Controversies have arisen as to whether adult stem cells or progenitor cells from bone marrow can engraft into nonhematopoietic tissues in vivo. To resolve some of the controversies, we developed a highly sensitive polymerase chain reaction-based single nucleotide polymorphism (PCR-SNP) assay for competitive engraftment of mixtures of stem/progenitor cells. We used the assay to follow engraftment in immunodeficient mice of subpopulations of the stem/progenitor cells from human bone marrow referred to as either mesenchymal stem cells or marrow stromal cells (MSCs). The engraftment into adult mice without induced tissue injury was low and variable, but there was preferential engraftment of a subpopulation of rapidly self-renewing MSCs (RS-MSCs) compared with a subpopulation of slowly renewing MSCs (SR-MSCs). After intravenous infusion, there was a tendency for the cells to engraft into the hippocampal region that was previously designated a “vascular niche.” Migration assays suggested that preferential engraftment of RS-MSCs was in part explained by their expression of CXCR4 and CX3R1, the receptors for SDF-1 and fractalkine.
Introduction
A series of reports have indicated that bone marrow contains several subpopulations of cells that engraft in multiple nonhematopoietc tissues after systemic or intracerebral infusion.1-13 It is not clear, however, which of the many subpopulations of cells from bone marrow can most effectively home to and engraft in nonhematopoietic tissues.10,11,13-19 Among the marrow subpopulations that have been shown to engraft widely are the plastic-adherent cells referred to as mesenchymal stem cells or marrow stromal cells (MSCs). MSCs are attractive candidates for cell and gene therapies,1,16,20,21 because they are readily obtained from the patient to be treated and therefore do not generate immune responses. Also, MSCs have a limited tendency to produce tumors, a prominent feature of embryonic stem cells.
Some of the interest in MSCs and other adult stem or progenitor cells is based on the observation that the cells home to injured tissues and repair them.16 The cells can repair tissues by several different mechanisms: the cells can differentiate into the phenotype of the damaged cells in the tissues, and the cells can secrete growth factors and cytokines that enhance repair of endogenous cells. The cells, like other stem/progenitor cells, can also undergo cell fusion, particularly if selective pressure for cell fusion is present.22-25 However, the extent of cell fusion is relatively low in most circumstances.26-28
A controversy concerning adult stem/progenitor cells has been generated by several recent publications reporting a failure to detect engraftment of cells from bone marrow into nonhematopoietic tissues in vivo.24,25,29 The controversy is largely explained by the complexity of experiments: (1) most of the stem/progenitor cells are either strongly committed to a lineage of differentiation or are poorly characterized so that cells prepared in different laboratories with similar protocols can differ markedly in subtle but important properties; (2) the cells are infused into animals in which the degree of engraftment is markedly influenced by factors such the rate of growth and the presence of different kinds and degrees of tissue injury; (3) the markers to label the cells can be lost as the cells differentiate in vivo; and (4) the analytic techniques used to detect the cells are frequently not quantitative or sensitive enough to detect low levels of engraftgment. In experiments with MSCs, some of the discrepancies are explained by the heterogeneity of the cells as they are expanded in culture.30,31 MSCs are readily prepared as clonal populations, but single-derived clones become heterogeneous as they expand so that they contain at least 2 subpopulations of cells: small, rapidly self-renewing MSCs (RS-MSCs) and larger, slowly renewing MSCs (SR-MSCs). The 2 subpopulations can in part be distinguished by differences in their potential for differentiation and their surface epitopes.30
To help resolve some of the current controversies, we have developed a sensitive polymerase chain reaction-based single nucleotide polymorphism (PCR-SNP) assay for the competitive engraftment of mixtures of subpopulations of MSCs from 2 different human donors. The assay demonstrated marked differences in the engraftment of human RS-MSCs and SR-MSCs after either intravenous or intracranial infusion into immunodeficient mice. Moreover, the results suggested that the more effective engraftment of RS-MSCs is explained by their expression of the receptors CXCR4 and CX3CR1, since RS-MSCs migrated more rapidly to murine neurospheres than SR-MSCs and their migration was selectively decreased by neutralizing antibodies to CXCR4 and CX3CR1.
Materials and methods
Isolation and culture of human MSCs
Frozen vials of human MSCs (hMSCs) from 2 healthy donors, prepared as described previously,30,32 were obtained from the Tulane Center for Distribution of Adult Stem Cells (Center for Gene Therapy, Tulane University Health Sciences Center, New Orleans, LA). The MSCs were prescreened to establish that the cells from the donors were distinguishable on the basis of SNPs in the first intron of the COL1A2 gene (Supplemental Table S1 and Figure S1; see the Supplemental Materials link at the top of the online article, at the Blood website). To expand the MSCs, a frozen vial of about 106 MSCs from Passage 2 was thawed, plated in a 175-cm2 dish, and incubated in 25 mL complete culture medium (CCM) composed of α-minimum essential medium (α-MEM; GIBCO/BRL, Grand Island, NY); 20% fetal bovine serum (FBS; lot-selected for rapid growth of MSCs; Atlanta Biologicals, Miami, FL); 100 U/mL penicillin; 100 μg/mL streptomycin; and 2 mM l-glutamine (GIBCO/BRL). After 1 to 2 days, the adherent cell layer was washed with phosphate buffered saline (PBS), and the viable adherent cells were harvested with 0.25% trypsin and 1 mM EDTA (ethylenediaminetetraacetic acid) for 5 minutes at 37°C. The cells were expanded by plating at initial densities of about 100 cells/cm2 in 175-cm2 dishes and in the CCM. The medium was replaced every 2 to 3 days. The cultures were lifted with EDTA/trypsin after they reached about 70% confluency in 5 to 7 days. Cell numbers were counted with a hemocytometer. To isolate samples enriched for RS-MSCs and SR-MSCs,30-32 MSCs from 5- to 7-day cultures were separated on the basis of forward scatter (FS) or side scatter (SS) of light using FACS Vantage SE flow cytometer (Becton Dickinson, San Jose, CA). The samples were sorted by gating for about 10% of the events in the lower left quadrant and about 10% of the cells in the upper right quadrant of the plot of FS/SS (Smith et al31 and Figure 1).
Cell cycle profile
From 2.5 × 105 to 5.0 × 105 MSCs in 100 μL PBS were mixed with 100 μL lysis buffer (DNA-Prep LPR; Beckman-Coulter, Fullerton, CA). The sample was gently vortexed and immediately afterward 500 μL of a propidium iodide solution containing a lysis buffer and a staining reagent (DNA-Prep Stain Reagent; Beckman-Coulter) was added. After gently vortexing for 5 seconds, the sample was incubated for 20 minutes at room temperature in the dark. The sample was assayed within 2 hours by flow cytometry (Cytomic FC 500; Beckman-Coulter), and cell cycle status was calculated by using software (ModFit 3.0; Beckman-Coulter).
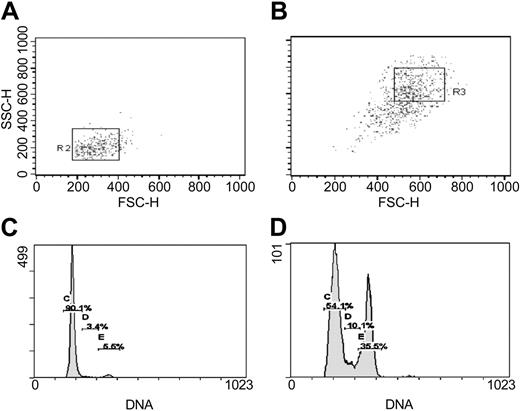
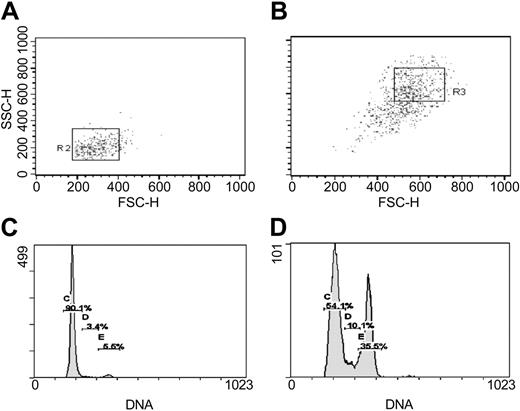
Separation by FACS and cell cycle assay of RS-MSCs and SR-MSCs from 5-day cultures of human MSCs initially plated at 100 cells/cm2. (A) Re-assay of FS/SS of RS-MSCs after sorting for low FS/low SS.31 (B) Re-assay of SR-MSCS after sorting for high FS/high SS. (C) Cell cycle analysis of RS-MSCs. (D) Cell cycle analysis of SR-MSCs.
Separation by FACS and cell cycle assay of RS-MSCs and SR-MSCs from 5-day cultures of human MSCs initially plated at 100 cells/cm2. (A) Re-assay of FS/SS of RS-MSCs after sorting for low FS/low SS.31 (B) Re-assay of SR-MSCS after sorting for high FS/high SS. (C) Cell cycle analysis of RS-MSCs. (D) Cell cycle analysis of SR-MSCs.
Intravenous and cerebral infusions
For intravenous infusion, 5- to 6-week-old severe combined immunodeficient (SCID)/Beige mice (Charles River, Wilmington, MA) were anesthetized with intraperitoneal injection of 80 mg/kg ketamine and 8 mg/kg xylazine. The tails were immersed in warm water for about 5 minutes and 200 μL of cell suspension in PBS was slowly infused into a tail vein with a 27-gauge needle. The cell suspension was a 1:1 mixture of 2.5 × 105 RS-MSCs from one human donor and 2.5 × 105 SR-MSCs from a second donor whose cells could be distinguished by SNP in the COL1A2 gene (Table S1 and Figure S1). Prior to injection, the cells were maintained at 4°C and they were gently resuspended with a pipette to ensure they did not aggregate before infusion.
For cerebral infusions, 5- to 6-week-old SCID/Beige mice were anesthetized with ketamine/xylazine and infused with either 100 000 RS-MSCs from one donor or with 1:1 mixtures of 50 000 RS-MSCs from one donor and 50 000 SR-MSCs from the second donor that could be distinguished by SNPs. The cells were injected through a 30-gauge needle over 5 minutes into the hippocampus of one hemisphere at -2.3 mm posterior, -2.0 mm lateral, and -2.5 mm ventral to bregma. The bevel of the needle was directed caudally.
Assays for Alu and SNPs by real-time PCR
For PCR assays, the mice were killed by CO2 gas and genomic DNA from tissues was extracted using DNeasy tissue kit (Qiagen, Chatsworth, CA). The amount of DNA was assayed first by absorbance and then by real-time PCR assays for the mouse albumin gene to normalize genomic DNA. PCR assays were performed in a volume of 50 μL that contained 25 μL Universal PCR Master Mix (Applied Biosystems, Foster City, CA), 900 nM each of the forward and reverse primers, 250 nM TaqMan probe, and 200 ng target template. Reactions were incubated at 50°C for 2 minutes for optimal uracil-N-glycosylase (UNG) enzyme activity and at 95°C for 10 minutes to activate AmpliTaq Gold enzyme followed by 40 cycles at 95°C for 15 seconds followed by 60°C for 1 minute. Standard curves were generated by serially diluting human genomic DNA prepared from MSCs into samples containing 200 ng genomic DNA from mouse brain. Primers for the mouse albumin gene were designed to amplify a specific intron sequence of the single copy serum albumin variant Alb1 on chromosomes (NCBI; Mus musculus genome view54 ). The mouse albumin forward primer was 5′-GAA AAC CAG GCG ACT ATC TCC A-3′; mouse albumin reverse primer was 5′-TGC ACA CTT CCT GGT CCT CA-3′. PCR assays for the mouse albumin gene were performed using SYBR Green PCR Master Mix (Applied Biosystems). The sequence of the PCR primers and the probe used for detection of human Alu repetitive sequences33 were as follows: Alu forward, 5′-CAT GGT GAA ACC CCG TCT CTA-3′; Alu reverse, 5′-GCC TCA GCC TCC CGA GTA G-3′; TaqMan probe, 5′-FAM-ATT AGC CGG GCG TGG TGG CG-TAMRA-3′ (Applied Biosystems). To express the results as human cells per organ, the detected level of human DNA in 200 ng mouse DNA was multiplied by total DNA per organ extracted with phenol/chloroform and assayed by absorbance. Assuming a value of 5 pg DNA per cell,34 the cellular contents were as follows: brain, 2.59 × 108 ± 0.43 (n = 5); heart, 4.21 × 107 ± 1.56 (n = 5); liver, 1.32 × 109 ± 0.605 (n = 4); kidney, 6.14 × 108 ± 1.00 (n = 5); spleen, 1.90 × 108 ± 0.797 (n = 5); and lung, 2.19 × 108 ± 0.56 (n = 5).
To assay SNPs, sequences in the first intron of the human COL1A2 gene region (Table S1 and Figure S1) were first amplified with primers flanking the SNPs.35 The PCRs were performed in a volume of 50 μL and contained 1 U/μL recombinant Taq DNA polymerase (Invitrogen, Carlsbad, CA), 1X PCR buffer, 0.2 mM each dNTP, 1.5 mM MgCl2, and 500 nM of each primer. The COL1A2 forward primer was 5′-CAT CCA CAC ACA TGC ACA GA-3′; the COL1A2 reverse primer was 5′-TTT CCC CTT TGT TGT TTC CA-3′. The amplification conditions were incubated at 95°C for 5 minutes, and then 25 cycles of 94°C for 1 minute, 58°C for 1 minute, and 72°C for 1 minute. After amplification of COL1A2 region, 2 μL of the PCR reaction was used for SNP assays by real-time PCR. The primers were designed to ensure specificity by introducing a mismatch into the penultimate 3′-base (see supplemental data and Maas et al36 ). The SNP forward primer for the A/A allele was 5′-GTA ATC ACA GCC TCC ATG AAA TAG A-3′; SNP forward primer for the G/G allele was 5′-GTAATC ACA GCC TCC ATG AAA TAT G-3′; SNP reverse primer for the A/A allele was 5′-ATA ACA TGG ATT TTA TCT AAA ATG TGT-3′; SNP reverse primer for the G/G allele was 5′-ATA ACA TGG ATT TTA TCT AAA ATG TGC-3′. The TaqMan probe was 5′-FAM-TGC CTA AAA AGC TAT TGT GAT GGA AAA GTG ACA GT-TAMRA-3′ (Applied Biosystems). The conditions for amplification were the same as for Alu sequences. All real-time PCR assays were performed in duplicate or triplicate and average values are presented.
Immunohistochemistry and immunocytochemistry
For immunohistochemistry of brain sections, mice were anesthetized with intraperitoneal injection of 80 mg/kg ketamine and 8 mg/kg xylazine, and perfused through the right atrium with 30 mL PBS followed by 10 mL of 4% paraformaldehyde. The tissues were excised, rinsed with PBS, fixed in 4% paraformaldehyde in PBS fixative overnight at 4°C, transferred to 30% sucrose solution overnight at 4°C, flash frozen, and 5-μm sections were cut in a cryostat. The sections were incubated for 18 hours at 4°C with antibody to a human-specific nuclear antigen (1:100, mouse anti-human nuclei monoclonal antibody; Chemicon, Temecula, CA). The slides were washed 3 times for 5 minutes with PBS and incubated for 1 hour at room temperature with secondary antibody (1:1000, Alexa-594; Molecular Probes, Eugene, OR). The slides were coverslipped with a DNA counterstain (DAPI; Vector Labs, Burlingame, CA). Controls included omitting the primary antibody. Slides were evaluated by epifluorescence (Eclipse 800; Nikon, Melville, NY) using a 60 × objective.
For immunocytochemistry of cells, RS-MSCs isolated by fluorescence-activated cell sorting (FACS) were plated at initial densities of about 1000 cells/cm2 in a slide chamber (LAB-TEK ll chamber slide; Nalge Nunc International, Rochester, NY). After incubation for 1 day, the cultures were rinsed with PBS and fixed in 4% paraformaldehyde in PBS for 20 minutes at room temperature. The slide chambers were incubated for 18 hours at 4°C with antibody to CXCR4 (1:200; Chemicon) and CX3CR1 (1:200; Abcam, Cambridge, MA). The slides were washed 3 times for 5 minutes with PBS and incubated for 1 hour at room temperature with secondary antibody (1:1000, Alexa-594; Molecular Probes). The slides were cover-slipped with a DNA counterstain (DAPI). Controls included omitting the primary antibody. Slides were evaluated by epifluorescence (Eclipse 800; Nikon) using a 40 × objective. Images were analyzed using SPOT-RT imaging software (Diagnostic Instruments, Sterling Heights, MI).
Migration assays
Isolated RS-MSCs and SR-MSCs were labeled by incubation at 37°C for 30 minutes in α-MEM containing a cellular dye (Tracker Green CMFDA; Molecular Probes) and washed 3 times by centrifugation with PBS. Migration assays were carried out in a 24-well transwell using opaque inserts with 8-μm pores (HTS FluoroBlok 24-Multiwell Insert System; BD Biosciences, San Jose, CA). Migration was followed either by a using a temperature-controlled fluorimeter (Fluostar Optima; BMG Labtechnologies, Offenburg, Germany) or by photomicrography of the underside of the opaque inserts. For the fluorometric assay, a standard curve was prepared with dye-labeled RS-MSCs and SR-MSCs placed in the bottom chambers of transwells with inserts. RS-MSCs or SR-MSCs at a concentration of 1 × 105 cells/mL in 300 μL of serum free medium were placed in the upper chamber. The bottom chambers were loaded with 1 mL serum-free medium containing 1 × 105 cells neural stem cells obtained from mouse brain of postnatal days 2 to 3,37 50 ng/mL SDF-1 (R&D Systems, Minneapolis, MN), or 10 ng/mL Fractalkine (Chemicon). For neutralization studies, MSCs were incubated for 30 minutes at room temperature with 10 μg/mL anti-human CXCR4 (clone 12G5; R&D Systems) or 5 μg/mL anti-human CX3CR1 (clone 2A9-1; MBL, Woburn, MA) before seeding. After loading both chambers, the transwells were incubated at 37°C for 16 hours. Values obtained with the fluorometric assay were confirmed by photomicroscopy using a 20 × objective of the underside of the inserts and counting cells in 2 random fields per filter. Alternatively, the assays were by photomicrography alone.
RT-PCR assays
RNA was isolated from 1 × 106 cells by the RNeasy RNA Isolation Kit (Qiagen, Valencia, CA) and 100 ng total RNA was used to perform reverse transcriptase (RT)-PCR assays with a commercial kit (M-MLV RT; Invitrogen). The samples were incubated at 37°C for 50 minutes followed by 15 minutes at 70°C to inactivate the RT. The cDNAs were amplified by PCR (Recombinant Taq DNA polymerase; Invitrogen) with 30 cycles at 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. PCR primers were CCR2 forward, 5′-CCA ACG AGA GCG GTG AAG AAG TC-3′, CCR2 reverse, 5′-TCC GCC AAA ATA ACC GAT GTG AT-3′; CXCR2 forward, 5′-CCG CCC CAT GTG AAC CAG AA-3′, CXCR2 reverse, 5′-AGG GCC AGG AGC AAG GAC AGA C-3′; CX3CR1 forward, 5′-TCC TTC TGG TGG TCA TCG-3′; CX3CR1 reverse 5′-TGT GCA TTG GGT CCA TCA-3′; CXCR4 forward, 5′-GGT GGT CTA TGT TGG CGT CT-3′; CXCR4 reverse, 5′-TGG AGT GTG ACA GCT TGG AG-3′; CCR5 forward, 5′-CTG GCC ATC TCT GAC CTG TTT TTC-3′, and CCR5 reverse 5′-CAG CCC TGT GCC TCT TCT TCT CAT-3′. The primers for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) used to control for efficiency of reverse transcription reaction were forward, 5′-TCA ACG GAT TTG GTC GTA TTG GG-3′; reverse, 5′-TGA TTT TGG AGG GAT CTC GC-3′.
Real-time RT-PCR assays for CXCR4 and CX3CR1
Total RNA from RS-MSCs, SR-MSCs, and total MSCs was extracted (RNeasy Mini Kit; Qiagen) and reverse transcribed (M-MLV RT; Invitrogen). cDNA was analyzed by real-time PCR (ABI 7700 Sequence Detector; Applied Biosystems) using the same primers as for the RT-PCR assays and a DNA dye (SYBR Green PCR Reagents; Applied Biosystems). Reactions were incubated at 50°C for 2 minutes, 95°C for 10 minutes, and then 40 cycles at 95°C for 15 seconds followed by 62°C for 1 minute.
Flow cytometry of CXCR4 and CX3R1 epitopes
RS-MSCs and SR-MSCs were isolated by FACS and about 17 500 cells plated in a 175-cm2 dish. The cells were incubated in CCM for 5 days and lifted with 1 mM EDTA for 10 minutes at 37°C. About 1 × 105 MSCs were stained by suspending the cells in 50 μL PBS containing 1% bovine serum albumin (BSA) and incubating for 30 minutes at 4°C with 10 μg/mL mouse monoclonal anti-human CXCR4-PE (clone 12G5; R&D Systems) or anti-human CX3CR1-FITC (clone 2A9-1; MBL). Cells were analyzed by flow cytometry (FACScalibur; BD Biosciences) with CellQuest software.
Results
Isolation and characterization of RS-MSCs and SR-MSCs
RS-MSCs and SR-MSCs were separated from early passage low-density cultures of the MSCs by FACS on the basis of forward scatter (FS) and side scatter (SC) of light (Figure 1). In initial reports,30 RS-MSCs were defined as the small subpopulation that clearly separated from the major body of the cells in plots of FS/SS. Subsequently, it was found that in order to obtain a large number of cells free of cellular debris, it was convenient to focus on a subpopulation of RS-MSCs that was recovered in the lower left quadrant of the distribution plot obtained by sorted unfractionated cultures of MSCs by FS/SS.31,32 The cells defined as RS-MSCs by this criterion were slightly larger but similar to the original subpopulation of RS-MSCs in that they expanded rapidly with doubling times of 10 to 12 hours and readily differentiated in culture.30,31 Also, up to 90% of the RS-MSCs generated single-cell-derived colonies in a clonogenic assay but the cells in each colony became morphologically heterogeneous as the colonies expanded.30,31
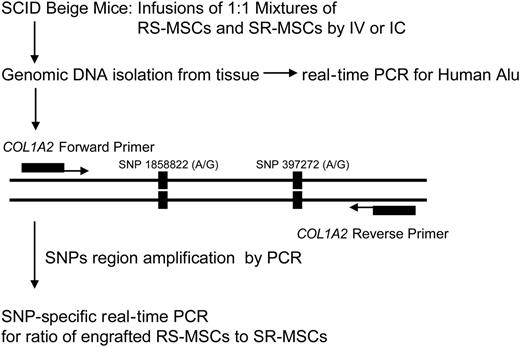
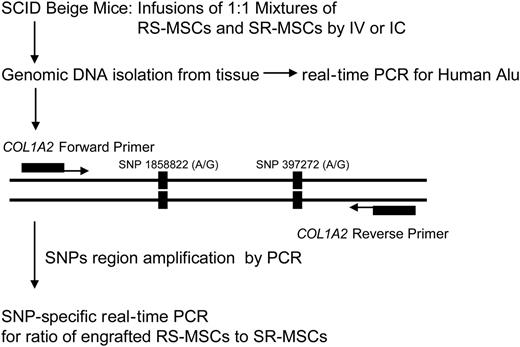
Experimental scheme for detection of human cells with allele-specific SNPs. The levels of engraftment were assayed by real-time PCR of Alu sequences. For the allele-specific SNP assay, a region of the COL1A2 gene containing a common polymorphism was amplified prior to the SNP assay (Figure S1).
Experimental scheme for detection of human cells with allele-specific SNPs. The levels of engraftment were assayed by real-time PCR of Alu sequences. For the allele-specific SNP assay, a region of the COL1A2 gene containing a common polymorphism was amplified prior to the SNP assay (Figure S1).
The relative homogeneity of RS-MSCs was demonstrated by re-assay of the cells by flow cytometry and cell cycle analysis (Figure 1). Essentially all of the initial sort of RS-MSCs were again recovered as RS-MSCs. About 90% of the RS-MSCs were in G1 (Figure 1A,C). In contrast, the fraction of SR-MSCs contained small and variable amounts of cells with low FS/SS, apparently because of adherence of some of the small cells to the larger SR-MSCs (Figure 1B). As expected, cell cycle assays indicated that the SR-MSCs were asynchronous (Figure 1D). RS-MSCs and SR-MSCs isolated as shown in Figure 1 were used in all subsequent experiments.
Real-time PCR-SNP assay for detection of low levels of engraftment of MSCs
Preliminary experiments (not shown) indicated that human MSCs infused into immunodeficient mice engrafted at very low levels. We employed 2 sensitive real-time PCR assays to compare the competitive engraftment of RS-MSCs and SR-MSCs (Figure 2). One assay was for the highly repetitive human Alu sequences, to determine the total level of engraftment of human cells. Standard curves demonstrated that the assay detected 1 pg human DNA in samples containing 200 ng mouse DNA (not shown). The second assay was an SNP-based assay that was used to determine the ratio of RS-MSCs to SR-MSCs in tissues of mice after 1:1 mixtures of cells from 2 different donors were infused. In the first step of the SNP assay, we amplified a 630-bp region of the COL1A2 gene for the proα2 (I) chain of type I collagen35 (Figure 2) that contains 2 high-frequency G/A SNPs (Table S1 and Figure S1). In the second step of the SNP assay, we used nested primers that specifically amplified either the allele with 2 G-G SNPs or the allele with the A-A SNPs. Experiments to standardize the assay indicated that it specifically detected either 1 pg of the G-G allele or 1 pg of the A-A allele in samples that contained 1 ng of the alternative allele and 200 ng of mouse DNA (Figure S3).
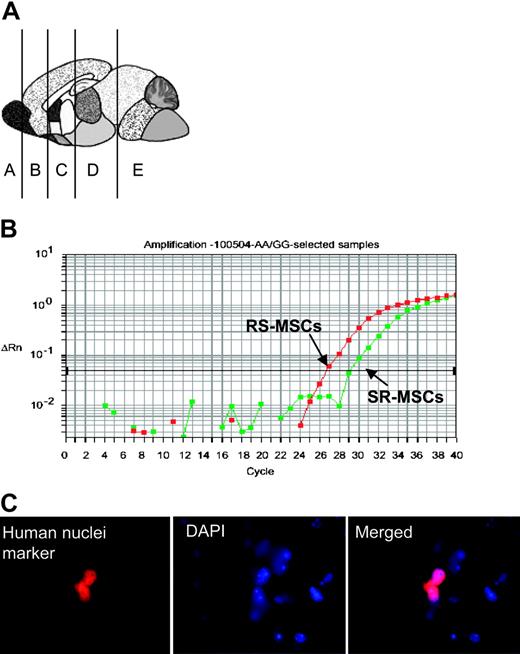
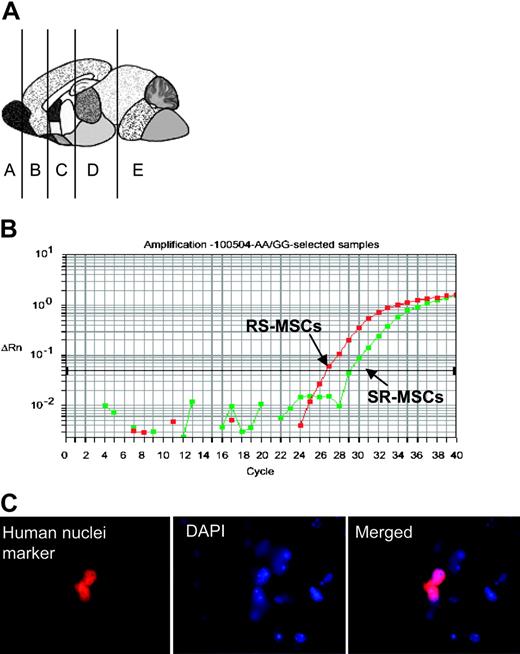
Engraftment assay after intravenous infusion of 1:1 mixtures of 2.5 × 105RS-MSCs and 2.5 × 105SR-MSCs from donors with different A/A and G/G alleles. (A) Schematic of brain regions assayed. (B) Example of competitive SNP assay. Genomic DNA isolated brain section D of mouse no. 3 was assayed. (C) Immunohistochemistry in section D of mouse no. 3 with an antihuman nuclear antigen (magnification, 600 ×).
Engraftment assay after intravenous infusion of 1:1 mixtures of 2.5 × 105RS-MSCs and 2.5 × 105SR-MSCs from donors with different A/A and G/G alleles. (A) Schematic of brain regions assayed. (B) Example of competitive SNP assay. Genomic DNA isolated brain section D of mouse no. 3 was assayed. (C) Immunohistochemistry in section D of mouse no. 3 with an antihuman nuclear antigen (magnification, 600 ×).
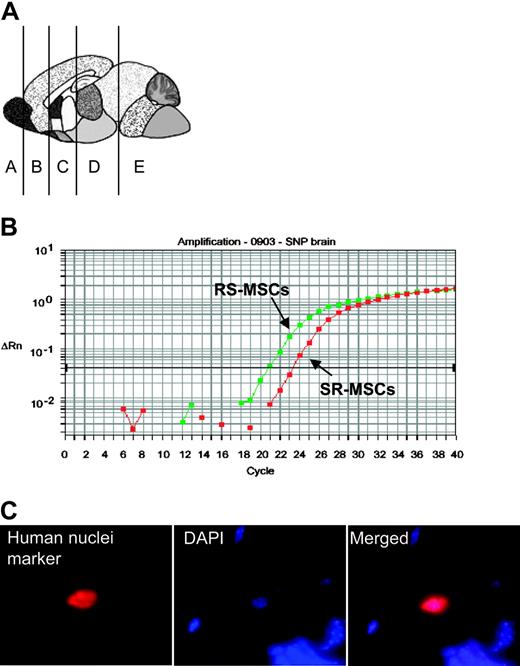
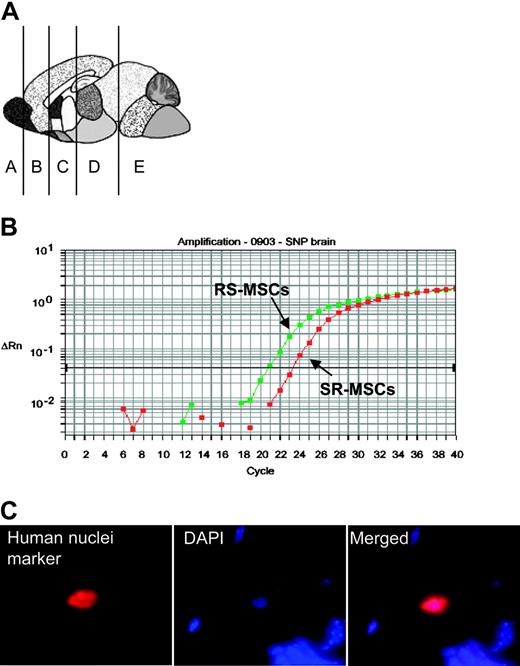
Engraftment after infusion into the hippocampus. Infusion into the hippocampus of 105 RS-MSCs from one donor or a 1:1 mixture of 2.5 × 104 RS-MSCs from one donor and 2.5 × 104 SR-MSCs from a second donor. (A) Schematic of brain regions assayed. Section D from mouse no. 1 was used for panels B and C (magnification, 600 ×).
Engraftment after infusion into the hippocampus. Infusion into the hippocampus of 105 RS-MSCs from one donor or a 1:1 mixture of 2.5 × 104 RS-MSCs from one donor and 2.5 × 104 SR-MSCs from a second donor. (A) Schematic of brain regions assayed. Section D from mouse no. 1 was used for panels B and C (magnification, 600 ×).
Competitive engraftment of RS-MSCs and SR-MSCs after intravenous infusion
To assay the competitive engraftment of RS-MSCs and SR-MSCs, 1:1 mixtures of RS-MSCs and SR-MSCs from 2 human donors with different SNP alleles were infused intravenously into immunodeficient mice without marrow ablation or other preconditioning. Real-time PCR assays for the human Alu sequences demonstrated low and variable levels of the engraftment in various tissues (Table 1, Figure 3A). The levels of engraftment varied from not detectable (< 1 human cell per 200 000 mouse cells) to several thousand per lung, spleen, kidney, heart, and selective regions of brain. All the mice assayed demonstrated a detectable level of engraftment in one or more of the organs assayed. However, none of the 8 mice assayed showed engraftment into all the organs examined. Also, there was no consistent pattern of organs engrafted at detectable levels, even in mice injected with the same preparations of cells on the same day and killed at the same time. The total number of human cells detected was a small fraction of the cells injected. In the mouse with the highest detected levels (mouse no. 6 in Table 1), the sum of all the organs assayed was about 78 000 human cells, or about 15% of the cells injected.
The allele-specific SNP assays were carried out on DNA samples that were identified by the assays for Alu sequences as containing at least 1 pg human DNA per 200 ng mouse DNA. The results of the allele-specific SNP assay indicated that with one exception (brain section D from mouse no. 5), the RS-MSCs had engrafted more efficiently than the SR-MSCs (Table 1, Figure 3B). In several samples, the allele-specific SNP assay detected RS-MSCs but no SR-MSCs (denoted as ratio of > 1000:1).
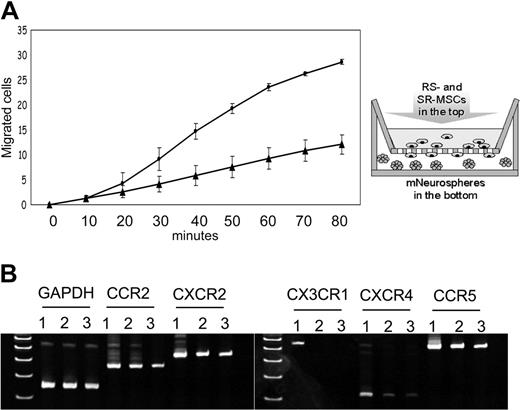
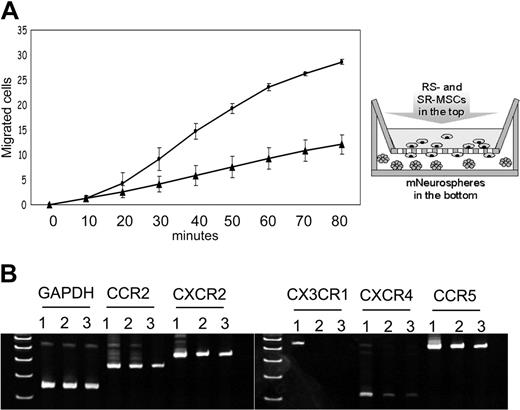
Migration to neurospheres and expression of chemokine receptors of RS-MSCs and SR-MSCs. (A) Migration of RS-MSCs (•) and SR-MSCs (▴) in response to neurospheres. Migration was followed by fluorimetry of underside of opaque inserts in the transwells. Data are expressed as mean and range of 2 values. (B) RT-PCR assays of total RNA for chemokine receptors. Lane 1: RS-MSCs. Lane 2: SR-MSCs. Lane 3: Unsorted MSCs.
Migration to neurospheres and expression of chemokine receptors of RS-MSCs and SR-MSCs. (A) Migration of RS-MSCs (•) and SR-MSCs (▴) in response to neurospheres. Migration was followed by fluorimetry of underside of opaque inserts in the transwells. Data are expressed as mean and range of 2 values. (B) RT-PCR assays of total RNA for chemokine receptors. Lane 1: RS-MSCs. Lane 2: SR-MSCs. Lane 3: Unsorted MSCs.
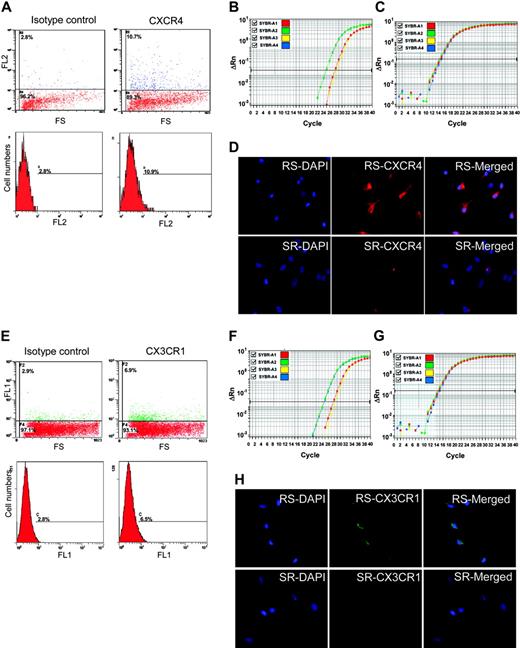
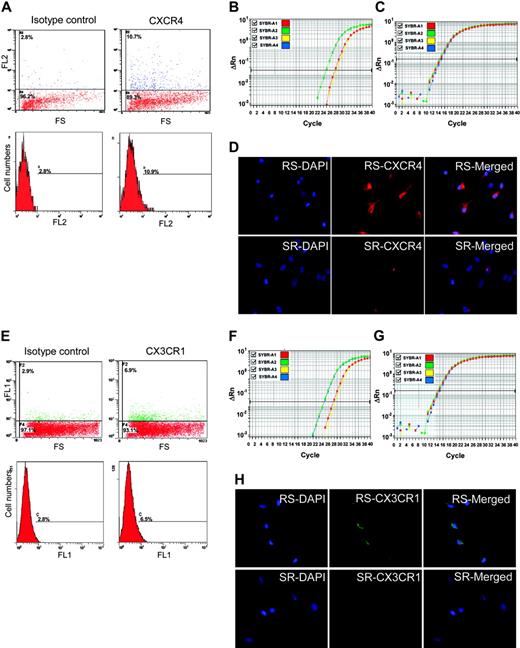
CXCR4 and CX3CR1 as epitopes on MSCs. (A) Flow cytometry of unsorted MSCs with anti-CXCR4-PE. The positive cells are primarily low in FS. (B) Real-time RT-PCR assay of CXCR4 in RS-MSCSs (green), SR-MSCs (red), and unsorted MSCs (yellow). (C) Real-time RT-PCR assay of GAPDH as a loading standard. (D) Immunocytochemistry of RS-MSCs and SR-MSCs cultured in chambered slides and stained with antibody to CXCR4 and DAPI (magnification, 400 ×). (E) Flow cytometry of unsorted MSCs with anti-CX3CR1-FITC. The positive cells are primarily low in FS. (F) Real-time RT-PCR assay of CX3CR1 in RS-MSCSs (green), SR-MSCs (red), and unsorted MSCs (yellow). (G) Real-time RT-PCR assay of GAPDH. (H) Immunocytochemistry of RS-MSCs and SR-MSCs cultured in chambered slides and stained with antibody to CX3CR1 and DAPI (magnification, 400 ×).
CXCR4 and CX3CR1 as epitopes on MSCs. (A) Flow cytometry of unsorted MSCs with anti-CXCR4-PE. The positive cells are primarily low in FS. (B) Real-time RT-PCR assay of CXCR4 in RS-MSCSs (green), SR-MSCs (red), and unsorted MSCs (yellow). (C) Real-time RT-PCR assay of GAPDH as a loading standard. (D) Immunocytochemistry of RS-MSCs and SR-MSCs cultured in chambered slides and stained with antibody to CXCR4 and DAPI (magnification, 400 ×). (E) Flow cytometry of unsorted MSCs with anti-CX3CR1-FITC. The positive cells are primarily low in FS. (F) Real-time RT-PCR assay of CX3CR1 in RS-MSCSs (green), SR-MSCs (red), and unsorted MSCs (yellow). (G) Real-time RT-PCR assay of GAPDH. (H) Immunocytochemistry of RS-MSCs and SR-MSCs cultured in chambered slides and stained with antibody to CX3CR1 and DAPI (magnification, 400 ×).
The presence of human cells in brain was confirmed by immunostaining of sections for the antibody for human nuclear specific protein (Figure 3C). The number of human cells detected was too low to develop definite data on the expression of neural proteins by double immunostaining with antibodies to the human nuclei antigen and neural proteins without assaying a prohibitive number of brain sections.
Competitive engraftment of RS-MSCs and SR-MSCs after intracerebral infusion
To further examine engraftment of the cells in brain, mixtures of RS-MSCs and SR-MSCs or RS-MSCs alone were injected directly into the hippocampus of the immunodeficient mice. After 2 weeks, human cells were detected by the assay for Alu sequences in brain section D containing the hippocampus in all 4 of the mice assayed (Table 2, Figure 4A). In 2 mice, the human cells were also detected in the adjacent section containing the cerebellum, an observation probably explained by the fact that the bevel of the needle was directed caudally during the injection.
The allele-specific SNP assay again indicated that the RS-MSCs engrafted more efficiently in the 2 mice in which a 1:1 mixture of RS-MSCs and SR-MSCs were injected (ratios presented in footnotes in Table 2). The presence of the human cells in the mouse brain was confirmed by staining of antibody to a human-specific nuclear protein (Figure 4C).
Comparison of RS-MSCs and SR-MSCs in migration assays to neurospheres
Because RS-MSCs were found to engraft more efficiently than SR-MSCs into the brain, the cells were compared in a migration assay in vitro with neurospheres. Murine neurospheres containing about 1 × 105 cells were placed in the lower chamber of transwells and the MSCs were placed in the upper chambers. The neurospheres increased the migration of both subpopulations, but the RS-MSCs migrated more rapidly than the SR-MSCs (Figure 5A).
RS-MSCs express higher levels of CXCR4 and CX3R1 than SR-MSCs
To search for a molecular basis for the preferential engraftment and migration ability of RS-MSCs, we examined expression patterns of several kinds of chemokine receptors in the RS-MSCs and SR-MSCs. Semiquantitative PCR assays detected the receptors CCR2, CXCR2, and CCR5 in both RS-MSCs and SR-MSCs (Figure 5B). However, only the RS-MSCs contained mRNA for CXCR4, the receptor for SDF-1, and CX3CR1, the receptor for fractalkine. Assays by flow cytometry of unfractionated MSCs demonstrated that about 8% of the cells expressed CXCR4 as a surface epitope (Figure 6A). The cells that expressed the receptor were cells with low FS corresponding to RS-MSCs. Real-time RT-PCR assays demonstrated that the levels of mRNA for CXCR4 were about 10-fold higher in RS-MSCs than in SR-MSCs (Figure 6B). The high expression of CXCR4 in RS-MSCs was confirmed by isolating RS-MSCs by FACS, culturing them for 1 day in chambered slides, and staining them with an antibody (Figure 6D).
Similar assays demonstrated that about 3% of unfractionated MSCs expressed CX3R1, the receptor for fractalkine (Figure 6E). The receptor was also found primarily on the cells with low FS of light corresponding to RS-MSCs. Real-time PCR assays indicated that the levels of mRNA for CX3R1 were about 10-fold higher in RS-MSCs than in SR-MSCs (Figure 6F-G). The higher expression of CX3R1 by RS-MSCs was confirmed by immunostaining of RS-MSCs isolated by FACS (Figure 6H).
Comparison of RS-MSCs and SR-MSCs in migration assays
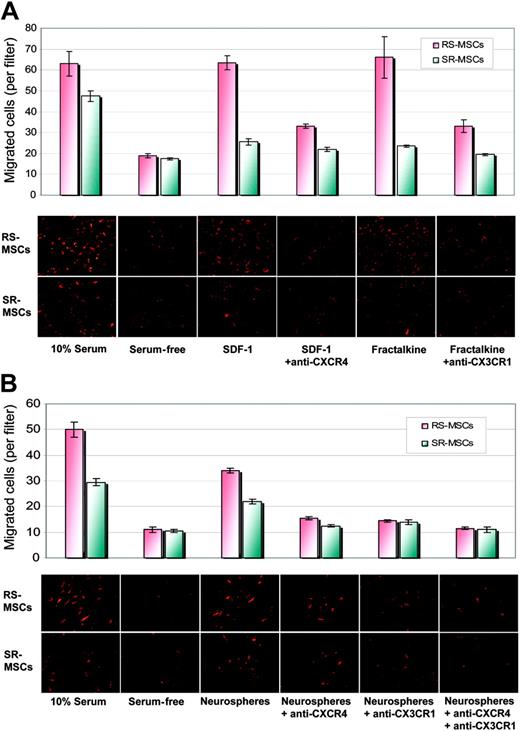
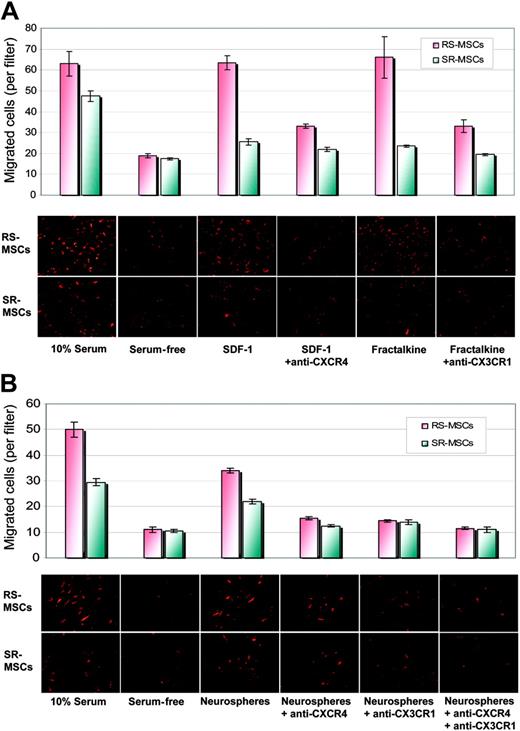
We then compared the migration of 2 subpopulations of MSCs to SDF-1, the ligand for CXCR4, and fractalkine, the ligand for CX3R1. SDF-1 increased the migration of RS-MSCs more than SR-MSCs relative to serum-free controls (Figure 7A). Antibodies to CXCR4 decreased the SDF-1-stimulated migration. Similar results were obtained with fractalkine and antibodies to CX3R1 (Figure 7A).
We also tested the effects of antibodies to CXCR4 and CX3R1 on the neurosphere-induced migration of the cells (Figure 7B). Antibodies to CXCR4 decreased the induced migration of RS-MSCs and SR-MSCs by about 40%. Antibodies to CX3CR1 had a similar effect and produced about a 40% decrease in migration of both cell types. The effects of the 2 antibodies were only partially addictive. Therefore, the results indicated the neurosphere-induced migration of the cells was partially but not completely dependent on CXCR4 and CX3R1 pathways.
Migration of RS-MSCs and SR-MSCs. (A) Migration of RS-MSCs and SR-MSCs induced by SDF-1 and fractalkine. The bottom wells contained either 20% fetal calf serum, serum-free medium, 50 ng/mL SDF-1, 50 ng/mL SDF-1 plus 10 μg/mL anti-CXCR4, 10 ng/mL fractalkine, or 10 ng/mL fractaline plus 5 μg/mL anti-CX3CR1. Migration was assayed by photomicrograph of the underside of the opaque inserts (magnification, × 200). Data are expressed as mean and range of 2 values. (B) Migration of RS-MSCs and SR-MSCs to neurospheres. Assays and values as in panelA, except SDF-1 and fractalkine were replaced with about 1 × 105 neural stem cells from neurospheres. Data are expressed as mean and range of 2 values.
Migration of RS-MSCs and SR-MSCs. (A) Migration of RS-MSCs and SR-MSCs induced by SDF-1 and fractalkine. The bottom wells contained either 20% fetal calf serum, serum-free medium, 50 ng/mL SDF-1, 50 ng/mL SDF-1 plus 10 μg/mL anti-CXCR4, 10 ng/mL fractalkine, or 10 ng/mL fractaline plus 5 μg/mL anti-CX3CR1. Migration was assayed by photomicrograph of the underside of the opaque inserts (magnification, × 200). Data are expressed as mean and range of 2 values. (B) Migration of RS-MSCs and SR-MSCs to neurospheres. Assays and values as in panelA, except SDF-1 and fractalkine were replaced with about 1 × 105 neural stem cells from neurospheres. Data are expressed as mean and range of 2 values.
The small size of the RS-MSCs may explain both the increased migration observed in the in vitro assays and their more efficient penetration into tissues in vivo. However, the more rapid migration of RS-MSCs was not explained by the smaller size of the cells, since RS-MSCs and SR-MSCs migrated at the same or similar rates when medium containing 10% FCS was placed in the lower well or in the presence of blocking antibodies (Figure 7A-B).
Discussion
The results presented here address several recent controversies about engraftment in vivo of stem cells/progenitor cells for nonhematopoietic tissues. The data confirm previous indications that only low levels of engraftment are seen after human MSCs are infused intravenously into naive adult animals that are not preconditioned by marrow ablation or injury to specific tissues.38,39 Also, for reasons that were not apparent, the degree of engraftment was highly variable even when the same preparations were infused at the same time into seemingly identical animals. Therefore, negative results are readily generated in such experiments unless adequate attention is paid to the animal-to-animal variations, the sensitivity of the assays, and the tendency of some markers to be lost during engraftment and differentiation. With one exception, RS-MSCs engrafted more effectively than SR-MSCs isolated from the same cultures. The more efficient engraftment of RS-MSCs may be explained by the observation that about 90% of the cells were in G0/G1, since previous reports indicated that hematopoietic stem cells engraft more effectively if they are in G0/G1.40-43 The appearance of the human cells in the hippocampus after intravenous infusion was unexpected, but the hippocampus was previously referred to as a “vascular niche” because of its abundant blood supply.44 Also, marrow cells were observed to serve as precursors to not only microglial cells but also a few astrocytes and neurons both in experimental animal models after direct injection4,45 and in some but not all patients who received sex mismatched bone marrow transplants.46,47
The observations presented here also suggested that the more efficient engraftment of MSCs was in part explained by their expression of CXCR4 and CX3CR1, receptors previously implicated in hematopoiesis, cardiogenesis, vasculogenesis, neuronal development, the trafficking of immune cells, and the trafficking of clonal cells from cancers.48-50 As the present work was being completed, several reports suggested CXCR4 and related receptors were important in the trafficking of MSCs.51-53 For example, Sordi et al53 demonstrated that a subpopulation of human MSCs expressed CXCR4, CX3R1, CXCR6, CCR1, and CCR7, that the cells are attracted to human pancreatic islets in culture, and that the attraction was principally mediated by the ligands CX3CL1 and CXCL12.
The PCR assays for human Alu sequences and SNPs that were employed here made it possible to detect the low levels of engraftment of human MSCs in nonhematopoietic tissues of normal adult mice. Also, the competitive assay based on the SNPs from 2 different donors and experimental strategies developed here should be useful in comparing the efficiencies of engraftment of the large number of different kinds of adult stem cells that have now been identified in different tissues and under different culture conditions.
Prepublished online as Blood First Edition Paper, November 8, 2005; DOI 10.1182/blood-2005-07-2701.
Supported in part by a grant from the W. M. Keck Foundation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.