Abstract
Gene products mutated in the inherited bone marrow failure syndromes dyskeratosis congenita (DC), cartilage-hair hypoplasia (CHH), Diamond-Blackfan anemia (DBA), and Shwachman-Diamond syndrome (SDS) are all predicted to be involved in different aspects of ribosome synthesis. At this moment, however, it is unclear whether this link indicates a causal relationship. Although defective ribosome synthesis may contribute to each of these bone marrow failure syndromes (and perhaps others), precisely which feature of each disease is a consequence of failure to produce adequate amounts of ribosomes is obscured by the tendency of each gene product to have extraribosomal functions. Delineation of the precise role of each gene product in ribosomal biogenesis and in hematopoietic development may have both therapeutic and prognostic importance and perhaps even direct the search for new bone marrow failure genes.
Introduction
Inherited bone marrow failure syndromes including dyskeratosis congenita (DC), cartilage-hair hypoplasia (CHH), Diamond-Blackfan anemia (DBA), and Shwachman-Diamond syndrome (SDS) have long fascinated and challenged geneticists and hematologists. This heterogeneous group of disorders is characterized by bone marrow failure and the variable presence of congenital anomalies and cancer predisposition (Table 1). While there has been rapid progress in identifying gene defects in nearly all the disorders, understanding the molecular pathophysiology has lagged behind in most instances. Intriguingly, genes mutated in many of these disorders encode factors involved in ribosome synthesis, suggesting a link between this fundamental cellular process and marrow failure. In this “Perspective,” we wish to define this association and critically assess its validity in the light of new research.
Broadly speaking, the genes mutated in the inherited marrow failure syndromes (reviewed in J. M. Lipton, J.M.L., and A. Vlachos, manuscript submitted) have widely disparate functions (Fanconi anemia genes, c-MPL, GFI1, ELA2, RUNX1, HOXA11, GATA1, and others) without an obvious molecular interrelationship. By contrast, genes mutated in X-linked DC,1 CHH,2 DBA,3 and SDS4 are all predicted to be involved in aspects of ribosome synthesis. Complicating this picture, however, is the fact that some of the factors encoded by these genes are multifunctional, influencing one or more critical cellular processes in addition to ribosome synthesis. The most extensively studied bone marrow failure syndrome from this perspective is X-linked DC, which is caused by mutations in DKC1 (its encoded protein also known as dyskerin).1 Dyskerin is a putative pseudouridyl synthase found in so-called H/ACA ribonucleoprotein complexes involved in ribosomal RNA (rRNA) modification. Since its discovery, dyskerin has also been shown to be a component of other ribonucleoprotein complexes including telomerase.5 A similarly complex picture is emerging for CHH. The gene affected in CHH is RMRP, which encodes the RNA component of ribonuclease mitochondrial RNA processing (RNase MRP).2 RNase MRP is a multifunctional ribonucleoprotein complex involved in cleaving rRNA precursors, processing of RNA primers used in mitochondrial DNA replication, and the turnover of certain cell cycle–regulated mRNAs. The only gene linked to DBA to date encodes ribosomal protein S19 (RPS19),3 which in yeast is required for the maturation of 40S ribosomal subunits.6 Here again, the human RPS19 protein has been proposed to have a function outside of this role.7 Finally, several studies have suggested that SBDS, mutations in which cause SDS,4 encodes a protein involved in ribosome synthesis.8,9 Thus, although defective ribosome synthesis may contribute to each of the bone marrow failure syndromes under discussion here, precisely which feature, if any, of each disease is a consequence of failure to produce adequate amounts of ribosomes is obscured by the tendency of each gene product to have extraribosomal functions (Figure 1).
Ribosome synthesis
The human ribosome is composed of 4 ribosomal RNAs and a minimum of 80 different ribosomal proteins. In addition to these structural components of the ribosome, there are well over 100 additional factors involved in its biosynthesis.10 These factors include components of the RNA polymerase I transcriptional machinery, small nucleolar ribonucleoprotein complexes involved in rRNA modification, a host of RNA helicases and other chaperone-like factors, nucleases, factors involved in nucleolar-cytoplasmic transport, and structural components of the nucleolus. In yeast, the genes encoding the majority of these factors are essential or cause significant growth defects when mutated, indicating that they play a critical role in cell growth and division. Collectively, these genes represent hundreds of potential target loci that when mutated may contribute to human disease, making it imperative that we understand the pathophysiology underlying the handful of human disorders currently linked to factors involved in ribosome synthesis.
Many of the structural components of the ribosome, as well as additional biosynthetic factors, have been shown to possess extraribosomal functions, ranging from roles in replication and DNA repair to serving as receptors on the cell surface.11 More recently, an emerging body of data has linked specific ribosomal proteins to active roles in cellular stress response,12,13 proliferation,14 and apoptosis.15 The challenge in studying these factors and their roles in human disease is sorting out their more general roles in modulating protein synthetic capacity from functions more specific to a given factor, a challenge further complicated by the diverse and variable array of clinical phenotypes observed in the bone marrow failure syndromes discussed. Here we summarize some of the current approaches being used to mechanistically dissect these disorders and suggest possible venues for future research.
DKC1 and X-linked DC: importance of additional disease-linked genes
We begin our analysis with X-linked DC, which may serve as a model wherein ribosomal dysfunction plays a role in bone marrow failure but is clearly not the sole explanation. Ectodermal dysplasia and hematopoietic failure characterize DC (reviewed in Liu and Dokal16 ). In addition to the classic triad of abnormal skin pigmentation, dystrophic nails, and leukoplakia of mucous membranes, there are a number of other somatic abnormalities.17 The most common of these are epiphora, developmental delay, pulmonary disease, short stature, esophageal webs, dental caries, tooth loss, premature gray hair, and hair loss. Pancytopenia is the hematologic hallmark of DC, and approximately 50% of patients develop severe aplastic anemia and greater than 90% of individuals at least a single cytopenia by 40 years of age.
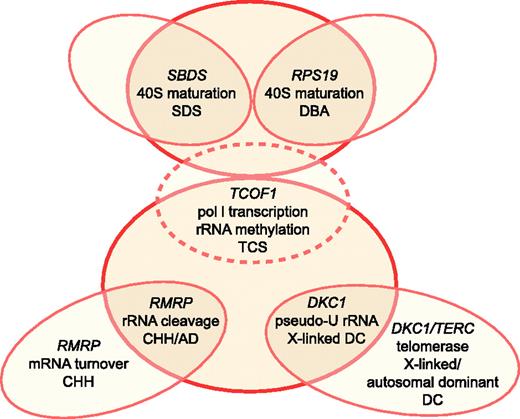
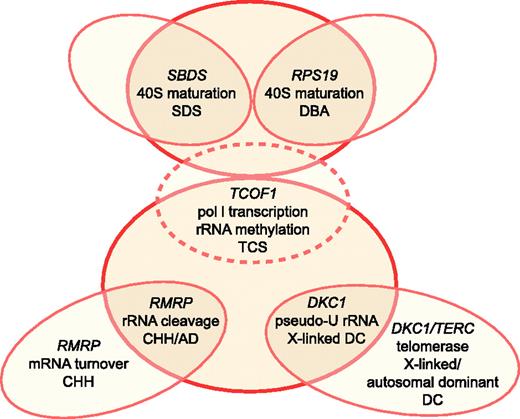
Schematic relationship between ribosome synthesis and SDS, DBA, TCS, CHH, AD, and DC. TCS is depicted by dotted lines, as it does not cause bone marrow failure. Putative functions in ribosomal biogenesis attributed to each gene product include 40S ribosomal subunit maturation, RNA polymerase I (pol I) transcription, rRNA methylation, rRNA cleavage, and pseudouridylation (pseudo-U) of rRNA. Illustration by Frank Forney.
Schematic relationship between ribosome synthesis and SDS, DBA, TCS, CHH, AD, and DC. TCS is depicted by dotted lines, as it does not cause bone marrow failure. Putative functions in ribosomal biogenesis attributed to each gene product include 40S ribosomal subunit maturation, RNA polymerase I (pol I) transcription, rRNA methylation, rRNA cleavage, and pseudouridylation (pseudo-U) of rRNA. Illustration by Frank Forney.
Dyskeratosis congenita is most commonly inherited as an X-linked recessive disorder, although autosomal dominant (ADO-DC) and autosomal recessive (AR-DC) forms exist. The gene responsible for the X-linked form was subsequently identified as DKC1.1 DKC1 encodes dyskerin, a nucleolar protein that is 1 of 4 proteins associated with small nucleolar ribonucleoprotein particles (snoRNPs) involved in rRNA maturation. Dyskerin associates with the H/ACA (referring to characteristic short snoRNA sequences) class of snoRNPs, which function in the pseudouridylation of rRNAs. Structural similarities between dyskerin and yeast and bacterial pseudouridine synthases suggested that dyskerin might be the active human pseudouridine synthase in the H/ACA complexes. At the time of its discovery, this was the only known function of dyskerin, thus leading to the inference that defects in ribosome synthesis, resulting from failure to properly modify rRNA, are responsible for the manifestations of X-linked DC (Figure 2).
Soon thereafter, however, it became clear that dyskerin had other functions within cells. Most importantly, dyskerin was found to be associated with the telomerase complex.5 The telomerase ribonucleoprotein complex, which is at times nucleolar, includes the telomerase reverse transcriptase TERT and the telomerase RNA component TERC, which acts as a template for the synthesis of the TTAGGG telomere repeats at the ends of chromosomes. Telomeres protect the ends of chromosomes from degradation and aberrant fusion events. TERC has an H/ACA-like domain and is associated with dyskerin, GAR1, NOP10, and NHP2. Here, dyskerin appears to serve as a structural component of the telomerase complex in contrast to its proposed role in the rRNA-modifying snoRNPs, as telomerase has no pseudouridylation target. With the discovery that cells from patients with X-linked DC and ADO-DC exhibit shortened telomeres18 and the subsequent identification of causative mutations in TERC in ADO-DC,19 it was presumed that defective telomerase activity, rather than impaired ribosomal maturation, underlies the bone marrow failure seen in DC. Mutations in the TERT reverse transcriptase gene have also been identified in aplastic anemia patients with short telomeres, further implicating the telomerase pathway.20
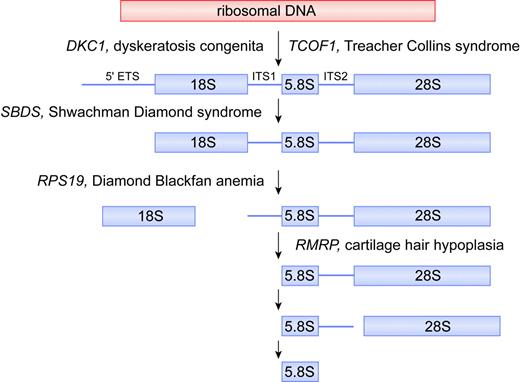
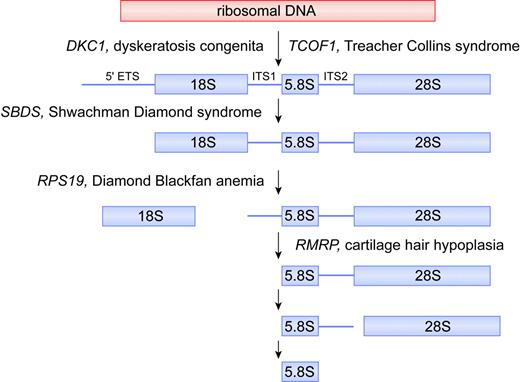
Putative biochemical function for DKC1, TCOF1, SBDS, RPS19, and RMRP in ribosomal biogenesis. For simplicity, the mammalian rRNA processing pathway is depicted, whereas the putative function of each gene product is deduced from data obtained from yeast and human orthologs. In the mammalian rRNA processing pathway, cleavages at the external transcribed sequence (5′ETS) and internal transcribed sequences (ITS1 and ITS2) lead to maturation of 18S, 5.8S, and 28S rRNA species. The 18S rRNA is a component of the 40S ribosomal subunit. The 5.8S and 28S rRNAs, together with the 5S rRNA (not shown), are components of the 60S ribosomal subunit. The 40S and 60S subunits are assembled into 80S ribosomes. Illustration by Frank Forney.
Putative biochemical function for DKC1, TCOF1, SBDS, RPS19, and RMRP in ribosomal biogenesis. For simplicity, the mammalian rRNA processing pathway is depicted, whereas the putative function of each gene product is deduced from data obtained from yeast and human orthologs. In the mammalian rRNA processing pathway, cleavages at the external transcribed sequence (5′ETS) and internal transcribed sequences (ITS1 and ITS2) lead to maturation of 18S, 5.8S, and 28S rRNA species. The 18S rRNA is a component of the 40S ribosomal subunit. The 5.8S and 28S rRNAs, together with the 5S rRNA (not shown), are components of the 60S ribosomal subunit. The 40S and 60S subunits are assembled into 80S ribosomes. Illustration by Frank Forney.
Although the pendulum has swung away from ribosome synthesis and toward telomere maintenance as explaining DC pathophysiology, the observation that the X-linked form of the disease is more severe than the autosomal dominant form leaves open the possibility that an effect on ribosome synthesis may contribute to phenotypic outcome. An extremely severe variant of DC known as Hoyeraal-Hreidarsson syndrome, which leads to aplastic anemia, immunodeficiency, and cerebellar hypoplasia, is also caused by mutations in DKC1, demonstrating that allelic differences in the same protein could lead to more severe phenotypes.21 We are also left with the puzzling features of the dyskerin hypomorphic mouse model,22 which displays some of the clinical features of DC prior to the onset of significant telomere shortening. That these mice display defects in ribosome synthesis suggests there may be some overlap in phenotype associated with defects in ribosome synthesis and telomere maintenance. In addition, 2 dyskerin patient mutations introduced into murine embryonic stem cells caused defective pseudouridylation and decreased pre-rRNA processing, but only 1 of the 2 mutations affected telomerase activity.23 Thus, in yeast,24 Drosophila,25 and 2 different mouse models defects in dyskerin orthologs lead to a reduced rate of production of mature rRNAs and accumulation of unprocessed precursor molecules. Collectively, these data argue for involvement of both ribosome deficiency and telomerase dysfunction in DC pathophysiology, with the contribution of each possibly dependent on the particular mutation site within dyskerin. The crystal structure of the dyskerin-NOP10-GAR1 complex has recently been deduced,26 identifying a dyskerin domain within which most DC mutations cluster. Current efforts employing detailed genotype/phenotype correlations may assist in clarifying the relative contributions of ribosome synthesis and telomere maintenance to the clinical features of DC.
RMRP and cartilage-hair hypoplasia: genotype/phenotype relationships
CHH, also known as metaphyseal chondrodysplasia McKusick type, was first described by McKusick et al27 as a form of dwarfism in the Amish. CHH is an autosomal recessive disorder characterized by cartilage/skeletal defects with short stature, fine sparse blond hair, and hypoplastic anemia. The anemia, while typically mild, can be persistent and severe as in DBA.28 CHH is also associated with defective cellular immunity,29 gastrointestinal dysfunction,30 and a predisposition to cancer (primarily lymphoma).31 Mutations in the untranslated RMRP gene were found to cause CHH, with an ancient founder mutation in Finland.2
RMRP encodes the RNA component of the RNase MRP complex and is classified as a small nucleolar RNA (RMRP and the previously mentioned TERC are the 2 only known nonmitochondrial examples of RNA genes that when mutated lead to disease). In yeast, the RMRP ortholog NME1 (nuclear mitochondrial endonuclease-1) has multiple functions and is essential for viability.32 First, it functions in mitochondrial DNA replication by cleaving RNA primers associated with nascent organelle DNA.33 Second, some nme1 mutants exhibit a late mitotic cell cycle arrest associated with increased cyclin B2 mRNA levels (normally, the RNase MRP complex cleaves the 5′ untranslated region of cyclin B2 mRNA).34 Third, RNase MRP plays a role in ribosomal RNA processing.35 In both yeast and humans, RNase MRP cleaves pre-rRNA within internal transcribed sequence 1 (ITS1), influencing the maturation of the 5′ end of 5.8S rRNA (Figure 2). Consistent with this latter function, RNase MRP is mainly located in the nucleoli packaged as one of the snoRNPs. In humans, the RNase MRP complex consists of the RNA component and at least 10 protein subunits,36 of which 8 are shared with ribonuclease P, involved in tRNA precursor maturation.
Recent work has suggested that it is the function of RMRP in cell cycle–regulated mRNA turnover that underlies the bone marrow failure seen in CHH.37 This conclusion is supported by the discovery of novel RMRP mutations that lead to another autosomal recessive growth disorder known as anauxetic dysplasia (AD), which is characterized by extreme short stature, hypodontia, and mild mental retardation. While the growth abnormalities in anauxetic dysplasia are reminiscent of those in CHH, hypoproliferative bone marrow dysfunction and cancer susceptibility are seen only in CHH and not in anauxetic dysplasia. In vitro studies have shown that human fibroblasts overexpressing the CHH founder mutation +70A > G exhibited both impaired rRNA endonucleolytic cleavage and cyclin B2 mRNA cleavage (and hence increased cyclin B2 mRNA levels), whereas cells overexpressing anauxetic dysplasia mutations exhibited only rRNA cleavage abnormalities.37 These genotype/phenotype studies imply that while RMRP mutations can disrupt 5.8S rRNA maturation (and hence 60S ribosomal assembly) resulting in some of the clinical features of CHH, it is likely the concurrent disruption of cyclin B2 mRNA degradation and mitotic delay that explains the bone marrow failure phenotype in CHH (Figure 1).
RPS19 and DBA: focus remains on ribosomes
We now turn our focus to DBA, currently the only known human disease caused by defects in a structural component of the ribosome, namely RPS19. DBA is a pure red cell aplasia predominantly seen in infancy and childhood. In addition to anemia, other significant cytopenias have been reported.38,39 An elevated erythrocyte adenosine deaminase activity (found in approximately 85% of patients), macrocytosis, and an elevated fetal hemoglobin level are supportive but not diagnostic of DBA. Congenital anomalies including thumb malformations and various others are seen in 50% of DBA patients and, in turn, half of these are craniofacial abnormalities.40
Approximately 45% of DBA cases are familial.41 The first DBA gene, mutated in about 25% of patients, was cloned and identified as RPS19, which encodes a protein component of the 40S ribosomal subunit.3,42 Patients are generally heterozygous with respect to RPS19 mutations, indicating that the disease likely results from protein haploinsufficiency.43 Studies of the yeast ortholog of RPS19 indicate it is required for the maturation of 40S ribosomal subunits6 (Figure 2). Yeast cells deficient in RPS19 have a reduction in the steady-state level of 40S ribosomal subunits and compromised protein synthetic capacity. Pre-40S particles that accumulate in cells depleted of RPS19 are retained in the nucleolus and fail to recruit factors needed for rRNA processing and transport to the cytoplasm.
If human RPS19, like its yeast ortholog, is required for the maturation of 40S ribosomal subunits, questions remain as to how haploinsufficiency for an essential ribosomal protein results in the predominantly hematopoietic manifestations of the disease. Ellis and Massey44 have proposed that differences in the level of individual ribosomal components can make proteins limiting for ribosome assembly in some tissues and not others, when present in a haploinsufficient state. As discussed later, in the section on Treacher Collins syndrome, this may explain the tissue specificity characterizing disorders such as DBA. Recent studies by other investigators showing that RPS19 interacts with the oncoprotein Pim1 have raised an alternative hypothesis, in which RPS19 links signal transduction pathways with the translational machinery.45 Finally, the observation that cross-linked dimers of RPS19 function as agonists and antagonists of the C5a complement receptors7 suggests other extraribosomal functions for RPS19 that may contribute to DBA pathophysiology in a manner not currently understood.
As the different functions for RPS19 are gradually clarified, ongoing efforts to genotype DBA patients and correlate clinical phenotypes with RPS19 mutations may assist in distinguishing between these alternative hypotheses. Likewise, identifying other genes responsible for DBA other than RPS19 will be critical in directing further research, as well as in either validation or questioning of the ribosome and marrow failure link.
SBDS and Shwachman-Diamond syndrome: new player on the block
Shwachman-Diamond syndrome (SDS), first described in 1964, is an autosomal recessive disorder characterized by neutropenia, exocrine pancreatic insufficiency, and metaphyseal dysostosis.46,47 Neutropenia, found in 88% to 100% of patients, is the hematologic hallmark of SDS, and in some patients there is also a defect in neutrophil chemotaxis (reviewed in Dror and Freedman48 ). Pancytopenia occurs in 10% to 65% of cases and may precede progressive bone marrow dysfunction characterized by severe aplastic anemia, myelodysplastic syndrome, or acute leukemia, usually of myeloid origin.49
Most cases of SDS are caused by mutations in the SBDS gene.4 While SBDS is highly conserved in species as divergent as archaebacteria, little is known regarding the precise function of its gene product. The localization of the archaeal SBDS in an operon with subunits of the exosome, a complex of nucleases involved in a number of different RNA processing reactions, initially hinted at a role for SBDS in RNA metabolism. These data are supported by studies in yeast, showing that depletion of the SBDS ortholog SDO1 (also known as YLR022c) interferes with the processing of the 35S rRNA primary transcript.50 In the initial screening experiments, which involved a panoramic view of yeast mutants involved in RNA metabolism using regulated promoter alleles to silence expression of selected genes, the SDO1 mutants exhibited a so-called processome-like phenotype.50 This is characterized by inefficient cleavage of the primary rRNA transcript within the 5′ external transcribed sequence (ETS) and ITS1 sites, which in turn should lead to a defect in 40S subunit maturation, similar to that seen with defects in RPS19 (Figure 2). Although our own preliminary experiments confirm inefficient cleavage of the primary rRNA transcript,51 the exact molecular function of the SBDS orthologs is likely to be more complex than initially believed and involve the 60S ribosomal subunit as well. In this regard, tagged Sdo1 copurified with over 20 proteins involved in ribosome biosynthesis, including components of the 60S subunit.8 The finding that the human SBDS protein is localized to the nucleolus further supports a role for this protein in ribosome synthesis.52 More detailed studies on the function of SBDS will be necessary to clarify this specific function. As noted for the other bone marrow failure gene products under discussion here, it would not be surprising to find extraribosomal functions for SBDS as well.
Treacher Collins syndrome: severing the ribosome synthesis/bone marrow failure link?
Treacher Collins syndrome (TCS), an autosomal dominant disorder of craniofacial development, is caused by mutations in the TCOF1 gene, which encodes a nucleolar phosphoprotein called treacle (Table 1). Treacle has been shown to affect ribosomal DNA transcription by interacting with upstream binding factor, an RNA polymerase I transcription factor53 (Figures 1, 2). In addition, treacle plays a role in pre-rRNA methylation.54 Haploinsufficiency of treacle in TCS disrupts ribosome synthesis in a manner reminiscent of that observed in dyskerin mutants, which also affect rRNA modification. Yet TCOF1 mutants lead to abnormal proliferation and/or differentiation of neural crest precursor cells involved in craniofacial development without interfering with hematopoiesis. A subset of DBA patients, who as a group appear to lack RPS19 mutations, presents with craniofacial abnormalities indistinguishable from those seen in TCS.55,56 Thus, mutations in as yet unidentified DBA genes may give rise to ribosomal defects reminiscent of TCOF1 haploinsufficiency, manifesting during embryogenesis and again during erythroid development.
These results, together with the foregoing discussion, suggest that defects in factors involved in ribosome synthesis affect tissues in different highly specific ways that are difficult to predict. Clearly, many questions remain unanswered regarding the bone marrow failure–ribosome synthesis link. It will be important to determine if the consequences of disrupting the function of ribosome synthesis factors result from quantitative changes in protein synthetic capacity in a tissue-specific manner or instead a differential sensitivity of certain cell lineages to a general reduction in protein synthetic capacity. Only further experiments will be able to resolve these critical issues.
Defects in ribosome synthesis: the cancer connection
Mutations in genes involved in each of the diseases discussed have the potential to perturb the synthesis of ribosomal subunits and, as such, fundamentally alter the protein synthetic capacity of cells. Such alterations may also sensitize cells to apoptotic stimuli, accelerating programmed cell death pathways. Recent evidence suggests that cytopenias in many, if not all, of the bone marrow failure syndromes may be a consequence of accelerated apoptotic death of hematopoietic progenitor cells.57-64 A possible explanation for this apoptotic sensitivity relates to the activation of the p53 tumor suppressor. It has been postulated that disturbances in ribosome synthesis must be detected and coupled with cell cycle arrest in order to prevent aberrant cell divisions. Hence, close coordination exists between p53-mediated cell growth regulation and cellular responses to malfunction of ribosome synthesis. Normally, ribosomal biogenesis is coordinated by growth signals.65 The tumor suppressors p53,66,67 the RB retinoblastoma protein,68-70 and p19ARF71 have all been shown to interfere with rRNA synthesis or processing, thereby controlling ribosomal biogenesis. Overproduction of these tumor suppressors inhibits protein synthesis while arresting cell growth in response to stress signals. Apparently, p53 can also sense perturbations in ribosomal biogenesis, as from defects in rRNA synthesis and processing or disruption of ribosome assembly.72,73 Therefore, disruption of ribosomal biogenesis (and resulting “nucleolar stress”13 ) should lead to p53 activation and oppose oncogenic transformation.74-76 While this may contribute to the proliferative defects in bone marrow failure, how might leukemia or nonhematopoietic cancer evolve? Presumably, for leukemia, there is strong selective pressure for clonal outgrowth of cells with mutations that can compensate for the “ribosome-depleted” state. One way to increase ribosome synthesis would be to overexpress the Myc oncoprotein,77 a major regulator of ribosome-related genes, or to inactivate RB, which dampens RNA polymerase I. For nonhematopoietic cancers as are seen in DBA,78 mutations in ribosomal genes may be directly oncogenic as suggested by the recent finding that heterozygous mutations in several zebrafish ribosomal protein genes induce tumors.79
Concluding remarks
In this “Perspective,” we have attempted to critically appraise the hypothesis that defects in ribosomal biogenesis underlie bone marrow failure. At present, the ribosome/marrow failure association does not seem to rise to the level of a molecular paradigm. Treacher Collins syndrome and anauxetic dysplasia demonstrate that defects in ribosome synthesis can cause clinical syndromes that do not include bone marrow failure. Moreover, the pathologies observed in X-linked DC and CHH appear to be predominantly consequences of extraribosomal functions of each affected gene product. In these latter examples, however, associated defects in ribosome synthesis may serve as important disease modifiers (Figure 1). In this context, defects in ribosome synthesis may enhance disease severity by compounding the effects mediated through the extraribosomal functions or by expanding the range of clinical manifestations by affecting additional tissue types. These data also do not exclude defects in ribosome synthesis as a primary cause for other bone marrow failure syndromes such as DBA and SDS. Regardless of whether factors that affect ribosome synthesis are a principal determinant or a modifier of disease severity in the bone marrow failure syndromes, novel therapeutic strategies aimed at restoring ribosomal function in hematopoietic cells may prove useful in treating cytopenias. Finally, components of the ribosomal pathway may represent targets for cancer treatment, which may break the vicious cycle between marrow failure and cancer predisposition.
Prepublished online as Blood First Edition Paper, February 28, 2006; DOI 10.1182/blood-2005-12-4831.
We thank Drs Jeffrey Lipton and Amy Massey for critically reviewing the manuscript.