and is up-regulated upon differentiation to more committed common myeloid progenitors (CMPs) and granulocyte/monocyte progenitors (GMPs), but is shut down in megakaryocyte/erythrocyte progenitors (MEPs) or in common lymphoid progenitors (CLPs).14 These observations suggested that the C/EBPα expression level determines the self-renewal function of HSCs. Indeed, Zhang et al15 recently demonstrated that the HSC repopulating capacity and self-renewal are strongly enhanced in the absence of C/EBPα. Reversibly, introduction of C/EBPα in human CD34+ cells resulted in a reduced long-term culture initiating cell (LTC-IC) frequency and facilitated granulocytic differentiation.16 It is therefore not surprising that disturbed C/EBPα signaling is often observed in hematological disorders such as acute myeloid leukemia (AML).17,18
Here, we demonstrate that C/EBPα is down-modulated by activated STAT5A in human cord blood–derived CD34+ cells. Introduction of a 4-hydroxytamoxifen (4-OHT)–inducible C/EBPα-ER fusion protein into STAT5A1*6 CB CD34+ cells reduced the stem and progenitor cell compartment and restored myeloid differentiation. These data indicate that down-modulation of C/EBPα expression is a critical event in the enhanced self-renewal and impaired myelopoiesis imposed on human CD34+ cells by active STAT5A.
Materials and methods
Cell culture and retroviral transductions
CD34+ cells were derived from neonatal cord blood from healthy full-term pregnancies from the obstetrics departments of the Martini Hospital and University Medical Center in Groningen, The Netherlands, after informed consent. The study was approved by the Ethics of the University Medical Center Groningen, The Netherlands. For all retroviral transduction experiments, the murine stem cell virus (MSCV) retroviral expression vector was used, which contained an encephalo-myelocarditis virus (EMCV)–derived internal ribosomal entry site (IRES2) in front of the enhanced green fluorescent protein (EGFP) (MiGR1 vector), DsRED2 (MiDR1 vector), or truncated neural growth factor receptor (NGF-R) (MiNR1 vector). The STAT5A1*6 retroviral vector was described previously,5 and the C/EBPα-ER vector was constructed by inserting a C/EBPα cDNA fused to the estrogen receptor domain (a kind gift from Dr J. Mulloy, Memorial Sloan-Kettering Cancer Center, New York, NY16 ) into the EcoRI site of MigR1, MiDR1, or MiNR1. Stable PG13 high-titer retroviral producer cell lines were generated as described previously. CB CD34+ cells were transduced in 3 consecutive rounds as described previously, and double transductions were performed by mixing viral supernatants prior to transduction of target cells.
Colony-forming cell (CFC), cobblestone area forming cell (CAFC), LTC-IC, and second CAFC assays
CFC, CAFC, and LTC-IC assays on MS5 stromal cells were performed as described previously.19 Briefly, CFC assays were performed in 1.2% methylcellulose containing 30% fetal calf serum (FCS), 57.2 μM β-mercaptoethanol, and 2 mM glutamine (StemCell Technologies, Grenoble, France), supplemented with 20 ng/mL interleukin-3 (IL-3), 20 ng/mL IL-6, 20 ng/mL G-CSF, 20 ng/mL c-kit ligand (KL), and 6 U/mL erythropoietin (EPO). Cells were either plated in bulk by 1000 transduced cells per plate in triplicate or in 96-well plates with 1, 3, 9, 27, 81, or 243 cells per well. LTC-IC assays were performed by plating transduced CB CD34+ cells in limiting dilutions in the range of 5 to 1000 cells per well on MS5 stromal cells in 96-well plates in LTC medium (α modified minimum essential media [MEM] supplemented with heat-inactivated 12.5% FCS; heat-inactivated 12.5% horse serum, penicillin, and streptomycin; 200 mM glutamine; 57.2 μM β-mercaptoethanol; and 1 μM hydrocortisone). After 5 weeks, methylcellulose was added to the wells. Two weeks later, wells containing CFCs were scored as positive. CAFC assays were performed as LTC-IC assays, but now CAFCs were counted at day 10 and at week 5 by microscopic evaluation of cocultures. Images were visualized using a Leica DM-IL microscope (Leica Microsystems, Rijswijk, The Netherlands) and a 40 × 0.60 numeric aperture (NA) objective. Only phase-dark colonies underneath the stroma were enumerated. For second CAFC assays, CAFCs were harvested by trypsinization of adherent cell populations, sorted on the basis of human CD45 expression (Miltenyi, Amsterdam, The Netherlands), and replated on fresh MS5 stroma or used for analysis.
Immunoblotting, histochemistry, and cytospins
Whole cell extracts were obtained by lysing 5 × 105 cells in boiling Laemmli sample buffer for 5 minutes prior to separation on 12% sodium dodecyl sulfate (SDS)–acrylamide gels. Proteins were transferred to nitrocellulose filters (Millipore, Etten Leur, The Netherlands) in Tris (tris(hydroxymethyl)aminomethane)-glycine buffer at 9 volts for 1.5 hours using a semidry electroblotter from Bio-Rad (Veenendaal, The Netherlands). Membranes were blocked in phosphate-buffered saline (PBS) containing 5% nonfat milk prior to incubation with antibodies. Binding of antibodies was detected by enhanced chemiluminescence (ECL) according to the manufacturer's instructions (Roche Diagnostics, Almere, The Netherlands). Antibodies against STAT5 (C17) and C/EBPα (N19) were obtained from Santa Cruz (Heerhugowaard, The Netherlands) and were used in dilutions of 1:1000. May-Grünwald Giemsa staining was used to analyze cytospins. Images were visualized using an Olympus BX50 microscope (Olympus Nederland, Zoeterwoude, The Netherlands) and a 100 × 1.3 NA oil objective.
PCR analysis
For reverse transcriptase–polymerase chain reaction (RT-PCR), total RNA was isolated from 1 × 106 cells using the RNeasy kit from Qiagen (Venlo, The Netherlands) according to the manufacturer's recommendations. Two micrograms of RNA was reverse transcribed with M-MuLV reverse transcriptase (Roche Diagnostics). For PCR, 2 μL cDNA was amplified using primers as indicated in the text (sequences and conditions are available on request) in a total volume of 50 μL using 2 units of Taq polymerase (Roche Diagnostics). As a negative control, RNA minus reverse transcriptase (–RT)–prepared cDNA was used in PCR reactions. Aliquots of 10 μL were run on 1.5% agarose gels. For real-time RT-PCR, cDNA was prepared by reverse transcribing the total RNA of 0.15 × 106 sorted cells using M-MuLV reverse transcriptase (Fermentas, St Leon-Roth, Germany) according to the manufacturer's instructions. Aliquots (2 μL) of cDNA were then real-time amplified using iQ SYBR Green supermix (Bio-Rad) on a MyIQ thermocycler (Bio-Rad) and quantified using MyIQ software (Bio-Rad). HPRT (hypoxanthine phosphoribosyl-transferase) expression was used to calculate relative expression levels.
Flow cytometry analysis
All antibodies were obtained from Becton Dickinson (Alphen a/d Rijn, The Netherlands). Cells were incubated with antibodies at 4°C for 45 minutes. For blocking nonspecific binding to Fcγ receptors, cells were blocked with anti-Fcγ antibodies for 15 minutes at 4°C. All fluorescence activated cell sorter (FACS) analyses were performed on a FACScalibur (Becton Dickinson), and data were analyzed using WinList 3D (Topsham, ME). Cells were sorted on a MoFLo (DakoCytomation, Carpinteria, CA).
EMSAs
Nuclear extracts of transiently transfected 293T cells were made according to the mini-scale procedure described by Schreiber et al.20 Electrophoretic mobility shift assay (EMSA) analysis was performed by incubating 5 μg nuclear extract with 5′-IRDye 700–labeled double-stranded oligonucleotides for 30 minutes at room temperature. Binding reactions were run on nondenaturing 4% acrylamide gels in 1 × TBE (Tris/borate/EDTA), and the gels were scanned using an Odyssey infrared scanner (Li-Cor Biosciences, Lincoln, NE). To check for specificity of the reactions, 50-fold molar excess of unlabeled oligonucleotide (either self or non-self) was added to the binding reactions. For supershift analysis, 1 μL C/EBPα-specific antibody (SC-61, Santa Cruz Biotechnology, Santa Cruz, CA) was added simultaneously with the probe.
Luciferase assays
For transactivation studies, 293T cells were transiently transfected in 12-well plates with the indicated constructs using Fugene6 (Roche, Basel, Switzerland) according to the manufacturer's instructions. Forty-eight hours after transfection, cells were stimulated for 8 hours with 500 nM 4-hydroxy tamoxifen to induce C/EBP binding, and cell extracts were made using luciferase lysis buffer (Promega, Leiden, The Netherlands). Luciferase expression was measured according to the manufacturer's protocol (Promega). β-galactosidase expression was measured to correct for differences in transfection efficiency.
Cell-cycle analysis
Cell-cycle analysis was performed by determining the DNA content of sorted cells by staining with 7-amino-actinomycin D in phosphate-citrate (PC) buffer (Sigma), supplemented with 0.15 M NaCl, 5 mM EDTA (ethylenediaminetetraacetic acid), 0.5% bovine serum albumin (BSA), and 0.02% Saponine for 10 minutes. After washing, the cells were analyzed on a FACSCalibur flow cytometer (Becton Dickinson), and DNA profiles were generated using Mod-Fit software (Topsham).
Results
Enforced activation of STAT5 in human CB CD34+ cells results in down-modulation of C/EBPα expression
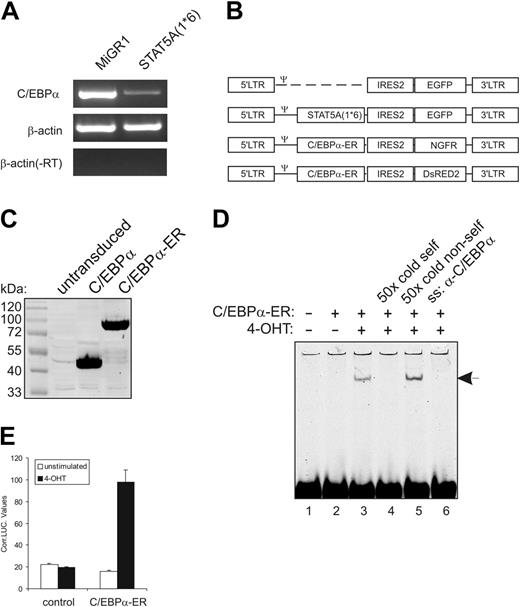
Previously, we have demonstrated that enforced activation of STAT5A in human CB-derived CD34+ stem/progenitor cells results in enhanced self-renewal and an impairment of myeloid differentiation.5 Affymetrix analysis of RNA isolated from MiGR1 versus STAT5A1*6-transduced cells revealed that C/EBPα was the most significantly down-modulated gene by activated STAT5A (–3.3-fold, P < .001). These data were confirmed by conventional (Figure 1A) and real-time RT-PCR (Figure 2D).
To determine whether the down-modulation of C/EBPα was involved in the phenotypes that were imposed on human CB CD34+ cells by STAT5A1*6, we generated retroviral expression vectors for C/EBPα-ER. These expression vectors contained either the truncated NGF receptor or DsRED2 as a marker gene for the identification of transduced cells (schematically depicted in Figure 1B). C/EBPα was fused to the estrogen receptor ligand-binding domain (ER), which allowed a 4-hydroxytamoxifen (4-OHT)–inducible activation of overexpressed C/EBPα. As control experiments, 293T cells were transduced with C/EBPα or C/EBPα-ER, and expression was verified by Western blotting (Figure 1C). The 4-OHT inducibility was tested by C/EBPα DNA-binding and transactivation assays. As shown in Figure 1D, in the absence of 4-OHT no C/EBPα DNA binding was observed (lane 2), while stimulation of cells with 1 μM 4-OHT for 1 hour resulted in the appearance of a DNA-binding complex (lane 3) that could be outcompeted by a 50-fold excess of unlabeled oligo (lane 4) but not by a 50-fold excess of random unlabeled oligo (lane 5). Addition of antibody against C/EBPα resulted in the disappearance of the C/EBPα-DNA complex (Figure 1D, lane 6). As shown in Figure 1E, stimulation with 1 μM 4-OHT also resulted in an increase in C/EBPα transactivation. These data show that C/EBPα-ER DNA binding and transactivation were induced only in the presence of 4-OHT.
Re-expression of C/EBPα in STAT5A1*6 CD34+ cells results in a restored myelopoiesis
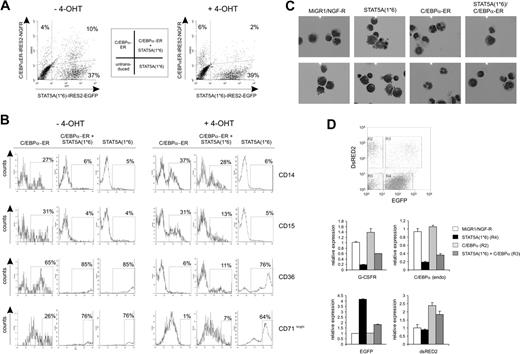
CB-derived CD34+ cells were transduced with both STAT5A1*6 and C/EBPα with efficiencies of 1% to 10% of double-transduced cells. A representative FACS plot is shown in Figure 2A (left panel) in which an efficiency of 10% was reached. Transduced cells were plated on MS5 stroma in the absence or presence of 4-OHT, and hematopoietic differentiation was monitored by FACS analysis, RT-PCRs, and cytospins. In control conditions—untransduced cells (not shown) or C/EBPα-ER without 4-OHT (Figure 2B, left panels)—27% of the cells were positive for CD14, 31% for CD15, 65% for CD36, and 26% for CD71bright. In agreement with data obtained previously,5,8 enforced activation of STAT5A resulted in a block in myeloid differentiation as determined by the percentage of cells positive for CD14 (5%) and CD15 (4%), while the number of cells that were positive for erythroid markers was significantly increased (85% were CD36+ and 76% were CD71bright (Figure 2B, left panels)). Re-activation of C/EBPα in STAT5A1*6 cells by administration of 1 μM 4-OHT to double-transduced cells resulted in a markedly restored myelopoiesis, as the number of CD14- and CD15-positive cells increased from 6% to 28% and 4% to 13%, respectively (representative data of 3 independent experiments are shown in Figure 2B, right panels). The imposed erythroid differentiation of CB CD34+ cells by STAT5A1*6 was reversed by reintroduction of C/EBPα, as the number of CD36+ and CD71bright cells was reduced from 85% to 11% and 76% to 7%, respectively. These results were further underscored by morphological analysis of MiGR1, STAT5A1*6, C/EBPα-ER, or double-transduced cells (Figure 2C). Cytospins from STAT5A1*6-transduced cells contained blasts, proerythroblasts, basophilic-, polychromatic-, and orthochromatic erythroblasts as well as erythrocytes, while MiGR1 control cells differentiated mostly along the myeloid lineage as cytospins revealed predominantly myeloblasts, monocytes, and granulocytes. Cytospins from C/EBPα-ER–transduced cells displayed even more mature granulocytic development without any signs of erythropoiesis. Double-transduced cells contained predominantly myeloblasts and monocytes. Real-time RT-PCR analyses on sorted transduced cells revealed that constitutive activation of STAT5A significantly down-modulated the expression of G-CSF-R and C/EBPα (Figure 2D). Reversibly, expression of C/EBPα resulted in an increase in G-CSFR expression. Also, endogenous C/EBPα expression was slightly up-regulated by C/EBPα-ER as determined by using primers that specifically recognize endogenous C/EBPα but not overexpressed C/EBPα-ER. Reintroduction of C/EBPα-ER into STAT5A1*6 cells partly restored G-CSFR and endogenous C/EBPα expression levels. Endogenous STAT5A expression levels were not affected by C/EBPα expression (data not shown). As controls, GFP+ and/or DsRED2+ sorted populations were analyzed for EGFP and DsRED2 RNA levels (Figure 2D, lower panels).
Enforced activation of STAT5 in human CB CD34+ cells results in down-modulation of C/EBPα expression. (A) CB-derived CD34+ cells were transduced with MiGR1 or STAT5A1*6, sorted on the basis of GFP expression, and total RNA was isolated and used in RT-PCR reactions with primers for C/EBPα and β-actin as indicated. (B) Schematic representation of the retroviral constructs that were used in these studies. (C) C/EBPα and C/EBPα-ER were overexpressed in 293T cells, and total cell lysates were Western blotted using anti-C/EBPα antibodies. (D) C/EBP-ER was overexpressed in 293T cells, and nuclear extracts were used in EMSA experiments as indicated, using a probe that contains the C/EBPα consensus binding sequence. 4-OHT induced DNA binding of C/EBPα-ER, which could be outcompeted with 50 × cold self-oligo (lane 4), while DNA binding was blocked by adding anti-C/EBPα (lane 6). (E) Luciferase assays were performed in 293T cells using luciferase reporters containing C/EBPα-binding sites. Cells were transfected with empty MiGR1 vector (control) or C/EBPα-ER expression vectors, and cells were either left unstimulated or were stimulated with 4-OHT. After 24 hours of stimulation, cells were harvested to perform luciferase and LacZ assays as described in “Materials and methods.”
Enforced activation of STAT5 in human CB CD34+ cells results in down-modulation of C/EBPα expression. (A) CB-derived CD34+ cells were transduced with MiGR1 or STAT5A1*6, sorted on the basis of GFP expression, and total RNA was isolated and used in RT-PCR reactions with primers for C/EBPα and β-actin as indicated. (B) Schematic representation of the retroviral constructs that were used in these studies. (C) C/EBPα and C/EBPα-ER were overexpressed in 293T cells, and total cell lysates were Western blotted using anti-C/EBPα antibodies. (D) C/EBP-ER was overexpressed in 293T cells, and nuclear extracts were used in EMSA experiments as indicated, using a probe that contains the C/EBPα consensus binding sequence. 4-OHT induced DNA binding of C/EBPα-ER, which could be outcompeted with 50 × cold self-oligo (lane 4), while DNA binding was blocked by adding anti-C/EBPα (lane 6). (E) Luciferase assays were performed in 293T cells using luciferase reporters containing C/EBPα-binding sites. Cells were transfected with empty MiGR1 vector (control) or C/EBPα-ER expression vectors, and cells were either left unstimulated or were stimulated with 4-OHT. After 24 hours of stimulation, cells were harvested to perform luciferase and LacZ assays as described in “Materials and methods.”
Re-expression of C/EBPα in STAT5A1*6 CD34+ cells results in a restored myelopoiesis. (A) Human CB CD34+ cells were transduced with STAT5A1*6-IRES2-EGFP and C/EBPα-ER-IRES2-NGF-R retroviral vectors. Representative transduction efficiencies are shown of cells grown for 1 week on MS5 stroma in the absence (left panel) or presence (right panel) of 4-OHT. (B) Cells shown in panel A were analyzed by FACS for the expression of CD14, CD15, CD36, and CD71 expression using allophycocyanin (APC)–conjugated antibodies. Data are representative of at least 3 independent experiments. (C) CB CD34+ cells were transduced with MiGR1/NGF-R empty vectors or double transduced with STAT5A1*6 and C/EBPa-ER, cocultured on MS5 for 1 week in the presence of 4-OHT, after which MiGR1 cells, STAT5A1*6 cells, C/EBPa-ER cells, and double-positive cells were sorted on the MoFlo and analyzed by cytospins and May-Grünwald Giemsa staining. (D) Experiment performed as described in panel C, but total RNA was isolated after MoFlo sorting for qRT-PCR analysis as indicated. The sorted cells for data in panels C and D were obtained from gates R2, R4, and R3, which represent C/EBPα-ER, STAT5A1*6, and double-transduced cells, respectively. Double-transduced empty control cells were sorted separately (data not shown).
Re-expression of C/EBPα in STAT5A1*6 CD34+ cells results in a restored myelopoiesis. (A) Human CB CD34+ cells were transduced with STAT5A1*6-IRES2-EGFP and C/EBPα-ER-IRES2-NGF-R retroviral vectors. Representative transduction efficiencies are shown of cells grown for 1 week on MS5 stroma in the absence (left panel) or presence (right panel) of 4-OHT. (B) Cells shown in panel A were analyzed by FACS for the expression of CD14, CD15, CD36, and CD71 expression using allophycocyanin (APC)–conjugated antibodies. Data are representative of at least 3 independent experiments. (C) CB CD34+ cells were transduced with MiGR1/NGF-R empty vectors or double transduced with STAT5A1*6 and C/EBPa-ER, cocultured on MS5 for 1 week in the presence of 4-OHT, after which MiGR1 cells, STAT5A1*6 cells, C/EBPa-ER cells, and double-positive cells were sorted on the MoFlo and analyzed by cytospins and May-Grünwald Giemsa staining. (D) Experiment performed as described in panel C, but total RNA was isolated after MoFlo sorting for qRT-PCR analysis as indicated. The sorted cells for data in panels C and D were obtained from gates R2, R4, and R3, which represent C/EBPα-ER, STAT5A1*6, and double-transduced cells, respectively. Double-transduced empty control cells were sorted separately (data not shown).
Re-expression of C/EBPα results in a loss of STAT5A1*6-induced proliferative advantage of human CD34+ cells
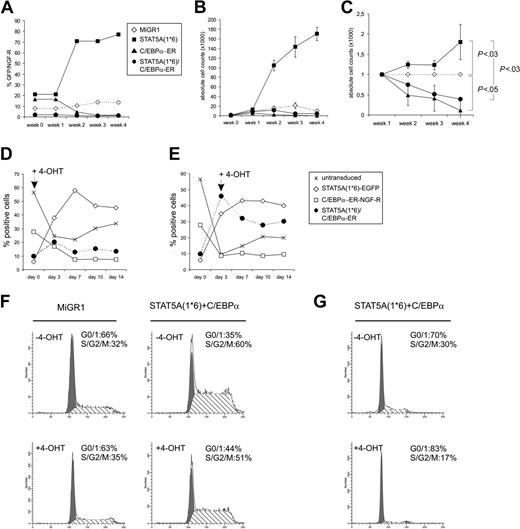
To study the proliferation of CB CD34+ cells that expressed STAT5A1*6, C/EBPα, or both, transduced cells were plated on MS5 stroma, and the expansion was monitored weekly. In experiments in which nonsorted transduced cells were used, the relative expansion could be determined by analyzing the percentage of GFP+ (STAT5A1*6), NGF-R+ (C-EBPα-ER), or double-positive cell populations. As shown in a representative set of experiments in Figure 3A-C, enforced activation of STAT5A in human CB CD34+ cells resulted in a proliferative advantage over MiGR1 control cells when expanded on MS5 stroma, as the percentage of GFP+ cells increased from 20% to more than 70% within 2 to 3 weeks (Figure 3A). In contrast, overexpression of C/EBPα-ER resulted in a proliferative disadvantage on MS5 stroma, as the percentage of NGF-R+ cells decreased from 18% to 2% within 3 weeks (Figure 3A). In Figure 3B, the absolute cell counts of individual cocultures are shown, and these experiments clearly indicate that expression of activated STAT5A1*6 results in enhanced expansion on MS5 stroma, while expression of C/EBPα represses the expansion on MS5 compared with controls. When C/EBPα was reintroduced into STAT5A1*6 cells, the proliferative advantage imposed on cells by activated STAT5A was completely reduced to below control levels (Figure 3B). Representative FACS profiles are shown in Figure 2A in which treatment of 4-OHT for 1 week resulted in a decrease of double-transduced cells from 10% to 2%. Data from 3 independent experiments are summarized in Figure 3C, in which the relative expansions of all groups are given compared to MiGR1 control cells, which was set to 1 at each time point. To further confirm that STAT5A1*6-induced proliferation depends on the down-modulation of C/EBPα, 4-OHT was administered to transduced cells at various time points. Data in Figure 3D-E is presented as the percentage of untransduced, STAT5A1*6 (EGFP+), C/EBPα-ER (NGF-R+), or double-transduced cells within 1 batch of transduced cells that was expanded on MS5 over a 2-week period. 4-OHT was added from the start of the experiment (Figure 3D) or at day 3 (Figure 3E), and only when 4-OHT was administered was the proliferative advantage of STAT5A1*6/C-EBPα-ER over controls reduced. Taken together, these data indicate that down-modulation of C/EBPα is a critical event for the enhancing effects of STAT5A1*6 on proliferation.
Re-expression of C/EBPα results in a loss of STAT5A1*6-induced proliferative advantage of human CD34+ cells. (A) Human CB CD34+ cells were transduced with MiGR1, STAT5A1*6-IRES2-EGFP, C/EBPα-ER-IRES2-NGF-R, or both STAT5A1*6 and C/EBPα-ER, as indicated. Cells were plated on MS5 stroma in the presence of 4-OHT, and the proliferation was monitored weekly by FACS analysis for GFP or NGF-R. (B) Experiment performed as described in panel A, but cells were sorted after transduction, and expansion was monitored by determining the cell counts weekly. (C) Combined data of 3 representative experiments in which the relative expansion is indicated as compared to the MiGR1 control that was set to 1 at each time point. Significant differences were determined using a Student t test. (D-E) Experiments as in panel B, but now 4-OHT was added at day 0 (D) or day 3 (E). Cultures were monitored for 14 days. Data are presented as the percentage of untransduced, STAT5A1*6 (EGFP+), C/EBPα-ER (NGF-R+), or double-transduced cells within 1 batch of transduced cells that was expanded on MS5 over a 2-week period. (F) Cells were transduced as indicated, plated onto MS5 stroma to allow expansion for 7 days, after which 4-OHT was added for an additional 24 hours. After 4-OHT stimulation, the cell-cycle distribution was determined by FACS. (G) Experiment as in panel F, but now cells were treated with 4-OHT for 48 hours.
Re-expression of C/EBPα results in a loss of STAT5A1*6-induced proliferative advantage of human CD34+ cells. (A) Human CB CD34+ cells were transduced with MiGR1, STAT5A1*6-IRES2-EGFP, C/EBPα-ER-IRES2-NGF-R, or both STAT5A1*6 and C/EBPα-ER, as indicated. Cells were plated on MS5 stroma in the presence of 4-OHT, and the proliferation was monitored weekly by FACS analysis for GFP or NGF-R. (B) Experiment performed as described in panel A, but cells were sorted after transduction, and expansion was monitored by determining the cell counts weekly. (C) Combined data of 3 representative experiments in which the relative expansion is indicated as compared to the MiGR1 control that was set to 1 at each time point. Significant differences were determined using a Student t test. (D-E) Experiments as in panel B, but now 4-OHT was added at day 0 (D) or day 3 (E). Cultures were monitored for 14 days. Data are presented as the percentage of untransduced, STAT5A1*6 (EGFP+), C/EBPα-ER (NGF-R+), or double-transduced cells within 1 batch of transduced cells that was expanded on MS5 over a 2-week period. (F) Cells were transduced as indicated, plated onto MS5 stroma to allow expansion for 7 days, after which 4-OHT was added for an additional 24 hours. After 4-OHT stimulation, the cell-cycle distribution was determined by FACS. (G) Experiment as in panel F, but now cells were treated with 4-OHT for 48 hours.
These data were further underscored in cell-cycle experiments of MiGR1/NGF-R and STAT5A1*6/C-EBPα-ER cells in the absence or presence of 4-OHT. Enforced activation of STAT5A resulted in an increase of the percentage of cells in S/G2/M phase from 32% to 60%, while the percentage of cells in G1 was reduced from 66% to 35% (Figure 3F). After 24 hours of treatment with 4-OHT, the percentage of cells in G1 was 44%, and the percentage of cells in S/G2/M was 51%. In a separate experiment, STAT5A1*6/C-EBPα-ER cells were treated with 4-OHT for 48 hours, which resulted in an even stronger decrease in the percentage of cells in S/G2/M phase (Figure 3G). No increase in apoptosis was observed upon stimulation with 4-OHT throughout all experiments (Figure 3F-G and data not shown), suggesting that the reduced expansion upon re-expression of C/EBPα into STAT5A1*6 cells does not involve elevated apoptosis.
Expression of C/EBPα results in a loss of STAT5A1*6-induced self-renewal and long-term expansion
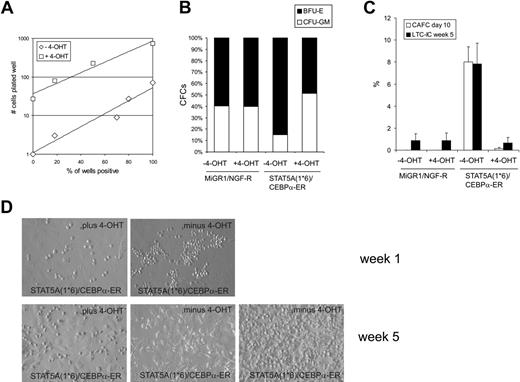
To enumerate the number of progenitors in transduced CB CD34+ cells, STAT5A1*6, C/EBPα-ER, or double-transduced cells were sorted and plated in methylcellulose under limiting dilution conditions in the absence or presence of 4-OHT. Administration of 4-OHT to MiGR1 or STAT5A1*6 cells did not affect the number of progenitors significantly (data not shown), although expression of STAT5A1*6 induced a shift toward significantly more erythroid burst-forming units (BFU-Es) at the expense of granulocyte macrophage colony-forming units (CFU-GMs) (Figure 4B). Reintroduction of C/EBPα into STAT5A1*6 cells by stimulation with 4-OHT significantly reduced the number of progenitors. To obtain 50% positive wells, 7.6 cells in the absence of 4-OHT and 176 cells in the presence of 4-OHT needed to be plated, resulting in a difference in CFC formation of 23.2-fold (Figure 4A). Also, the shift toward more BFU-E–type colonies induced by activated STAT5A was reversed to a normal CFU-GM versus BFU-E distribution when C/EBPα was reintroduced in STAT5A1*6 cells (Figure 4B).
The number of hematopoietic stem cells was enumerated by CAFC and LTC-IC assays on MS5 stroma under limiting dilution conditions. As shown in Figure 4C, enforced activation of STAT5A1*6 resulted in the appearance of early CAFCs within 10 days after plating at a frequency of 8.2%. These early CAFCs were not observed when C/EBPα-ER was co-expressed in STAT5A1*6 cells (Figure 4C). Representative examples of cocultures at week 1 are shown in Figure 4D. After 5 weeks, methylcellulose was added to the wells to determine the LTC-IC frequencies. Enumeration of week 5 LTC-IC frequencies revealed that STAT5A1*6 enhanced the frequency from 0.8% ± 0.6% to 7.8% ± 1.9% compared with MiGR1 control cells. Importantly, this elevated LTC-IC frequency required the down-modulation of C/EBPα, as reintroduction of C/EBPα-ER in STAT5A1*6 cells significantly reduced the LTC-IC frequency to 0.7% ± 0.5% at week 5 (Figure 4C-D). While the STAT5A1*6 week 5 CAFCs could be harvested and passaged onto new MS5 to generate second CAFCs, no long-term cultures could be established from the double-transduced cells in the presence of 4-OHT (data not shown).
Expression of C/EBPα results in a loss of STAT5A1*6-induced self-renewal and long-term expansion. (A) Human CB CD34+ cells were transduced with both STAT5A1*6-IRES2-EGFP and C/EBPα-ER-IRES2-NGF-R, and cells in gate R3 were sorted on the MoFlo. Cells were plated in the presence or absence of 4-OHT in methylcellulose assays in 96-well plates in limiting dilutions of 1, 3, 9, 27, 81, and 243 cells per well. Two weeks later, CFC counts were scored. (B) Cells were transduced with both MiGR1/NGF-R empty vectors or STAT5A1*6-IRES2-EGFP and C/EBPα-ER-IRES2-NGF-R as indicated. Cells were sorted and plated in bulk methylcellulose cultures, and the ratio between BFU-E and CFU-GM type colonies is shown in the absence or presence of 4-OHT. (C) Experiment performed as described in panel B, but sorted cells were plated onto MS5 stroma in 96-well plates in limiting dilutions of 5 to 1000 cells per well. After 5 weeks, methylcellulose was added to the wells, and 2 weeks later wells were scored as positive or negative to determine LTC-IC frequencies. Data from a representative experiment are shown. (D) Representative microscopy images of several MS5 cocultures at weeks 1 and 5.
Expression of C/EBPα results in a loss of STAT5A1*6-induced self-renewal and long-term expansion. (A) Human CB CD34+ cells were transduced with both STAT5A1*6-IRES2-EGFP and C/EBPα-ER-IRES2-NGF-R, and cells in gate R3 were sorted on the MoFlo. Cells were plated in the presence or absence of 4-OHT in methylcellulose assays in 96-well plates in limiting dilutions of 1, 3, 9, 27, 81, and 243 cells per well. Two weeks later, CFC counts were scored. (B) Cells were transduced with both MiGR1/NGF-R empty vectors or STAT5A1*6-IRES2-EGFP and C/EBPα-ER-IRES2-NGF-R as indicated. Cells were sorted and plated in bulk methylcellulose cultures, and the ratio between BFU-E and CFU-GM type colonies is shown in the absence or presence of 4-OHT. (C) Experiment performed as described in panel B, but sorted cells were plated onto MS5 stroma in 96-well plates in limiting dilutions of 5 to 1000 cells per well. After 5 weeks, methylcellulose was added to the wells, and 2 weeks later wells were scored as positive or negative to determine LTC-IC frequencies. Data from a representative experiment are shown. (D) Representative microscopy images of several MS5 cocultures at weeks 1 and 5.
Discussion
The results presented in this manuscript allow us to reach 3 main conclusions. First, enforced activation of the transcription factor STAT5A in human CD34+ cells results in a down-modulation of C/EBPα. Second, STAT5A1*6-induced self-renewal and long-term expansion of human stem/progenitor cells on stroma requires the down-modulation of C/EBPα. Finally, the impaired myelopoiesis and the bias toward erythropoiesis that is imposed upon human CD34+ cells by activated STAT5A also involves down-modulation of C/EBPα.
So far, little information is available on the molecular mechanisms that are involved in self-renewal of normal and leukemic stem/progenitor cells. Our previous studies have indicated that activating mutations in the tyrosine kinase receptor FLT321 and particularly enforced activation of STAT5 result in enhanced long-term self-renewal of early stem/progenitor cells.5 Affymetrix microarray analysis revealed C/EBPα as one of the most prominent down-regulated genes in human CB-derived CD34+ cells transduced with activated STAT5A1*6 or the FLT3-W51 internal tandem duplication (Schuringa et al5 and Ki Young Chung, Giovanni Morrone, J.J.S., and M.A.S.M., unpublished observations, June 2003). Hematopoietic stem cell self-renewal, repopulating capacity, and myeloid differentiation have been closely linked to C/EBPα expression. In mouse models, loss of C/EBPα in the HSC resulted in elevated competitive reconstitution activity, while the transition from the common myeloid progenitor to the granulocyte/monocyte progenitor was blocked.15 Also, in mouse models, C/EBPα deficiency22 as well as dominant-negative mutations in C/EBPα23 have been shown to increase the capacity of myeloid progenitors to proliferate. Reversibly, overexpression of C/EBPα has been associated with cell-cycle arrest and the onset of myeloid differentiation.16,24-26 Our data indicate that the down-modulation of C/EBPα expression is a key event in the STAT5A-mediated long-term self-renewal and impaired myeloid differentiation of human CB-derived CD34+ cells.
Our analyses at the stem and progenitor levels revealed that introduction of an activated mutant of STAT5A, STAT5A1*6,27 resulted in the formation of early CAFCs with elevated frequencies compared with controls (Figure 4C and Schuringa et al5 ). These early CAFCs persisted for more than 5 weeks and could be passaged onto new stroma to give rise to second and third CAFCs for up to a period of 18 weeks. It has been shown that these CAFCs are a representation of the number of in vivo long-term repopulating HSCs as measured in the NOD-SCID transplantation model.5,28,29 These effects of active STAT5A on human stem and progenitor cells can be reversed by overexpression of C/EBPα. Both in LTC-IC assays as well as in CFC assays in methylcellulose we consistently observed a distinct reduction in stem and progenitor frequency, and no long-term cultures could be established from STAT5A1*6 cells when C/EBPα was re-introduced. It has been demonstrated that in C/EBPα-deficient murine HSCs Bmi-1 expression is elevated, suggesting that one of the normal functions of C/EBPα might be to inhibit HSC self-renewal by down-modulating Bmi-1.15 Future studies will be aimed at elucidating whether Bmi-1 expression and function is altered in human CD34+ cells that express activated STAT5A.
Our observations at the stem/progenitor level correlate with data from hematopoietic differentiation and expansion studies. Overexpression of C/EBPα in human CD34+ cells resulted in a block in proliferation and enhanced myeloid differentiation. While expression of activated STAT5A1*6 in CB CD34+ cells resulted in a proliferative advantage compared with control cells, this was dramatically down-modulated by reintroduction of C/EBPα. These observations coincided with an increased percentage of cells in S/G2/M phase when STAT5A was activated in CB CD34+ cells, which could be reduced by re-expression of C/EBPα. These data clearly indicate that down-modulation of C/EBPα is required for the proliferative advantage imposed on human CB CD34+ cells by activated STAT5A. C/EBPα has been identified as a potent suppressor of cell proliferation, and several mechanisms have been proposed. C/EBPα interacts with and inhibits cdk2/4,30 induces expression of the cell-cycle inhibitor p21WAF-1,31 and represses the E2F complex of cell-cycle regulatory transcription factors.32 Thus, one or more of these mechanisms could account for the STAT5A1*6-induced proliferative advantage of human CB CD34+ cells by down-modulation of C/EBPα, thereby alleviating the block on cell-cycle progression. Furthermore, C/EBPα has been recognized as a key transcription factor that determines lineage commitment from the HSC toward the GM progenitor.15 We observed that enforced activation of STAT5A in human CD34+ cells resulted in a potent block in myeloid differentiation and diverted differentiation toward erythropoiesis. These lineage fate decisions induced by STAT5 could be reprogrammed by reintroduction of C/EBPα into STAT5A1*6 cells.
Various lines of evidence suggest that activation of STAT5A is a critical event in leukemic transformation. Constitutive activation of STAT5A has been observed in the majority of acute and chronic leukemias. A number of genetic abnormalities, such as Bcr-Abl,33,34 Tel-Jak2,35 and FLT3-ITDs36,37 induce constitutive activation of STAT5, and particularly in the case of Tel-Jak2 it has been demonstrated that STAT5 is necessary for Tel-Jak2–induced myeloproliferative disease.35 Mutations in JAK2 that result in hyperactivation of STAT5 have been identified in polycythemia vera.38-41 More recently, a single activating mutation in STAT5A (cS5F) has been shown to induce multilineage leukemia in mice.42 Nevertheless, no cell biological mechanisms have been identified by which STAT5A might induce leukemic transformation of human cells. Our data are the first to link STAT5 signal transduction to C/EBPα expression in STAT5A-mediated long-term expansion of human hematopoietic stem/progenitor cells. Also, the disturbed myelopoiesis that is induced by activated STAT5A requires down-modulation of C/EBPα. It is intriguing that these phenotypes directly parallel those observed in AML patients. Deregulation of C/EBPα expression has been observed in a variety of hematological malignancies, including in AMLs with t(8;21),43 inv16,44 or Flt3 mutations.45 In addition, mutations in C/EBPα have been identified that either affect the transactivation potential or block the translation of the full-length 42-kDa form (p42), which leads to overexpression of the shorter 30-kDa form (p30) that acts as a dominant-negative transcription factor.17,18,46 Further studies will define whether re-introduction of C/EBPα in these AML cells will restore the differentiation process of the affected cells only or whether it will also restrict self-renewal capacity of the leukemic stem cell pool.
Prepublished online as Blood First Edition Paper, February 2, 2006; DOI 10.1182/blood-2005-11-4608.
Supported by Dutch Cancer Society (KWF) grant RUG 2003-2316 (E.V.) and a Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO)–VENI 2004 grant (J.J.S.).
A.T.J.W. performed research and analyzed data; H.S. contributed analytic tools; M.A.S.M. analyzed data; E.V. analyzed data; J.J.S. designed research, analyzed data, and wrote the paper.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors would like to acknowledge Geert Mesander and Henk Moes for help with Flow Cytometry, and Amgen (La Jolla, CA) and Kirin (Kirin Brewery, Japan) for providing cytokines. The authors greatly appreciate the help of Dr A. van Loon and Dr J. J. Erwich and colleagues (obstetrics departments from the Martini Hospital and University Medical Center Groningen) for collecting cord blood.