Abstract
A gene expression signature of tumor proliferation rate in mantle cell lymphoma (MCL) is an overriding molecular predictor of the length of survival following diagnosis. Many strongly proliferative MCL tumors have exceptionally high cyclin D1 mRNA levels and preferentially express short cyclin D1 mRNA isoforms. We demonstrate here that these short mRNAs are cyclin D1a isoforms with truncated 3′UTRs, not alternatively spliced cyclin D1b mRNA isoforms. Among 15 MCL tumors with truncated cyclin D1 mRNAs, 7 had genomic deletions in the CCND1 3′UTR region. In 3 others, CCND1 contained point mutations that created premature polyadenylation signals, giving rise to 1.5-kb mRNAs lacking most of the 3′UTR. Both types of genomic alteration created transcripts lacking mRNA destabilization elements present in the wild-type cyclin D1a mRNA. Premature polyadenylation due to a 3′UTR mutation also was present in the Z-138 MCL cell line, which expressed both truncated and full-length cyclin D1a mRNAs. In these cells, the half-life of the short cyclin D1a mRNA was much longer than that of the full-length mRNA. We conclude that alterations of CCND1 3′UTR structure can significantly increase its oncogenic effect and worsen the clinical course of MCL patients.
Introduction
Mantle cell lymphoma (MCL) comprises about 6% of all non-Hodgkin lymphoma and is considered incurable with standard chemotherapy.1,2 Median survival is approximately 3 years, but survival ranges from less than one year to > 6 years. The hallmark genetic feature of MCL is the t(11;14) translocation that leads to misexpression of cyclin D1 in the malignant cells.3–5 The t(11;14) is not unique to MCL and occurs also in multiple myeloma.6 Cyclin D1 is a member of the D-type cyclins that regulate the transition from G0/G1 phase to S phase of the cell cycle.7 Cyclin D1 is not normally expressed at high levels in lymphoid cells, and its expression from the t(11;14) translocated allele is driven by enhancer elements in the immunoglobulin heavy chain locus. Most t(11;14) translocations take place at the 5′ end of the cyclin D1 locus, but translocations at the 3′end of the gene also have been described in some cases.8
CCND1 has 5 exons, which can be alternatively spliced to create 2 major isoforms, cyclin D1a and D1b. The cyclin D1a isoform is 4.5 kb in length, with a coding region of only 882 bp. The majority of this mRNA consists of 3′UTR sequences containing mRNA destabilizing elements. The cyclin D1b isoform lacks exon 5 but retains intron 4, which contains a translation stop codon after 99 bp and a polyadenylation signal less then 300 bp 3′ from this stop codon. The 1.7-kb cyclin D1b mRNA is found in most tumors and cell lines that express cyclin D1 and encodes a 274 amino acid protein that differs at the C terminus from the 294 amino acid protein encoded by the cyclin D1a mRNA.9–13 In contrast to cyclin D1a, cyclin D1b is potently transforming in experimental models.13,14
The relative abundance of the cyclin D1b isoform is reportedly affected by a G/A single nucleotide polymorphism at the last base of exon 4 (position 870, codon 241), which is the −1 position of the intron 4 splice donor consensus.9,15,16 The G allele is thought to favor correct splicing, whereas the A allele is thought to impair splicing and thereby increase the relative expression of cyclin D1b isoform.9,10 In healthy individuals, the G allele is slightly more common, with reported allele frequencies of 51% to 61%.12,17,18 Homozygosity for the A allele has been associated with an increased risk of cancer and at times with more aggressive disease.9 However, a recent study found no influence of the G/A870 polymorphism on prognosis in 42 patients with MCL.18
The clinical course of MCL is highly variable: some patients succumb rapidly to the disease, while others have a more chronic course and may not require treatment for prolonged periods of time. Using gene expression profiles of 92 MCL lymph node biopsies, a gene expression–based measure of tumor proliferation rate was identified that was the dominant prognostic factor predicting the length of survival following diagnosis.19 This gene expression “signature” of proliferation consists of 20 genes involved in various aspects of cell cycle control, DNA synthesis and repair, and nucleotide metabolism. Based on the average expression of these proliferation signature genes, the MCL patients were divided into 4 quartile groups that differed dramatically in overall survival. Patients in the quartile with the highest expression of the proliferation signature had a median survival of 10 months, whereas patients in the lowest proliferation signature quartile had a median survival of 6.7 years.19
Interestingly, many of the MCL tumors that were the most proliferative appeared to express cyclin D1 mRNA species that contained the 5′ coding region but lacked the 3′UTR region.19 The overall cyclin D1 mRNA abundance was higher in these tumors than in those whose cyclin D1 mRNAs possessed the 3′UTR region. Higher levels of cyclin D1 mRNA were quantitatively related to higher proliferation rate and shorter survival. Deletions of the gene encoding the cyclin-dependent kinase inhibitor p16 also were associated with a higher proliferation rate and were statistically independent of cyclin D1 mRNA expression in a multivariate model of the proliferation rate. These findings suggested a model in which quantitative differences in the efficiency/probability of the G1/S phase transition in MCL tumors are dictated by the integrated effect of multiple alterations in genes that control this cell-cycle transition.19
We postulated that genetic events at the CCND1 locus besides the t(11;14) are responsible for alterations in the structure of cyclin D1 mRNAs in MCL and their relative abundance. In the present study we demonstrate that the observed variability in cyclin D1 mRNA expression is due to truncation of the cyclin D1a mRNA in some tumors and not to the expression of the cyclin D1b isoform by alternative splicing. We find that cyclin D1 mRNA stabilization is due to genomic deletions and point mutations that lead to premature polyadenylation and removal of destabilization elements from the mRNA.
Patients, materials, and methods
Patients
Lymph node biopsies from 92 previously untreated patients with cyclin D1–positive MCL were included in the study, according to a protocol approved by the National Cancer Institute institutional review board. The biopsy samples were the same as in a prior study,19 and the gene expression profiling data and the gene expression–based measure of tumor proliferation rate was described previously.19 RNA for the further analyses presented in this study was available from 84 of these cases, and genomic DNA was available from 86 cases.
Real-time quantitative PCR
To measure cyclin D1 mRNA isoforms by real-time quantitative reverse transcriptase–polymerase chain reaction (RT-PCR), an aliquot of mRNA was diluted to approximately .5 ng/μL and 2-5 μL was used in 10-25 μL reactions using TaqMan One-step RT-PCR Master Mix and analyzed on an Applied Biosystems Thermal Cycler (Applied Biosystems, Foster City, CA). To test for genomic deletions in CCND1, genomic DNA was diluted to 50 ng/μL, and 2-5 μL was used in 10-25 μL reactions using the TaqMan Universal PCR Master Mix and analyzed on an Applied Biosystems Thermal Cycler. All samples were run in triplicate, and samples with more than 15% variance between replicates were repeated. Standard curves were generated using RNA and DNA from the KMS-12 myeloma cell line, which has a t(11;14) translocation and expresses full-length 4.5 kb cyclin D1a mRNA.20 A probe for the beta-2-microglobulin locus was chosen as a reference for RNA expression.
Primers and probes for the beta-2-microglobulin (B2M) locus and the cyclin D1 Exon1/Exon2 junction have been described previously.21 Additional primers and probes were as follows: intron 4: 5′-CCCAGGCCACAGACTGACA-3′ (sense); 5′-CCCAGGCCACAGACTGACA-3′ (antisense); 5′-Fam-CACCGGCTTCTTCCACTGCTCCTAGA-Tamra-3′ (probe); proximal UTR: 5′-GGAAAGCTTCATTCTCCTTGTTG-3′ (sense); 5′-TTCTTTTGCTTAAGTCAGAGATGGAA-3′ (antisense); 5′-Fam- TGGTTGTTTTTTCCTTTGCTCTTTCCCC-Tamara-3′ (probe); distal UTR: 5′-TCTCAATGAAGCCAGCTCACA-3′ (sense); 5′-CTTTTGGTTCGGCAGCTTG-3′ (antisense); 5′-FAM-TGCTGTGTGCCCCGGTCACCTA-TAMRA-3′ (probe).
Analysis of restriction fragment length polymorphism for A870G
The 870G allele gives rise to a restriction site for the enzyme NciI that is absent in the 870A allele.12 We determined the NciI restriction fragment length polymorphism (RFLP) at this position in PCR products as previously described with minor modifications.18 Briefly, using genomic DNA or cDNA a 199-bp PCR product was generated with a primer 5′-AGTTCATTTCCAATCCGCCC-3′ (sense) in exon 4 and a primer 5′-AGGTGTCTCCCCCTGTAAGCC-3′ (antisense) in intron 4. This was digested with NciI for 4 hours at 37°C, resulting in 176 bp and 23 bp fragments for an A at position 870 and in 141-bp, 35-bp, and 23-bp fragments for a G at this position. Digested products were separated by gel electrophoresis on 3% agarose gels.
Amplification and sequencing of CCND1 3′UTR
To sequence the proximal 3′UTR, we amplified the region from genomic DNA and/or cDNA using primers 5′-ATCGAAGCCCTGCTGGAGTCAAG-3′ (sense), 5′-CGTAAAGGATGGAACCTAATCCTCTC-3′ (antisense) and 5′-CATTAACACAAAGGAGGCGTCTC-3′ (sense) and 5′-CCTCCACTGGATGGTTTGTCACTG-3′ (antisense). PCR products were purified through a spin column (Qiagen, Valencia, CA) and directly sequenced by cycle sequencing (Applied Biosystems). Some products were subcloned using TOPO-TA cloning (Invitrogen, Carlsbad, CA) before sequencing. Sequencing electropherograms were analyzed with ChromasPro software (Technelysium Pty Ltd, Tewantin Qld, Australia). All sequence positions are given in reference to accession number X59798.1 for cyclin D1 mRNA.
Analysis of cyclin D1 mRNA half-life and protein expression
The Z-138 MCL cell line was grown in RPMI media supplemented with 10% bovine serum. To inhibit transcription, cells were treated with actinomycin D (10 μg/mL; EMD Biosciences, San Diego, CA) and harvested at the indicated time points. RNA was extracted using Trizol (Invitrogen). Quantitative RT-PCR to measure the decay in RNA abundance was carried out as described under “Real-time quantitative PCR.” To analyze cyclin D1 protein, 2 × 106 cells were lysed in 1 mL buffer containing 1% Triton X-100. Western blots were probed with antibodies to cyclin D1 (clone DCS-6, Santa Cruz Biotechnology, Santa Cruz, CA) and to α-tubulin (DM1A, Sigma, St Louis, MO).
Statistical methods
To test whether there were any differences between patient quartiles for the expression of truncated mRNAs or the genotype of the translocated alleles, the Fisher exact test was used. A t test was used to determine the extent to which cyclin D1 mRNA isoforms were differentially expressed between patient subgroups. Survival curves were estimated using the Kaplan-Meier method and compared by the log-rank test.
Results
Cyclin D1 mRNAs lacking full-length 3′UTRs are common in highly proliferative MCL tumors and are not due to differential splicing
We used 2 qRT-PCR assays to measure the relative expression of cyclin D1 mRNAs possessing an intact 3′UTR region compared with cyclin D1 mRNAs lacking this region (Figure 1A). One qRT-PCR assay used primers spanning the exon 1-2 junction, and the second used primers from the distal end of the 3′UTR. Previously, the MCL tumors in this study were assigned to 4 quartile groups that differed in the expression of the proliferation gene expression signature and hence in their proliferation rates.19 Using DNA microarrays, those MCL tumors belonging to the quartile with the lowest proliferation rate expressed predominantly the full-length cyclin D1 mRNA.19 Using the qRT-PCR assay described under “Patients, materials, and methods,” the MCL tumors in this quartile had an average distal UTR/Exon1-2 ratio of 1.62. It is likely that this ratio differs from the ideal ratio of 1 due to different amplification efficiencies of the 2 primer pairs.
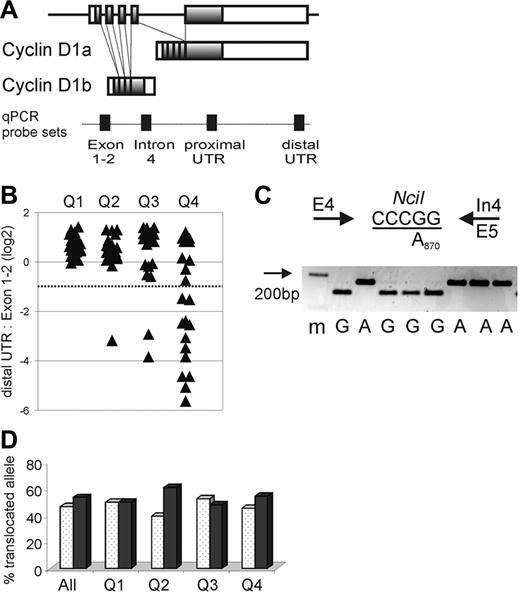
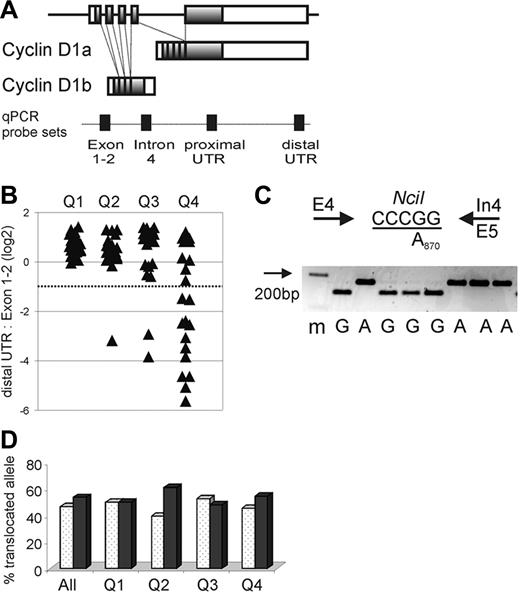
CCND1 locus and expression of cyclin D1 mRNA isoforms in MCL. (A) Schematic representation of the CCND1 locus and alternatively spliced cyclin D1 mRNA transcripts cyclin D1a and D1b. Shaded boxes represent coding sequences, open boxes represent noncoding exon sequences, and filled boxes on the dotted line indicate the location of qPCR probes. (B) The relative amount of cyclin D1 mRNA containing full-length 3′UTR in relation to all cyclin D1 mRNA transcripts measured by the exon 1-2 probe is shown on a log2 scale. The dotted line indicates the cutoff, below which more than 50% of transcripts lack full-length 3′UTR. Quartiles of proliferation as determined in a prior study19 are given on the x-axis with tumors in quartile 1 having the lowest and those in quartile 4 having the highest proliferation (P < .001 for any difference between quartiles). Number of samples analyzed were for Q1 n = 22, Q2 n = 20, Q3 n = 22, Q4 n = 20. (C) The A870G polymorphism at the last nucleotide of exon 4 (codon 241) was determined using an NciI RFLP. PCR primers are indicated by arrows, the NciI restriction site is underlined, and a representative analysis of cDNA samples separated on a 3% agarose gel with a size marker (m) in the first lane is shown. (D) The frequency of cyclin D1 G870 alleles (white columns) or A870 alleles (black columns) involved in the t(11;14) translocation is summarized for all patients as well as for each proliferation quartile. There was no significant difference in genotype of the translocated alleles between quartiles (P = .84). Number of samples analyzed were for Q1 n = 22, Q2 n = 23, Q3 n = 21, Q4 n = 22.
CCND1 locus and expression of cyclin D1 mRNA isoforms in MCL. (A) Schematic representation of the CCND1 locus and alternatively spliced cyclin D1 mRNA transcripts cyclin D1a and D1b. Shaded boxes represent coding sequences, open boxes represent noncoding exon sequences, and filled boxes on the dotted line indicate the location of qPCR probes. (B) The relative amount of cyclin D1 mRNA containing full-length 3′UTR in relation to all cyclin D1 mRNA transcripts measured by the exon 1-2 probe is shown on a log2 scale. The dotted line indicates the cutoff, below which more than 50% of transcripts lack full-length 3′UTR. Quartiles of proliferation as determined in a prior study19 are given on the x-axis with tumors in quartile 1 having the lowest and those in quartile 4 having the highest proliferation (P < .001 for any difference between quartiles). Number of samples analyzed were for Q1 n = 22, Q2 n = 20, Q3 n = 22, Q4 n = 20. (C) The A870G polymorphism at the last nucleotide of exon 4 (codon 241) was determined using an NciI RFLP. PCR primers are indicated by arrows, the NciI restriction site is underlined, and a representative analysis of cDNA samples separated on a 3% agarose gel with a size marker (m) in the first lane is shown. (D) The frequency of cyclin D1 G870 alleles (white columns) or A870 alleles (black columns) involved in the t(11;14) translocation is summarized for all patients as well as for each proliferation quartile. There was no significant difference in genotype of the translocated alleles between quartiles (P = .84). Number of samples analyzed were for Q1 n = 22, Q2 n = 23, Q3 n = 21, Q4 n = 22.
Importantly, among all 84 MCL biopsy samples, the distal UTR/Exon1-2 ratio was < 0.5 in 15 tumors (Figure 1B). This finding suggested that more than half of all cyclin D1 transcripts in these cases did not contain an intact 3′UTR. For 9 of these cases, the exceedingly low distal UTR/Exon1-2 ratios suggested that < 10% of the cyclin D1 transcripts were full length. Of the 15 cases with low distal UTR/Exon1-2 ratios, 12 belonged to the quartile with the highest expression of the proliferation signature (Q4), and none of the cases belonged to the quartile with the lowest tumor proliferation (Q1; Figure 1B).
The preferential expression of cyclin D1 mRNAs without a full-length 3′UTR could be due to increased levels of the alternatively spliced cyclin D1b isoform, which lacks exon 5 and the 3′UTR sequences contained therein (Figure 1A). To address this possibility, we separately measured the expression of the cyclin D1a isoform (using a qRT-PCR assay based on exon 5 sequences) and the expression of the cyclin D1b isoform (using a qRT-PCR assay based on sequences in the proximal region of intron 4). In tumors with apparently normal 3′UTR structure, the average cyclin D1a/cyclin D1b ratio was 0.86, whereas in tumors with predominantly non–full-length mRNAs, this ratio was 1.82. Thus, tumors with truncated cyclin D1 mRNAs had lower, not greater, relative expression of cyclin D1b mRNA (P < .001). Therefore, the preferential expression of short cyclin D1 mRNAs in some highly proliferative MCL tumors cannot be explained by an overrepresentation of the cyclin D1b splice variant.
The A870G polymorphism at the −1 position of the splice donor consensus sequence of intron 4 has been reported to affect the relative abundance of the cyclin D1a and D1b splice variants. To test whether this polymorphism influenced cyclin D1 mRNA expression in our MCL cohort, we created an RFLP assay to determine the A870G genotype in all 86 MCL patients for whom genomic DNA was available as well as in 38 healthy control individuals. In this MCL cohort, there was a slight overrepresentation of the A870 allele (50% of cases as compared to 38% in the controls). Because only the translocated cyclin D1 allele is expressed in the tumor cells, we repeated the RFLP analysis using cDNA (Figure 1C,D). The genotype of the translocated allele was A870 in 47% and G870 in 53% of cases, and the relative frequencies of the 2 genotypes were comparable among tumors in all 4 proliferation quartiles, arguing against an effect of this polymorphism on MCL tumor proliferation (Figure 1D). Furthermore, there was no detectable influence of the A870G polymorphism on the relative expression of the cyclin D1 splice variants (data not shown).
Truncation of the 3′UTR in cyclin D1a mRNA is associated with inferior outcome
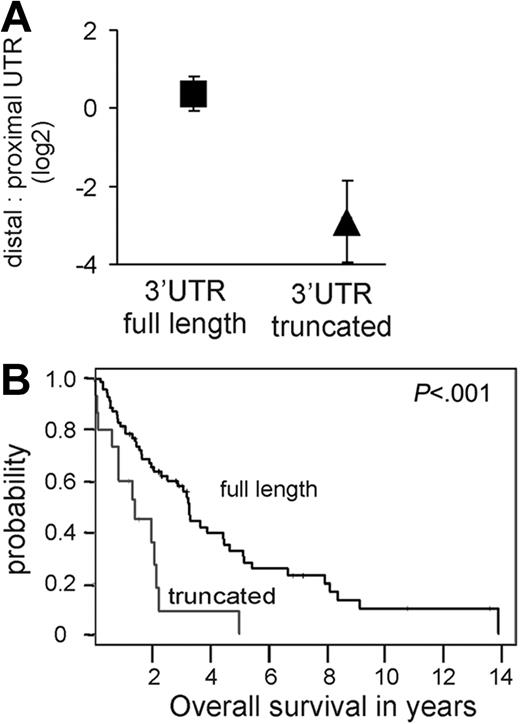
The above analysis suggested that, in some MCL cases, aberrant cyclin D1a transcripts are expressed that lack all or part of the 3′UTR. To map these putative 3′UTR truncations more precisely, we used qRT-PCR assays targeting the proximal end of the 3′UTR (proximal UTR probe set; Figure 1A) or the distal end of the 3′UTR (distal UTR probe set; Figure 1A). The relative abundance of transcripts measured with the distal compared to the proximal UTR probe sets was 1.25 in tumors with apparent full-length cyclin D1 transcripts (ie, those with high distal UTR/Exon1-2 ratios; Figure 1B). By contrast, all cases with low distal UTR/Exon1-2 ratios (Figure 1B) expressed truncated cyclin D1a transcripts, with average distal UTR/proximal UTR ratios of 0.17 (range, 0.04-0.56). Many of these tumors had distal UTR/proximal UTR ratios of < 0.1, indicating that almost all cyclin D1a transcripts in these tumors had a truncation of the 3′UTR (P < .001; Figure 2A). Of the 15 biopsies with truncated cyclin D1a expression, 12 were from the 23 most highly proliferative tumors, and these patients had a significantly shorter survival than patients without this abnormality (Figure 2B).
Cyclin D1a transcripts with a truncated 3′UTR are associated with inferior survival. (A) The relative amount of cyclin D1a transcripts with a truncated 3′UTR compared to full-length form was determined with probes in the proximal UTR and the distal UTR (see map in Figure 1). The mean and standard deviation for this ratio is given for tumors expressing full-length transcripts (square; a representative subset of 32 samples from Figure 1B was analyzed) as compared to transcripts with truncated 3′UTR (triangle; n = 15). (B) Kaplan-Meier estimates of overall survival according to predominant type of cyclin D1a transcript. The median survival for patients whose tumors expressed full-length cyclin D1 mRNA (n = 69) was 3.28 years, compared to 1.38 years for tumors expressing truncated cyclin D1a mRNAs (n = 15).
Cyclin D1a transcripts with a truncated 3′UTR are associated with inferior survival. (A) The relative amount of cyclin D1a transcripts with a truncated 3′UTR compared to full-length form was determined with probes in the proximal UTR and the distal UTR (see map in Figure 1). The mean and standard deviation for this ratio is given for tumors expressing full-length transcripts (square; a representative subset of 32 samples from Figure 1B was analyzed) as compared to transcripts with truncated 3′UTR (triangle; n = 15). (B) Kaplan-Meier estimates of overall survival according to predominant type of cyclin D1a transcript. The median survival for patients whose tumors expressed full-length cyclin D1 mRNA (n = 69) was 3.28 years, compared to 1.38 years for tumors expressing truncated cyclin D1a mRNAs (n = 15).
Truncated cyclin D1 mRNA due to genomic deletion of 3′UTR sequences
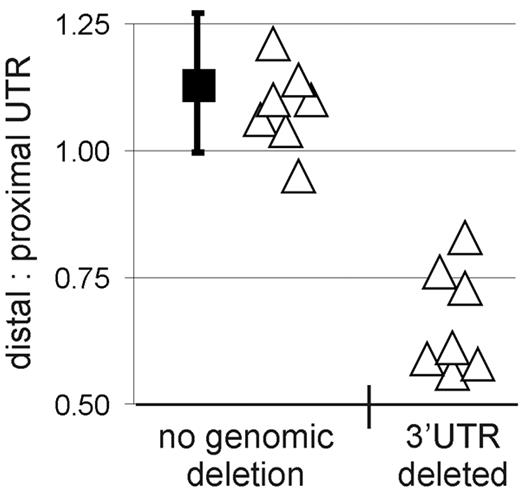
We next investigated whether genomic deletions in the exon 5 region of CCND120,22,23 could explain the expression of truncated cyclin D1a mRNA in some MCL cases. To detect genomic deletions in CCND1, we analyzed genomic DNA from MCL cases by quantitative PCR using distal and proximal UTR and intron 4 probe sets (Figure 1A). In samples expressing full-length cyclin D1 mRNA, the amplification signals obtained using these 3 probes were comparable. Specifically, the average distal UTR/proximal UTR ratio was 1.13, consistent with a normal genomic locus for both alleles in these tumors (Figure 3). By contrast, the distal UTR/proximal UTR ratio was between 0.56 and 0.61 in 4 of the 15 samples expressing truncated cyclin D1a mRNA, consistent with a genomic deletion of distal 3′UTR sequences. Three more samples had a distal UTR/proximal UTR ratio ranging from 0.73 to 0.83; one scenario that may explain these intermediate ratios is amplification of the CCND1 locus with deletion in the 3′UTR region in one amplified copy of the gene, as described previously.24 In summary, all 7 cases had distal UTR/proximal UTR ratios below the 5th percentile (0.93) of the normal data and are therefore considered to have a genomic deletion (P < .001). Interestingly, no genomic deletions were detected by qPCR in 7 cases expressing truncated cyclin D1a mRNA, implying that other molecular mechanisms were responsible for the aberrant cyclin D1a transcripts in these cases.
Genomic deletions cause truncation of cyclin D1a mRNA. Proximal and distal 3′UTR probes were used to amplify genomic DNA, and the ratio of the respective amplification strength was determined. The black square indicates the mean in tumors with normal cyclin D1a mRNA transcripts (n = 69), and the bars represent the standard deviation of this mean estimate. Individual tumor samples expressing truncated cyclin D1a mRNA transcripts are represented by triangles (n = 14, for one patient no DNA was available). Samples with a ratio of distal/proximal UTR below the 5th percentile of the samples with normal cyclin D1 locus (n = 69, black squares) were considered to have a genomic deletion of the 3′UTR.
Genomic deletions cause truncation of cyclin D1a mRNA. Proximal and distal 3′UTR probes were used to amplify genomic DNA, and the ratio of the respective amplification strength was determined. The black square indicates the mean in tumors with normal cyclin D1a mRNA transcripts (n = 69), and the bars represent the standard deviation of this mean estimate. Individual tumor samples expressing truncated cyclin D1a mRNA transcripts are represented by triangles (n = 14, for one patient no DNA was available). Samples with a ratio of distal/proximal UTR below the 5th percentile of the samples with normal cyclin D1 locus (n = 69, black squares) were considered to have a genomic deletion of the 3′UTR.
Mutations in the proximal 3′UTR region of CCND1 cause premature polyadenylation
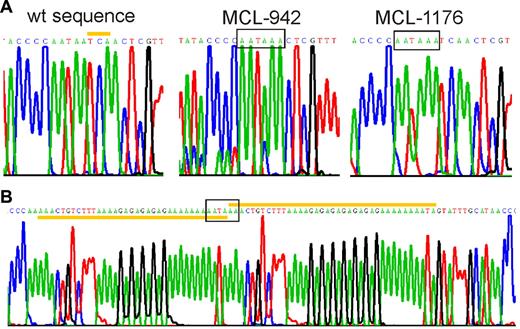
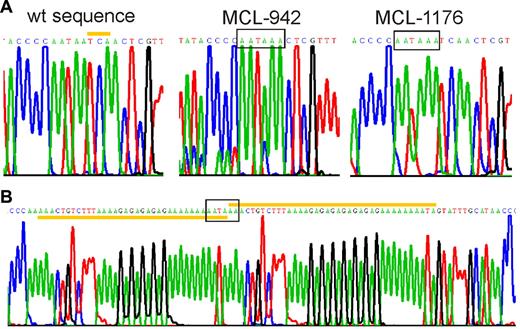
To determine whether genomic mutations could account for cyclin D1mRNA truncations, we sequenced approximately 700 bp of the proximal 3′UTR region, from the stop codon to the start of the AU-rich mRNA destabilizing element. In 3 patients, we identified mutations that created novel polyadenylation signals within 300 bp from the stop codon (Figure 4). All mutations were unique but were located within 100 bp of each other in a region rich in simple sequence repeats. MCL case 942 had a 3 base pair deletion, and MCL case 1176 had an A insertion, both of which created a polyadenylation signal at the identical site, 304 bp 3′ of the stop codon. In MCL case 970, a partial duplication of an adenin-rich sequence resulted in a novel polyadenylation sequence. The MCL cell line Z-13825 also expressed a truncated cyclin D1a transcript, and carried the same mutation as MCL case 1176. All 3 patients and the Z-138 cell line were heterozygous for the mutations as shown by direct sequencing of PCR-amplified genomic DNA as well as by sequencing of individual cloned PCR products.
Mutations in the 3′UTR generate premature polyadenylation signals. Sequencing chromatograms showing premature polyadenylation signals (boxed). (A) Wild-type and mutated cyclin D1a alleles from MCL-tumors 942 and 1176 are shown. The yellow bar highlights the 3 nucleotides at position 1344-46 of wild-type cyclin D1a mRNA that were deleted in MCL-942. In MCL-1176 and Z-138, there was an A insertion before position 1250. (B) The mutated allele of MCL-970 contained a partial duplication (yellow bars), starting at position 1233.
Mutations in the 3′UTR generate premature polyadenylation signals. Sequencing chromatograms showing premature polyadenylation signals (boxed). (A) Wild-type and mutated cyclin D1a alleles from MCL-tumors 942 and 1176 are shown. The yellow bar highlights the 3 nucleotides at position 1344-46 of wild-type cyclin D1a mRNA that were deleted in MCL-942. In MCL-1176 and Z-138, there was an A insertion before position 1250. (B) The mutated allele of MCL-970 contained a partial duplication (yellow bars), starting at position 1233.
In summary, among tumors expressing short cyclin D1a transcripts, we identified mutations creating premature polyadenylation signals in 3 and genomic deletions of the 3′UTR region in 7. In the 5 remaining samples, we have no explanation for the expression of truncated cyclin D1 mRNAs at present; in 4 tumors for which material was available, we failed to identify premature polyadenylation signals despite extensive sequencing of genomic DNA and 3′ end amplification of mRNA.
As expected, in tumors with polyadenylation mutations, the abundance of short cyclin D1 mRNAs was greater than the abundance of full-length mRNAs: the amplification signal obtained using the proximal UTR probe was 2.5- to 5-fold higher than that obtained using the distal UTR probe. However, these ratios were lower than those observed in cases harboring genomic deletions in the 3′UTR region (10-fold average ratio; range, 7- to 30-fold). This suggested that the efficiency of polyadenylation and cleavage caused by the polyadenylation mutations might not be 100% and that truncated mRNAs and full-length mRNAs could coexist in such cases. In keeping with this hypothesis, distal 3′UTR primers were able to amplify cyclin D1 mRNA from the Z-138 cell line despite the presence of a cyclin D1 polyadenylation mutation in these cells (Figure 5B). The amplification signal using exon 1-2 primers was roughly 6-fold higher than that obtained with the distal UTR primers, demonstrating that the premature polyadenylation signal resulted in significant but incomplete premature polyadenylation and cleavage. To confirm this view, we amplified the proximal 3′UTR region from mRNA derived from the 3 cases with the polyadenylation mutations and from the Z-138 cell line; in each instance, the polyadenylation mutation was present in the full-length mRNA, demonstrating that the polyadenylation mutations were only partially effective in terminating transcription prematurely.
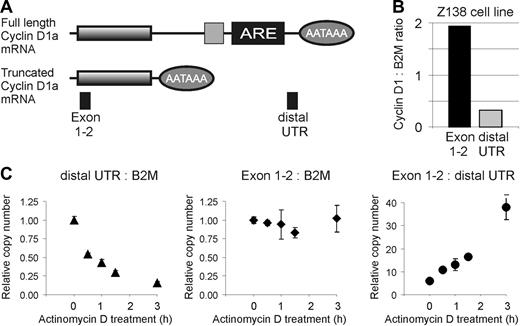
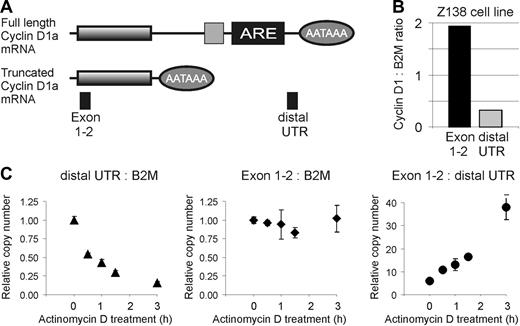
Cyclin D1a transcripts with full-length and truncated 3′UTRs are expressed in Z-138, but the transcript with a truncated 3′UTR is more stable. (A) Schematic representation of the 2 cyclin D1a transcripts found in Z-138. Lines represent the mRNA, with shaded boxes indicating the coding sequence. Ovals indicate the position of the polyadenylation signal (AATAAA), while the light gray square and the black box represent mRNA destabilizing elements, including an AU-rich element (ARE). PCR probes are represented by filled boxes. (B) Expression of cyclin D1 transcripts in Z-138 cells in relation to beta-2 microglobulin (B2M). (C-E) Relative expression levels of cyclin D1 and B2M transcripts in Z-138 cells after inhibition of transcription with 10 μg/mL actinomycin D.
Cyclin D1a transcripts with full-length and truncated 3′UTRs are expressed in Z-138, but the transcript with a truncated 3′UTR is more stable. (A) Schematic representation of the 2 cyclin D1a transcripts found in Z-138. Lines represent the mRNA, with shaded boxes indicating the coding sequence. Ovals indicate the position of the polyadenylation signal (AATAAA), while the light gray square and the black box represent mRNA destabilizing elements, including an AU-rich element (ARE). PCR probes are represented by filled boxes. (B) Expression of cyclin D1 transcripts in Z-138 cells in relation to beta-2 microglobulin (B2M). (C-E) Relative expression levels of cyclin D1 and B2M transcripts in Z-138 cells after inhibition of transcription with 10 μg/mL actinomycin D.
Truncated cyclin D1 mRNAs are more stable than the full-length mRNA
Full-length cyclin D1a mRNA contains mRNA destabilizing elements, including an AU-rich element that is known to shorten mRNA half-life,26,27 and the truncated cyclin D1 mRNAs described above lack this region. To investigate whether the truncated cyclin D1 mRNAs are more stable than the full-length transcript, we took advantage of the fact that the Z-138 cell line expresses both mRNA species. To measure the decay rate of the full-length and truncated mRNAs, we treated Z-138 cells with actinomycin D and measured the change in cyclin D1 mRNA over time using 2 different primer pairs: distal UTR primers were used to measure the abundance of full-length mRNA and exon 1-2 primers were used to measure the combined abundance of the full-length and truncated mRNAs. However, since the truncated mRNA is roughly 6-fold more abundant than the full-length mRNA (Figure 5B), the signal from the exon1-2 primers primarily reflected the abundance of the truncated mRNA species. In all experiments, the abundance of cyclin D1 mRNA was compared to the abundance of beta-2- microglobulin (B2M) mRNA, which is a relatively stable mRNA species. The full-length cyclin D1 mRNA decayed much more rapidly than the B2M mRNA and was reduced by 50% at 30 minutes (Figure 5C). By contrast, the truncated cyclin D1 mRNA was much more stable (Figure 5D). Consequently, the exon 1-2/distal 3′UTR amplification ratio increased from 5:1 to 36:1 over the 3 hours period of actinomycin D treatment. These findings confirm that truncated cyclin D1mRNA lacking the mRNA destabilizing elements has an extended half life.
To assess whether the presence of the truncated cyclin D1a mRNA isoform could lead to prolonged high cyclin D1 protein expression, we analyzed cells treated with actinomycin D by Western blotting. KMS-12 cells showed moderate cyclin D1 protein expression at baseline, and as expected, cyclin D1 protein became undetectable within 3 hours of actinomycin D treatment (Figure 6). In contrast, Z-138 cells showed high cyclin D1 protein expression for more than 8 hours of actinomycin D treatment, demonstrating that the truncated cyclin D1a mRNA leads to persistently high cyclin D1 protein expression long after transcription has been shut off.
Cyclin D1 protein expression is higher and persists longer after inhibition with actinomycin D in cells expressing a truncated cyclin D1a transcript than in cells expressing a normal cyclin D1a transcript. Western blots of cyclin D1 protein expression in KMS-12 (normal cyclin D1a transcript) and Z-138 cells (truncated cyclin D1a transcript) treated with actinomycin D for the indicated time points. Antibodies against alpha tubulin (loading control at 55 kDa) and cyclin D1 (38 kDa) were used simultaneously. Vehicle (dimethyl sulfoxide [DMSO]) only treated cells at 12 hours are included as control (far right lane).
Cyclin D1 protein expression is higher and persists longer after inhibition with actinomycin D in cells expressing a truncated cyclin D1a transcript than in cells expressing a normal cyclin D1a transcript. Western blots of cyclin D1 protein expression in KMS-12 (normal cyclin D1a transcript) and Z-138 cells (truncated cyclin D1a transcript) treated with actinomycin D for the indicated time points. Antibodies against alpha tubulin (loading control at 55 kDa) and cyclin D1 (38 kDa) were used simultaneously. Vehicle (dimethyl sulfoxide [DMSO]) only treated cells at 12 hours are included as control (far right lane).
Discussion
The translocation of cyclin D1 plays an initiating role in MCL. In addition, cyclin D1 apparently plays an ongoing role in the biology of MCL since higher expression of cyclin D1 in some MCL tumors has been associated with higher tumor proliferation rate and shorter length of survival following diagnosis.19 Here we demonstrate that some of the most proliferative MCL tumors express high levels of cyclin D1 transcripts that lack a full-length 3′UTR. These transcripts are truncated variants of cyclin D1a mRNA, not the alternatively spliced cyclin D1b mRNA. We identified 2 distinct genetic events that can cause the expression of truncated cyclin D1 transcripts: genomic deletions of the 3′UTR region and point mutations in the 3′UTR region that create alternative polyadenylation signals. Both genetic events generate short cyclin D1a mRNAs that are more stable than the full-length mRNA since they lack mRNA destabilizing elements. Our data support a model in which these genetic events enhance the influence of the t(11;14) by generating higher levels of cyclin D1 mRNA and protein, thereby augmenting cell cycle progression through the G1/S transition.
The previously observed differences in cyclin D1 mRNA levels among MCL tumors19 could have been explained by differential expression of the alternatively spliced cyclin D1b isoform, since this isoform lacks the mRNA destabilizing elements in the 3′UTR region. However, using splice form–specific quantitative PCR, we determined that cases with high proliferation and inferior survival expressed variants of the cyclin D1a splice isoform that are truncated in the 3′UTR. The common polymorphism A870 at the exon 4/intron 4 boundary has been associated with preferential splicing to the cyclin D1b form and adverse clinical outcome in solid tumors, but we found no correlation between the G870A polymorphism and either tumor proliferation rate or the relative abundance of the cyclin D1a or D1b splice isoforms.
The expression of truncated cyclin D1 mRNA was strongly associated with high tumor proliferation rate: 48% of the tumors in the highest quartile of the proliferation signature expressed these truncated mRNA species, whereas all of the tumors in the lowest quartile of the proliferation signature expressed full-length cyclin D1a mRNA. Patients whose tumors expressed short mRNAs had a median survival significantly shorter than patients without this additional change. In half of the tumors that expressed truncated cyclin D1 mRNA, we identified corresponding genomic deletions in the 3′UTR, the exact extent of which remains to be defined but which could include internal deletions within a larger cyclin D1 locus or a genomic rearrangement that replaces the cyclin D1 3′UTR with another sequence as described before.22,28 In addition, we also identified 3 tumors and one MCL cell line in which truncations of the 3′UTR were due to premature polyadenylation signals generated by a single base insertion, a small deletion, or the duplication of a repetitive element in the genomic DNA. The premature polyadenylation signals occur within the first 320 bp of the 3′UTR, leading to a predicted mRNA of approximately 1.5 kb.
Previous studies have described short cyclin D1 mRNAs in MCL or in other tumors and tumor cell lines. These short mRNAs typically have been estimated to be 1.5 to 1.7 kb in length and were often coexpressed with the full-length cyclin D1a transcript of 4.4-4.8 kb.6,8,29 One previous study reported on 22 cases with cyclin D1 expression, 3 of which showed predominantly short transcripts of 1.5 kb, and these cases had very high total cyclin D1 levels.5 The nature of these short cyclin D1 mRNAs cannot be determined with certainty because most of these studies failed to distinguish between cyclin D1a and D1b transcripts. Apparently truncated cyclin D1a mRNAs were described in 3 MCL cases, but none of these transcripts contained a polyadenylation signal, and thus the molecular mechanism accounting for these short mRNAs remained unclear.28 Similarly, many cyclin D1a sequences published or deposited in public databases are not full length and do not contain a polyadenylation signal, indicating that many of these cDNA sequences were likely the result of internal priming by oligo dT in A-rich regions of the proximal 3′UTR. This interpretation also has been entertained by other investigators.28
Interestingly, one previous study identified an MCL sample with a new polyadenylation signal that was due to the same 3-bp deletion in the proximal 3′UTR that we identified in MCL 940. Further, we identified an insertion of an A at position 1344 (309 bp 3′ of the stop codon) in MCL case 1176 as well as in the MCL cell line Z-138. Corresponding germ-line DNA was not available for any of these tumor samples, leaving open the possibility that these sequence changes in cyclin D1 are actually polymorphisms within the human population. A public database (http://www.ncbi.nlm.nih.gov/projects/SNP/) lists 106 polymorphisms in the human cyclin D1 gene, 33 of which affect the 3′UTR. None of these creates a polyadenylation consensus sequence, and none is found in the vicinity of the premature polyadenylation site identified here. Of course, it is impossible to rule out the possibility that these sequence changes are rare polymorphisms that have not been encountered previously. However, it is striking that the 2 recurrent changes described above occur in the near vicinity of the same AATAA sequence in the germ-line CCND1 sequence. Thus, it seems more likely that these sequence changes are somatic mutations in the tumor that conferred a selective advantage to the malignant clone due to the creation of a short cyclin D1 mRNA species.
Both genomic deletions and premature polyadenylation create cyclin D1 mRNAs that lack the long 3′UTR. Using the Z-138 MCL cell line, we demonstrated that the half life of the truncated cyclin D1 mRNA (> 3 hours) is considerably longer than that of the full-length mRNA (∼30 minutes), in agreement with previous reports.28,30 One main regulator of mRNA stability is the adenylate- and uridylate-rich, so called AU-rich, element (ARE), which is typically located in the 3′UTR and contains several copies of the pentamer AUUUA (reviewed in Bevilacqua et al26 and Guhaniyogi et al27 ). ARE elements are commonly found in short-lived transcripts of growth factors, cytokines, cell cycle proteins, and a wide variety of genes that are induced rapidly by signaling through cell surface receptors.31 The cyclin D1 3′UTR contains an ARE element with 4 AUUUA pentamers within a stretch of 100 bp.
ARE binding proteins (AUBP) attach to ARE elements and recruit an exosome that degrades the transcript.32 The ARE of cyclin D1 binds the AUBP tristetraprolin (TTP).33,34 Interestingly, the effect of TTP on cyclin D1 was observed to be modulated by mTOR inhibitors such as rapamycin.33 Under certain conditions, rapamycin treatment could induce rapid degradation of cyclin D1 transcripts containing the AREs but not of transcripts with 3′UTR deletion or mutated AUUUA pentamers. Of note, rapamycin analogs are currently in clinical trials in MCL and have been reported to have activity in some patients.35 In this regard it will be interesting to test whether differential expression of cyclin D1 mRNA isoforms correlates with clinical response to mTOR inhibitors.
An additional 390-bp sequence element in the proximal 3′UTR of cyclin D1 has been identified that binds the RNA binding protein AUF-1, another member of the AUBP family known to mediate mRNA decay.36 Treatment of cells with the experimental chemotherapeutic agent prostaglandin A2 (PGA2) triggered AUF-1 binding to cyclin D1 mRNA, which resulted in rapid mRNA degradation.36 As expected, the 3′UTR truncated cyclin D1 form expressed by the breast cancer cell line MDA-MB-453 was resistant to AUF-1–triggered decay. Based on these findings, MCL tumors with truncated cyclin D1a mRNA expression would be predicted to be resistant to PGA2 treatment.
An additional putative consequence of the cyclin D1 3′UTR deletions might be deletion of target sequences for microRNAs. Recently, it has been appreciated that a large number of conserved elements in mammalian 3′UTR regions are binding sites for microRNAs.37–39 MicroRNAs have the ability to decrease mRNA expression as well as decrease mRNA translation into protein.40 The cyclin D1 3′UTR region has a predicted match to the “seed” regions of several mammalian microRNAs (hsa-miR-15a, hsa-miR-15b, hsa-miR-16, and hsa-miR-195),41 which is notable since the seed region is thought to direct interaction of a microRNA with a target gene. Thus, the genetic alterations of cyclin D1 that result in 3′UTR truncations might make the mRNA insensitive to the effect of microRNAs. It will therefore be of interest to test whether these microRNAs control the expression of cyclin D1 mRNA and/or protein.
Premature polyadenylation and mRNA stabilization represents a novel gain of function mechanism for oncogene activation. We could account for most MCL cases with truncation of cyclin D1 mRNA by the genomic deletions and mutations described above. However, in some tumors we failed to identify a premature polyadenylation signal, and the mechanism underlying the cyclin D1 mRNA truncation remains unknown. One possibility is that mutations in the distal region of the 3′UTR might activate cryptic upstream polyadenylation sites, as has been described for the DHFR gene in a Chinese hamster cell line.42 Alternatively, these tumors may have as yet undefined abnormalities that affect the regulation of polyadenylation and cleavage of all genes, allowing cryptic polyadenylation signals to be used.
In summary, 2 types of genetic alteration in MCL yield more stable cyclin D1 mRNAs that lead to higher or more prolonged cyclin D1 mRNA and protein expression, resulting in higher tumor proliferation and shorter survival. Numerous genes involved in the control of cell growth, cell cycle progression, and cell signaling have complex 3′UTRs that harbor mRNA destabilizing elements. Indeed, mutations or deletions affecting AREs can activate the oncogenic potential of growth factors and proto-oncogenes.43,44 Therefore, a systematic screen for 3′UTR alterations in malignancies could uncover important control mechanisms and identify new therapeutic targets.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health, the National Cancer Institute and the National, Heart, Lung and Blood Institute.
Authorship
Contribution: A.W. and L.M.S. designed the study, carried out the analysis, and wrote the paper. A.W., M.T., M.C., F.G., K.N., and H.L. performed the molecular assays and performed data analysis. G.W. did the statistical analysis. A.R. contributed the gene expression data. H.K.M.-H., G.O., W.C.C, T.C.G., D.D.W., J.V., J.O.A., R.D.G., J.M.C., E.C., E.M., F.B., E.B.S., S.K., H.H., J.D., R.I.F., T.M.G., T.P.M., W.H.W., and E.S.J. contributed the clinical samples, reviewed the pathology of cases included in the study, and assisted in the design of the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of the members of the Lymphoma/Leukemia Molecular Profiling Project appears as a data supplement to the online version of this article.
Correspondence: Louis M. Staudt, Metabolism Branch, CCR, NCI, Bldg 10, Rm 4N114, NIH, 9000 Rockville Pike, Bethesda, MD 20892; e-mail: lstaudt@mail.nih.gov.


![Figure 6. Cyclin D1 protein expression is higher and persists longer after inhibition with actinomycin D in cells expressing a truncated cyclin D1a transcript than in cells expressing a normal cyclin D1a transcript. Western blots of cyclin D1 protein expression in KMS-12 (normal cyclin D1a transcript) and Z-138 cells (truncated cyclin D1a transcript) treated with actinomycin D for the indicated time points. Antibodies against alpha tubulin (loading control at 55 kDa) and cyclin D1 (38 kDa) were used simultaneously. Vehicle (dimethyl sulfoxide [DMSO]) only treated cells at 12 hours are included as control (far right lane).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/11/10.1182_blood-2006-08-039859/4/m_zh80110704180006.jpeg?Expires=1763834817&Signature=mq2jVfjSEne6SmKHPMYa2J6H2Xy8C6zFTZmxF~t~Xh6SedqJXAFXqXCuiCwvAvphiZpGQ99Yj30VSCgq~Tt7XmP920NWgHWVRJKZOu3t~HYIIEwondxIQ2H-Sw9K83k0oNjfHIRDL4BpGRJPBEEe6Gwxlg1H67L1~gQvVRkzUgKyx-gc66pngCXZChmhFvtVvesSz-zIUAv44SwzV8RYJNoR3YbOzK9y6nqib1AfszbML65GFM7CX4curFmGE15WgJjbC6XhYv9~SymJn9lVALejuHK90robfT7-Z5WIf1R~kS104Sx0kYyzl9aSS5PGPjlPWmAG~tkZcM~lXeaM8A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 6. Cyclin D1 protein expression is higher and persists longer after inhibition with actinomycin D in cells expressing a truncated cyclin D1a transcript than in cells expressing a normal cyclin D1a transcript. Western blots of cyclin D1 protein expression in KMS-12 (normal cyclin D1a transcript) and Z-138 cells (truncated cyclin D1a transcript) treated with actinomycin D for the indicated time points. Antibodies against alpha tubulin (loading control at 55 kDa) and cyclin D1 (38 kDa) were used simultaneously. Vehicle (dimethyl sulfoxide [DMSO]) only treated cells at 12 hours are included as control (far right lane).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/11/10.1182_blood-2006-08-039859/4/m_zh80110704180006.jpeg?Expires=1763834818&Signature=Orq2yIXLRq~ROUoyco0BzVkI10ypou2cr3~8Y1~GHVVHr-rQW7xjkAVvPWiMuo~ZWmpnc~tM2kzeEaDsZqiAKCz0QS4DbePRRY8kLdEhQgVdEHqc6HArohDG8OMkMHpNaP78bUwSUyBtcKx9233SUe1mRjHxCo7VbBsUktdE5NsTOseee0EEZbU99RMHDrCWWmvrHScp3ZeaxOPyW9W3N6~fZFPh4kUmpavgckJFFXNdF3IYoCqOH02IIH4vkrJff6hQLzJRB4O-3-k27fZSVQQCXjHVRX5282wkllxs5j~SaFJjpdDaypEk2NMGJxkxdl83yf-p-jlZlfRcXlbyXQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)