Abstract
The Homeobox (Hox) transcription factors are important regulators of normal and malignant hematopoiesis because they control proliferation, differentiation, and self-renewal of hematopoietic cells at different levels of the hematopoietic hierarchy. In transgenic mice we show that the expression of HOXA10 is tightly regulated by doxycycline. Intermediate concentrations of HOXA10 induced a 15-fold increase in the repopulating capacity of hematopoietic stem cells (HSCs) after 13 days of in vitro culture. Notably, the proliferation induction of HSC by HOXA10 was dependent on the HOXA10 concentration, because high levels of HOXA10 had no effect on HSC proliferation. Furthermore, high levels of HOXA10 blocked erythroid and megakaryocyte development, demonstrating that tight regulation of HOXA10 is critical for normal development of the erythroid and megakaryocytic lineages. The HOXA10-mediated effects on hematopoietic cells were associated with altered expression of genes that govern stem-cell self-renewal and lineage commitment (eg, hepatic leukemia factor [HlF], Dickkopf-1 [Dkk-1], growth factor independent-1 [Gfi-1], and Gata-1). Interestingly, binding sites for HOXA10 were found in HLF, Dkk-1, and Gata-1, and Dkk-1 and Gfi-1 were transcriptionally activated by HOXA10. These findings reveal novel molecular pathways that act downstream of HOXA10 and identify HOXA10 as a master regulator of postnatal hematopoietic development.
Introduction
Hematopoiesis is a lifelong, dynamic process, originating from a low number of hematopoietic stem cells residing in the adult bone marrow. Hematopoietic stem cells (HSCs) have the ability to self-renew and generate all blood lineages throughout life. The machinery regulating HSCs has been shown to include both external and internal signals. The external signals from the bone marrow niche where postnatal HSCs reside can be mediated by cell-cell interactions (eg, Notch signaling and integrins1,2 ), cell–extracellular matrix–HSC interactions (eg, osteopontin and collagen I3,4 ), classical cytokines (eg, TPO and SCF5 ), and developmental fate determinants (eg, Wnt, BMP, TGF-β, and Notch ligands6–12 ). The internal signals are represented by the gene expression pattern of transcription factors, transcriptional repressors, and cell-cycle regulators.13,14 Cell-cycle regulators like the cyclin-dependant kinase inhibitors play crucial roles in regulating the self-renewal of HSCs (p21, p18) and primitive hematopoietic progenitors (p27),15–18 and reports demonstrate that the transcriptional repressors Gfi-1 and Bmi-1 are powerful HSC regulators.19,20 A number of transcription factors have been shown to act as critical intrinsic factors for specification of HSCs during ontogeny, and many of these factors are also important for normal regulation of HSC fate postnatally (eg, Runx-1, Pu.1, Scl, and Lmo2).21–23 Knockout mouse models have also demonstrated crucial roles for several transcription factors in early hematopoietic commitment steps and lineage outcome, for example, Gata-1 for erythrocyte and megakaryocyte development and Pax-5 for B-cell development.24–27
The Homeobox (Hox) genes encode transcription factors that are expressed in specific compartments of the hematopoietic hierarchy,28–30 and functional studies demonstrate their role in the regulation of hematopoietic proliferation, differentiation, and lineage commitment.29,31–33 Hox proteins contain a highly conserved DNA-binding domain (homeodomain) and variable sequences flanking the homeodomain that influence the DNA-binding specificity, by coordinating interactions to the cofactors Pbx and Meis.34 Lack-of-function studies demonstrate a physiologic role for Hox genes in the regulation of hematopoietic stem cell fate, although the hematopoietic phenotypes of Hox gene knockout mice have, with the exception of Hoxa9, usually been mild.33,35–37 However, the clearest indications for a role of Hox transcription factors in the regulation of hematopoietic progenitor and stem cells have come from overexpression studies. HOXA10 is expressed in the most primitive hematopoietic cell compartment, and overexpression of HOXA10 leads to impaired lymphoid development in vitro.38–40 In addition, enforced expression of HOXA10 affects myelopoiesis which can ultimately lead to myeloid leukemia.41 Notably, overexpression of HOXB4 expands the stem-cell pool in vitro without affecting stem-cell pluripotency and lineage choice.42 However, new findings indicate that high levels of HOXB4 may perturb lineage choice in primitive human hematopoietic progenitors, suggesting a dosage effect.43,44 Similarly, the transcription factors PU.1 and Gata-1 have concentration-dependent effects on differentiation and lineage choice,45–47 and graded expression of PU.1 can induce acute myeloid leukemia (AML) in mice.47
Here, we asked whether tight regulation of HOXA10 is essential to control normal fate decisions in hematopoietic progenitors and stem cells. To address this question we used transgenic mice, where the human cDNA, encoding HOXA10 is driven by an inducible promoter (tetO) that can be regulated by tetracycline transactivators (tTA).48 In the present study, we took advantage of the recently developed Rosa26-rtTA-nls mouse. The Rosa26-rtTA-nls mouse model was generated by knocking in the reverse tetracycline transactivator (rtTA) with a nuclear localization sequence (nls) into the ROSA26 locus.49–51 This locus has been shown to be transcriptionally active in all hematopoietic tissues.51,52 HOXA10 transgenic mice were crossed with Rosa26-rtTA-nls mice to generate a mouse model in which the expression level of HOXA10 can be tightly regulated using doxycycline. The expression of HOXA10 induced an increase in HSC repopulating capacity in vitro and identified the stem-cell regulators Hlf, Dkk1, and Gfi-1 as downstream targets of HOXA10. Notably, the proliferation induction of HSC by HOXA10 was dependent on the HOXA10 concentration, because high levels of HOXA10 had no effect on HSC proliferation. Furthermore, high levels of HOXA10 lead to an accumulation of MEPs and blocked erythroid and megakaryocyte development. Interestingly, in a mouse model lacking Hoxa7, So et al53 show that the level of MEPs was reduced, suggesting that correct Hox-gene regulation is important for erythrocyte/megakaryocyte differentiation. We further found that the block in erythropoiesis and megakaryopoieis is accompanied by down-regulation of Gata-1, suggested here as a direct downstream target of HOXA10. Our findings demonstrate that HOXA10 acts as an important regulator of hematopoiesis, governing both proliferation and differentiation of hematopoietic progenitor and stem cells, where distinct fate outcomes depend on the HOXA10 concentration.
Materials and methods
Generation and screening of transgenic mice
The generation of the tetO-HOXA10 mouse strain and its characterization has been described previously.48 To generate rtTA-HA10, tetO-HOXA10 mice were mated with the Rosa26-rtTA-nls mice (provided by Dr R. Jaenisch, Whitehead Institute for Biomedical Research, Cambridge, MA). Genomic DNA was isolated from tail biopsy and analyzed by polymerase chain reaction (PCR) for detection of tetO-HOXA10 (see Bjornsson et al48 ) and for Rosa26-rtTA-nls Neo2, CATCCTGATCGACAAGACC, and Neo3, CTATTCGGCTATGACTGG. Background level, induction, and the reversibility of HOXA10 were tested by reverse transcriptase (RT)–PCR. For HOXA10 primers, see Björnsson et al.48 RNA was extracted from cultured whole bone marrow cells (RNeasy Mini kit; Qiagen, Stockholm, Sweden), and cDNA was transcribed using Supercript III (Invitrogen, Stockholm, Sweden) and random hexamers. PCR with HPRT-specific primers48 was performed to evaluate genomic DNA contamination and the presence of cDNA. Lund University's Ethical Committee approved all animal experiments.
Clonogenic assays
For CFU-GM colonies, 1000 cultured bone marrow cells were cultured in 1 mL methylcellulose medium (M3534; Stem Cell Technologies, Vancouver, BC) containing 50 ng/mL mSCF, 10 ng/mL mIL-3, and 10 ng/mL hIL-6 (all from R&D System, Abingdon, United Kingdom). Colonies were scored on day 6.
Fluorescence-activated cell sorting (FACS) analysis
Cells cultured for 12 days were stained with fluorescein isothiocyanate (FITC)–conjugated rat anti–mouse Gr1 antibody and phycoerythrin (PE)–conjugated anti-Mac1 antibody for analysis of myeloid cells. For analysis of reconstituted mice, PE-conjugated anti-CD45.1 (Ly5.1) and allophycocyanin (APC)–biotin–anti-CD45.2 (Ly5.2) antibodies were used. For detection of B, T, myeloid, and erythroid cells PE-conjugated anti-CD3, -CD4, -B220, -CD71, -Mac, and FITC-conjugated anti-CD8, -Mac1, -Gr-1, -Ter119 were used (all antibodies from BD Pharmingen, Stockholm, Sweden, unless otherwise indicated). To identify the CMP, GMP, and MEP, lineage staining was performed as previously described.54
Electrophoretic mobility shift assay (EMSA)
Histology and blood analysis
Organs were processed for routine histology by fixation in PBS-buffered 4% parafomaldehyde followed by paraffin embedding and sectioning. Sections were stained with Erlisch eosin for microscopic examination. Blood analysis was performed on peripheral blood using the Sysmex machine KX-21N.
Single-cell and ex vivo expansion cultures
Lineage negative (Lin−), Sca1+, c-kit+ (LSK) cells were sorted by FACS as previously described directly as single cells or 1000 cells/well into 96-well plates.33 Cells were cultured in serum-free medium (X-vivo 15; BioWhittaker, Verviers, Belgium) supplemented with 1% bovine serum albumin (Stem Cell Technologies), 100 IU penicillin and 100 μg streptomycin/mL, 2 mM L-glutamine (Gibco-BRL, Stockholm, Sweden), 10−4 M 2-mercaptoethanol (Sigma-Aldrich, Stockholm, Sweden) (hereafter denoted as complete serum-free medium) with 25 ng/mL SCF, TPO, and Flt-3 ligand in 0, 0.1, 0.2, or 0.5 μg doxycycline/mL (Sigma-Aldrich). Fresh medium with doxycycline was added every fourth day, and cells were scored on day 12.
In vivo reconstitution experiments
rtTA-HA10 or control littermate LSK cells (Ly5.2/5.2) were used as donor cells. C57BL/6-B6SJL (Ly5.2-Ly5.1) recipient mice received transplants of 200 freshly isolated LSK cells or the expansion equivalence, together with 200 000 competitor cells (Ly5.1/5.1). Recipient mice were lethally irradiated (9.5 Gy [950 rad]) 6 hours before transplantation. For in vivo induction of HOXA10 5 × 105 to 5 × 106 rtTA-HA10 whole bone marrow cells were transplanted with or without Ly5.1 competitor bone marrow cells into lethally irradiated recipients receiving 0.2 mg/mL doxycycline and 3% sucrose in the drinking water.
Q-RT-PCR
RNA was extracted from cultured LSK cells or whole bone marrow cells using RNeasy Mini kit (Qiagen). cDNA was transcribed (Supercript III; Invitrogen) and quantitive PCR (Q-PCR) was performed using TaqMan probes from Applied Biosystems (Stockholm, Sweden) and analyzed in an ABI Prism 7700 Sequence Detection System. Values for each PCR were normalized against HPRT giving a relative intensity (RI).
Affymetrix gene expression and data analysis
LSK cells were sorted from rtTA-HA10 and cultured for 72 hours in complete serum-free medium in 0, 0.1, and 0.5 μg/mL doxycycline. Labeling, amplification, hybridization, and scanning was done according to Affymetrix (Swegene, Lund, Sweden) standard protocol. The GC-RMA method was used for normalization, and values below 0.01 were set to 0.01. All genes in each sample were divided by the median of the specified list of 100 positive control genes present on the MOE430 v2 chip. All samples were then normalized against the median of the untreated control samples. Measurements for each gene in the treated samples were divided by the median of that gene's measurements in the corresponding control samples. We judged genes to be differentially expressed when the difference in expression between 2 conditions was at least 2-fold. Microarray data are available at the GEO website with GEO accession no. GSE 3861.56
5′ Rapid amplification of cDNA ends (RACE) analysis
5′ RACE was performed as previously described.55 RACE primers were as follows: Gata-1r1, GCCTCAGCTTCTCTGTAGTAGG; Gata-1r2, AGTAGGCCACTGCTGATGCTGC; Gata-1r3, AGTGGAAGAAGATGCTGC ATCC; Dkk1r1, GGCAGGTTCTTGATCGCGTTGG; Dkk1r2, TTGATGAGA ACTGAGTTCAAGG; Dkk1r3, AGTTCAAGGTGGCACTGGCTCC; gfi1r1, AGATCATCACTCTCCTGAGAGC; gfi1r2, GAGAGCAGTAGTCCACAACTGG; gfir3, GGTCCGTTATCCCAAGTCAACC; HLFr1, TGCCTCCAACACTCAGTC TTGC; HLFr2, GTTCAAAATTGCAAGGAGGTGG; HLFr3, ACGTAGACGGCTG CAGATGAGC.
Transient transfections and luciferase assays
Transient transfections and luciferase assays were performed as previously described by Smith et al.55
Statistics
Results are presented as mean ± SEM. The Student t test or Mann-Whitney rank sum test was applied to determine significant differences (P < .05).
Results
A mouse model with regulated expression of HOXA10
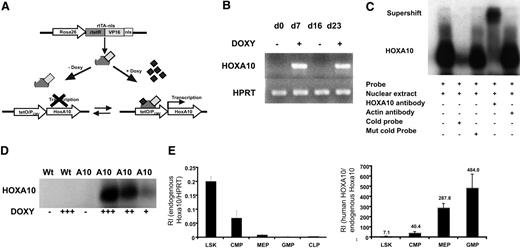
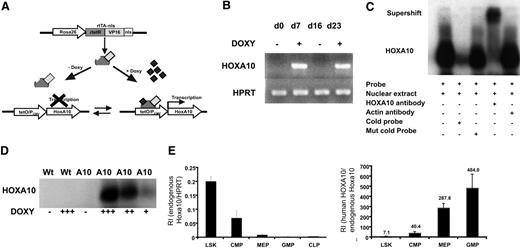
To enable regulated expression of HOXA10 in mice by adding or removing doxycycline, we mated the tetO-HOXA10 transgenic mouse line with Rosa26-rtTA-nls mice to generate rtTA-HA10 mice (Figure 1A). rtTA-HA10 mice were born at normal Mendelian ratios with normal hematologic parameters (data not shown). One mouse line showing no leakiness and high inducibility was chosen for these experiments.48 To test the inducibility of HOXA10, rtTA-HA10 bone marrow cells were harvested and cultured with doxycycline for 7 days. High induction of HOXA10 was demonstrated by RT-PCR. Furthermore, by removing doxycycline from the medium we could terminate the expression of HOXA10, and by subsequent re-addition of doxycycline the expression was restored, demonstrating reversibility of induction (Figure 1B). To verify production of the HOXA10 protein, nuclear extracts were generated from rtTA-HA10 bone marrow cells cultured for 5 days in different concentrations of doxycycline. Using EMSA we demonstrated induction of functional HOXA10 protein, with DNA binding capacity that was tightly regulated by doxycycline in vitro and without any leakiness (Figure 1C-D). Q-RT-PCR detected high induction of HOXA10 mRNA in whole bone marrow cells from in vivo–induced rtTA-HA10 mice receiving 0.2 mg/mL doxycycline in the drinking water for 7 days (RI in induced rtTA-HA10, 6.65 ± 2.3; RI in uninduced rtTA-HA10, 0; n = 2), which correlates to the highest doxycycline dose in vitro (RI in induced LSK cells in vitro with 0.5 μg/mL doxycycline = 5; see Figure 2A). In addition, FACS analysis of whole bone marrow cells cultured in different concentrations of doxycycline showed increasing proportion of Mac-1 single-positive cells with increasing concentration of doxycycline, confirming previous findings showing that HOXA10 promotes myeloid differentiation (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).38–40 We further verified that the endogenous hoxa10 is expressed in the most primitive hematopoietic cell population and then down-regulated on differentiation in wild-type mice (Figure 1E). As expected the difference between in vivo–induced HOXA10 and endogenous hoxa10 expression increases with differentiation (Figure 1F). These findings show that we have developed an animal model to study regulated expression of HOXA10.
Generation of mice with regulated expression of HOXA10. (A) The rtTA is a fusion protein, composed of the rtetR repressor protein fused to the VP16 activation domain and the nls, driven by the Rosa26 locus. In the absence of doxycycline, rtTA will not recognize the tetO sequence and no expression of HOXA10 will occur. Addition of doxycycline results in binding of the rtTA to the tetO sequence and transcriptional activation of HOXA10. The inducible expression system is reversible and withdrawal of doxycycline terminates the expression. (B) HOXA10 expression was measured in cultured inducible whole bone marrow cells at indicated time points by RT-PCR. Doxycycline was added and removed from the medium (off-on-off-on). (C) Functional HOXA10 protein was detected in protein extracts from inducible whole bone marrow cells cultured with doxycycline for 5 days by EMSA. (D) Inducible bone marrow cells were cultured for 5 days in different doses of doxycycline, and the level of functional protein expression was detected by EMSA. (E) Endogenous hoxa10 expression was measured by Q-RT-PCR in different hierarchical compartments of the hematopoietic system using sorted WT bone marrow cells, and relative intensity (RI) was calculated by comparing results using the formula 2−(CtHOXA10 − CtHPRT). (F) RI of endogenous murine hoxa10 expression and induced human HOXA10 expression was measured in sorted bone marrow cells 7 days after in vivo induction. Bars show relative HOXA10 expression in induced cells compared with endogenous hoxa10 (2−(CtInducedHOXA10 − CtEndougenushoxa10)). Data represent mean ± SEM.
Generation of mice with regulated expression of HOXA10. (A) The rtTA is a fusion protein, composed of the rtetR repressor protein fused to the VP16 activation domain and the nls, driven by the Rosa26 locus. In the absence of doxycycline, rtTA will not recognize the tetO sequence and no expression of HOXA10 will occur. Addition of doxycycline results in binding of the rtTA to the tetO sequence and transcriptional activation of HOXA10. The inducible expression system is reversible and withdrawal of doxycycline terminates the expression. (B) HOXA10 expression was measured in cultured inducible whole bone marrow cells at indicated time points by RT-PCR. Doxycycline was added and removed from the medium (off-on-off-on). (C) Functional HOXA10 protein was detected in protein extracts from inducible whole bone marrow cells cultured with doxycycline for 5 days by EMSA. (D) Inducible bone marrow cells were cultured for 5 days in different doses of doxycycline, and the level of functional protein expression was detected by EMSA. (E) Endogenous hoxa10 expression was measured by Q-RT-PCR in different hierarchical compartments of the hematopoietic system using sorted WT bone marrow cells, and relative intensity (RI) was calculated by comparing results using the formula 2−(CtHOXA10 − CtHPRT). (F) RI of endogenous murine hoxa10 expression and induced human HOXA10 expression was measured in sorted bone marrow cells 7 days after in vivo induction. Bars show relative HOXA10 expression in induced cells compared with endogenous hoxa10 (2−(CtInducedHOXA10 − CtEndougenushoxa10)). Data represent mean ± SEM.
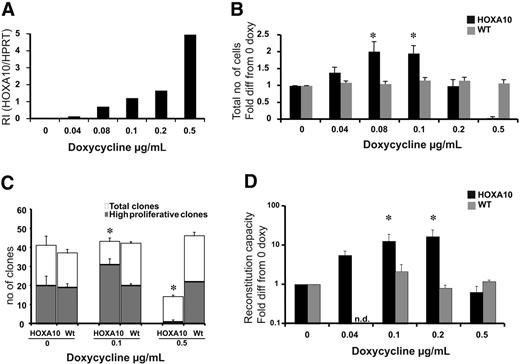
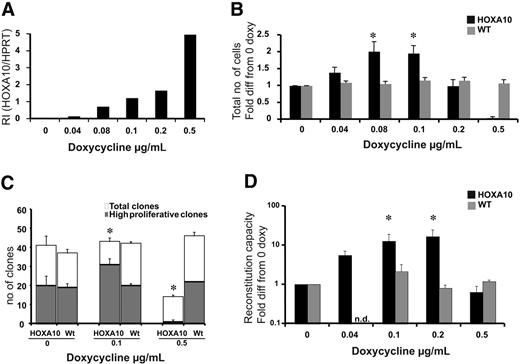
Low levels of HOXA10 expand stem cells in vitro, whereas high levels block expansion. LSK cells from rtTA-HA10 mice and WT mice were purified and cultured for 13 days in TPO, FLT-3, and SCF with 6 different concentrations of doxycycline. (A) HOXA10 mRNA was detected using Taq-man PCR, and RI was calculated by comparing results using the formula 2−(CtHOXA10 − CtHPRT). (B) The diagram shows fold expansion of total cells after 13 days of liquid culture with SCF, FLT-3, and TPO in 6 different concentrations of doxycycline. Black bars represents inducible HOXA10 LSK cells and gray bars represent WT LSK cells (n = 7) (C) LSK cells were cultured as single cells in 3 different concentration of doxycycline to measure recruitment into proliferation. Clones were scored at day 12 as high proliferative clones (> 50 cells) or as low proliferative clones (< 50 cells); n = 3. (D) Two hundred freshly sorted LSK cells or the expansion equivalent (Ly5.2/5.2) after 13 days of culture from panel A were transplanted with 200 000 unfractionated (Ly5.1/5.1) competitor bone marrow cells into lethally irradiated recipients. Peripheral blood cells were analyzed by flow cytometry 16 to 20 weeks after bone marrow transplantation to evaluate reconstitution of transplanted cells. Shown here is the reconstitution in comparison to uninduced HOXA10 LSK cells cultured for 13 days. Black bars represent inducible HOXA10 LSK cells, and gray bars represent WT LSK cells. Nd indicates not done (n = 7); *P < .05. Data represent mean ± SEM.
Low levels of HOXA10 expand stem cells in vitro, whereas high levels block expansion. LSK cells from rtTA-HA10 mice and WT mice were purified and cultured for 13 days in TPO, FLT-3, and SCF with 6 different concentrations of doxycycline. (A) HOXA10 mRNA was detected using Taq-man PCR, and RI was calculated by comparing results using the formula 2−(CtHOXA10 − CtHPRT). (B) The diagram shows fold expansion of total cells after 13 days of liquid culture with SCF, FLT-3, and TPO in 6 different concentrations of doxycycline. Black bars represents inducible HOXA10 LSK cells and gray bars represent WT LSK cells (n = 7) (C) LSK cells were cultured as single cells in 3 different concentration of doxycycline to measure recruitment into proliferation. Clones were scored at day 12 as high proliferative clones (> 50 cells) or as low proliferative clones (< 50 cells); n = 3. (D) Two hundred freshly sorted LSK cells or the expansion equivalent (Ly5.2/5.2) after 13 days of culture from panel A were transplanted with 200 000 unfractionated (Ly5.1/5.1) competitor bone marrow cells into lethally irradiated recipients. Peripheral blood cells were analyzed by flow cytometry 16 to 20 weeks after bone marrow transplantation to evaluate reconstitution of transplanted cells. Shown here is the reconstitution in comparison to uninduced HOXA10 LSK cells cultured for 13 days. Black bars represent inducible HOXA10 LSK cells, and gray bars represent WT LSK cells. Nd indicates not done (n = 7); *P < .05. Data represent mean ± SEM.
HOXA10 induces proliferation of primitive hematopoietic cells
To ask how increasing levels of HOXA10 affect the proliferative capacity of primitive hematopoietic cells, LSK cells were sorted and cultured for 13 days in different concentrations of doxycycline. To verify that the dose of doxycycline was directly correlated to the expression level of HOXA10 in LSK cells, Q-RT-PCR was performed. We found that the expression of HOXA10 was highly controllable in this primitive progenitor population, and no leakiness was detected (Figure 2A). After 13 days of culture, the cells were counted, revealing that lower concentrations of doxycycline lead to a 2-fold increase in proliferation, whereas increasing levels of doxycycline resulted in impaired proliferation (Figure 2B). Importantly, no difference in viability was detected using trypan blue staining.
To address whether the increased proliferation of low-to-intermediate levels of HOXA10 where due to the expansion of the LSK cells at the single-cell level, LSK cells from rtTA-HA10 mice were plated as single cells to measure proliferation recruitment. The findings show an increased number of high-proliferative clones at lower concentrations of doxycycline and a reduced number of high-proliferative clones as well as total number of clones at high concentrations of doxycycline (Figure 2C). Therefore, low-to-intermediate levels of HOXA10 promote proliferation of LSK cells in vitro.
HOXA10 promotes the proliferation of repopulating HSCs
Because HOXA10 increases the proliferation of the LSK cells in vitro, we asked how graded expression of HOXA10 in LSK cells affects the repopulation ability of HSCs after 13 days of liquid culture. LSK cells from rtTA-HA10 mice (Ly5.2/5.2) were cultured in different concentrations of doxycycline and transplanted with a fixed number of competitive bone marrow cells from WT donors (Ly5.1/5.1) into lethally irradiated recipients (Ly5.1/5.2) without administration of doxycycline to the transplant recipients to eliminate further effects of HOXA10 overexpression in vivo. Sixteen weeks after transplantation, rtTA-HA10 HSCs exhibit a 15-fold increase in multilineage repopulation capacity induced with the intermediate concentration of doxycycline compared with uninduced HOXA10 LSK cells cultured for 13 days (Figure 2D). In contrast, rtTA-HA10 cells cultured in high concentrations of doxycycline had normal viability but exhibited no repopulating advantage (Figure 2D). These data are consistent with the findings from the culture of LSK cells in vitro and indicate that tight regulation of HOXA10 governs proliferation of repopulating HSCs. Despite this decisive effect on proliferation of HSCs, the progeny of the transplanted cells were represented in all lineages (Table S1).
High levels of HOXA10 block murine erythroid development and cause an accumulation of hematopoietic progenitors in vivo
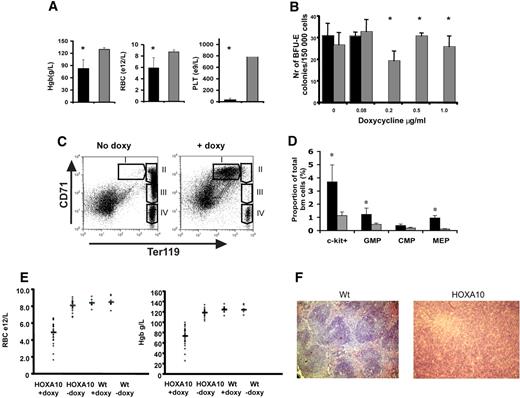
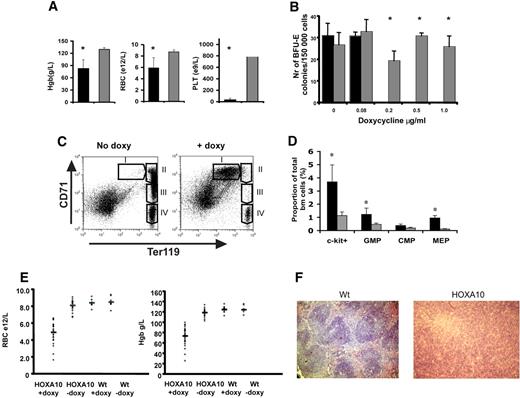
Next, we wanted to study the role of HOXA10 in vivo by administrating doxycycline to the rtTA-HA10 mice via the drinking water. Strikingly, rtTA-HA10 mice receiving doxycycline developed a dramatic weight loss and died 10 to 20 days after induction. The mice displayed hemorrhages in the skin, clearly visible on the ears. Blood analysis showed extremely low platelet counts as well as a significant reduction in red blood cells and hemoglobin levels (Figure 3A). These in vivo findings were also confirmed in vitro, because high levels of HOXA10 inhibited both the development of megakaryocyte and BFU-E colonies in vitro (Figure 3B; data not shown). To determine the precise stage at which HOXA10 affects erythropoiesis, bone marrow and spleen from induced rtTA-HA10 mice were stained with markers for erythroid development (anti-CD71, anti-Ter119) 2 weeks after induction.57 FACS analysis showed an accumulation of early precursor cells (CD71+Ter119+) and a block in the erythroid development at the proerythroblast stage (Figure 3C).
High levels of HOXA10 block erythroid development in vivo and lead to accumulation of hematopoietic progenitors. (A) Analysis of blood from rtTA-HA10 mice 2 weeks after induction of HOXA10 expression in vivo revealed reduced levels of hemoglobin (Hgb), red blood cells (RBCs), and platelets (PLTs) (n = 6). (B) Bone marrow cells were plated in methylcellulose supporting the formation of BFU-E colonies, with 4 different concentrations of doxycycline. Colonies were scored after 6 days of culture. Shown are the numbers of BFU-E colonies/100 000 input cells. Black bars represent inducible HOXA10 bone marrow (n = 4), and gray bars represent WT bone marrow (n = 4); *P < .05. (C) FACS analysis of uninduced bone marrow using anti-CD71/anti-Ter119 displaying normal erythropoietic development (I, proerythroblasts; II, basophilic erythroblast; III, late basophilic and chromatophilic erythroblast; IV, orthochromatophilic erythroblast and nonerythroblast) next to a representative bone marrow from a rtTA-HA10 mouse 14 days after induction, showing a block in the erythroid development. Gate I uninduced = 0.7% ± 0.1%, rtTA-HA10 = 23.1% ± 2.3% (n = 4). (D) Bone marrow from the mice above where analyzed for early progenitors using FACS showing an accumulation of c-kit+, GMP, and MEP cells. (n = 4) (E) Mice receiving transplants of inducible HOXA10 bone marrow developed anemia with low RBC counts and low Hgb levels. (F) Spleen sections from representative WT and anemic mouse (same mouse as in panel C) stained with hematoxylin-eosin displaying the disrupted spleen structure replaced by immature erythrocytes (magnification, ×100); n = 4; *P < .05. Images were obtained using an Olympus BX50 microscope equipped with a 100×/1.25 NA objective (LR1, Lund, Sweden). Images were acquired using an Olympus C-3040 digital camera (LR1) and were processed using Iphoto 2.0.1 (LDC, Lund, Sweden).
High levels of HOXA10 block erythroid development in vivo and lead to accumulation of hematopoietic progenitors. (A) Analysis of blood from rtTA-HA10 mice 2 weeks after induction of HOXA10 expression in vivo revealed reduced levels of hemoglobin (Hgb), red blood cells (RBCs), and platelets (PLTs) (n = 6). (B) Bone marrow cells were plated in methylcellulose supporting the formation of BFU-E colonies, with 4 different concentrations of doxycycline. Colonies were scored after 6 days of culture. Shown are the numbers of BFU-E colonies/100 000 input cells. Black bars represent inducible HOXA10 bone marrow (n = 4), and gray bars represent WT bone marrow (n = 4); *P < .05. (C) FACS analysis of uninduced bone marrow using anti-CD71/anti-Ter119 displaying normal erythropoietic development (I, proerythroblasts; II, basophilic erythroblast; III, late basophilic and chromatophilic erythroblast; IV, orthochromatophilic erythroblast and nonerythroblast) next to a representative bone marrow from a rtTA-HA10 mouse 14 days after induction, showing a block in the erythroid development. Gate I uninduced = 0.7% ± 0.1%, rtTA-HA10 = 23.1% ± 2.3% (n = 4). (D) Bone marrow from the mice above where analyzed for early progenitors using FACS showing an accumulation of c-kit+, GMP, and MEP cells. (n = 4) (E) Mice receiving transplants of inducible HOXA10 bone marrow developed anemia with low RBC counts and low Hgb levels. (F) Spleen sections from representative WT and anemic mouse (same mouse as in panel C) stained with hematoxylin-eosin displaying the disrupted spleen structure replaced by immature erythrocytes (magnification, ×100); n = 4; *P < .05. Images were obtained using an Olympus BX50 microscope equipped with a 100×/1.25 NA objective (LR1, Lund, Sweden). Images were acquired using an Olympus C-3040 digital camera (LR1) and were processed using Iphoto 2.0.1 (LDC, Lund, Sweden).
Because high levels of HOXA10 blocked erythropoiesis and megakaryocyte development, we next asked whether primitive progenitors accumulate in vivo by using flow cytometry to purify common myeloid progenitors (CMPs), megakaryocyte/erythroid progenitors (MEPs), and granulocyte/macrophage progenitors (GMPs).58 No difference in total bone marrow cellularity was seen when comparing bone marrow of inducible HOXA10 and WT control mice 2 weeks after induction (data not shown), whereas the portion of c-kit+ cells in the induced mice were increased 3.2-fold (Figure 3D). Interestingly, no significant difference in the proportion of phenotypic CMPs was detected; however, the proportion of GMPs was elevated 2.5-fold (Figure 3D). Moreover, the proportion of MEPs was increased almost 5-fold, supporting the notion that the anemia and thrombocytopenia seen in vivo as well as the absence of megakaryocytes in the in vitro cultures at high concentration of doxycycline (data not shown) is caused by a differentiation block (Figure 3D).
To eliminate potential effects derived from stroma and other organs, rtTA-HA10 bone marrow cells (Ly5.2/5.2) were transplanted to lethally irradiated WT (Ly5.1/5.1) mice that received doxycycline in the drinking water immediately after transplantation. After 4 weeks of induction the recipients that had received rtTA-HA10 bone marrow began to lose weight and appeared hunch-backed. Peripheral blood analysis of the sick recipients showed a severe reduction in hemoglobin as well as in their red blood cell count, and several recipients died with severe anemia (Figure 3E). When the remaining mice were killed, we found that all mice that received rtTA-HA10 bone marrow and were given doxycycline displayed different degrees of splenomegaly, clearly correlating with the anemia (Figure S2). Histology of the enlarged spleens showed disruption of the normal spleen structure attributable to the presence of large numbers of normoblastic erythrocytes (Figure 3F). Furthermore, FACS analysis for anti-CD71/anti-Ter119 of the bone marrow and spleen in the anemic recipients recapitulated the erythroid block seen in the systemic-induced mice (data not shown). To be able to study long-term effects of HOXA10 induction, irradiated mice received transplants of wt and HOXA10-inducible bone marrow at a 1:1 ratio and were given doxycycline in the drinking water from the day of transplantation. All mice survived long term and appeared well. FACS analysis of peripheral blood at 16 weeks revealed reduced levels of CD4+ and CD8+ T cells in induced mice, verifying previously published data on retroviral gene transfer of HOXA10 to human HSCs38 (Figure S3). We hereby show that continued high HOXA10 expression blocks erythropoiesis at an intermediate stage of erythroid development, resulting in an accumulation of erythroid progenitor cells at the proerythroblast stage and that continued expression of HOXA10 hampers the T-lymphoid lineage differentiation. These data demonstrate the importance of regulated HOXA10 expression in primitive hematopoietic progenitors to allow normal lymphoid and erythroid development.
HOXA10 binds with high affinity to HLF, Dkk1, and Gata-1
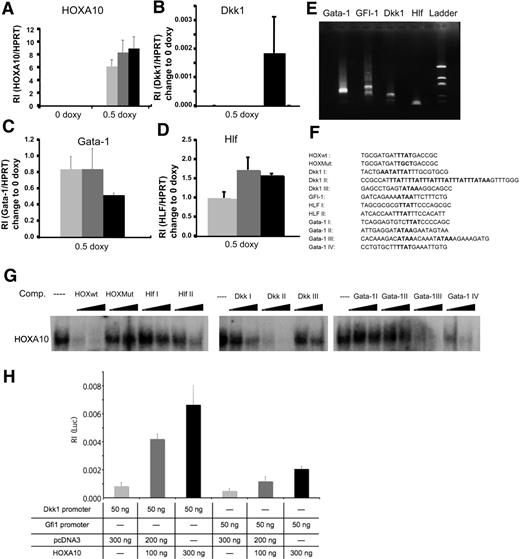
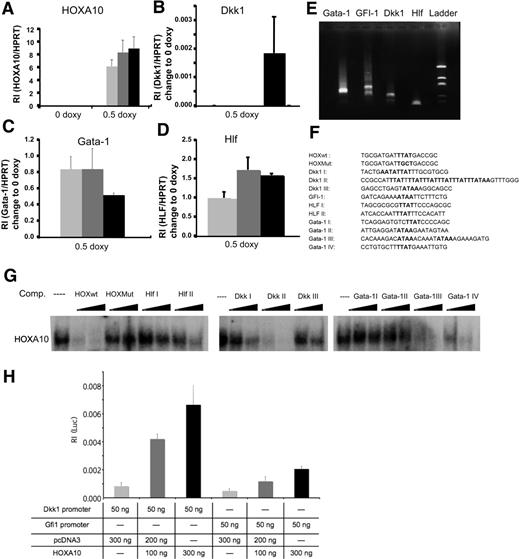
To investigate what mechanisms are involved in the phenotype caused by overexpression of HOXA10, we performed global gene expression analysis using the Affymetrix platform. We used RNA from sorted rtTA-HA10 LSK cells cultured for 72 hours with or without doxycycline. The analysis was performed by Swegene (www.swegene.com) and showed that overexpression of HOXA10 up-regulated 1214 genes in comparison to uninduced LSK cells, whereas 874 genes were down-regulated. Interestingly, we found a number of dysregulated genes that might explain the hematopoietic phenotypes generated by overexpression of HOXA10. Dickkopf 1 (Dkk1), growth factor independent-1 (Gfi-1), and hepatic leukemia factor (HLF) are genes important for stem-cell self-renewal that were all up-regulated by HOXA10 (Table 1). In addition, Gata-1, essential for the development of red blood cells and platelets, was down-regulated by HOXA10 (Table 1). We also found up-regulation of Hoxa5 and Hoxb3 by HOXA10, demonstrating cross-regulation within the Hox family (Table 1). Although the time span of 72 hours used for the gene expression analysis is unlikely to result in major changes in the cellular composition of the cultures, these experiments described above do not address whether the induced or repressed genes are directly regulated by HOXA10. Thus, to further elucidate whether Hlf, Dkk1, Gfi-1, and Gata-1 are direct targets for HOXA10 we wanted to investigate the modulation of expression shortly after HOXA10 induction. To this end, Lin− bone marrow cells were cultured for 16 hours with 0.5 μg/mL doxycycline. Subsequent Q-RT-PCR showed that Hlf, Dkk1, and Gata-1 were all significantly modulated compared with uninduced cells 16 hours after induction, suggesting that these genes rapidly respond to HOXA10 induction and that their altered expression is not a result of altered cellular composition in the cultures (Figure 4A-D). To find putative binding sites for HOXA10 in the promoter elements of the responding genes, we identified the transcriptional start sites by amplifying the 5′ end of cDNA of Hlf, Dkk1, Gfi-1, and Gata-1 from Lin− bone marrow cells. The PCR products were sequenced, and by alignment to genomic DNA sequences retrieved from NCBI were used for identification of putative promoter sequences (Figure 4E; Figure S4). This analysis indicated that the mRNAs were mainly initiated in the regions previously suggested to compose the immediate 5′ regions of the investigated genes (data not shown). Visual inspection of the genomic DNA sequence revealed that the Hlf promoter contained 2 potential binding sites aligning to the core sequence for a HOXA10 binding site (TTAT) (Figure 4F). The Dkk1 promoter contained a more complex array of potential binding sites with one single site (Dkk1 III), one duplex site (Dkk1 I), and one region composed of an array of 6 consensus-binding sites (Dkk1 II) (Figure 4F). In addition, we found one consensus site in the GFI-1 promoter, whereas the proximal Gata-1 promoter lacked consensus sequences for HOXA10 binding sites. However, investigating the sequence of intron I in the Gata-1 gene, we were able to find several potential binding sites for HOXA10 (Gata-1 I-IV) (Figure 4F). To investigate whether the putative binding sites had the ability to interact with HOXA10 in vitro, we used double-stranded unlabeled nucleotides covering these regions (Figure 4F) to compete for complex formation between a P32 -labeled known Hox binding site and HOXA10 in nuclear extracts from induced lineage-depleted cells from rtTA-HA10 transgenic mice using EMSA analysis (Figure 4G). This revealed that sites from the HLF and Dkk1 promoters and Gata-1 intron I were all able to compete for complex formation, whereas no functional binding site could be found in the Gfi-1 5′ sequence (Figure 4G). To address whether HOXA10 binding lead to the transcriptional activation of the promoters, the identified sequences from Gfi-1 and Dkk1 were cloned into a Luciferase-reporter plasmid. Luciferase assay in HeLA cells revealed that HOXA10 induced expression of the reporter gene in plasmids containing both sequences, Dkk1 8-fold and Gfi-1 4-fold as shown in Figure 4H. Taken together, these data suggest that Dkk1, Hlf, Gata-1, and Gfi-1 are direct downstream targets of HOXA10.
HOXA10 binds with high affinity to Hlf, Dkk1, and Gata-1. Lin− rtTA-HA10 bone marrow cells were cultured in 0 or 0.5 μg/mL doxycycline and the expression of (A) HOXA10, (B) Dkk1, (C) Gata-1, and (D) Hlf was quantified by Q-RT-PCR 2 hours, 6 hours, and 16 hours after induction and compared with the value of 0 μg/mL doxycycline sample at each time point; n = 3; *P < .05. (E) Shown are the products of the amplified 5′ cDNA ends from Lin− bone marrow cells for each gene that was sequenced. (F) Using the sequences from panel E, we identified possible binding sites for HOXA10 in the promoter region of Dkk1, Gfi-1, Hlf, and in the intron-1 of Gata-1. (G) EMSA showing the sequences from panel F as competitors to binding of HOXA10 in nuclear extracts from induced transgenic lineage-negative cells to a known HOXA10 probe in 100- and 300-fold excess of unlabeled duplex oligo-nucleotide. (H) Luciferase assay in HeLA cells transfected with 100 or 300 ng HOXA10 expression plasmid reveal induction of luciferase when driven by promoter regions for Dkk1 and Gfi-1. Increasing concentrations of HOXA10 expression results in increasing luciferase activity. The luciferase values were normalized to Renilla, used as an internal transfection control to assess relative intensity (RI) (n = 6). Data represent mean ± SD.
HOXA10 binds with high affinity to Hlf, Dkk1, and Gata-1. Lin− rtTA-HA10 bone marrow cells were cultured in 0 or 0.5 μg/mL doxycycline and the expression of (A) HOXA10, (B) Dkk1, (C) Gata-1, and (D) Hlf was quantified by Q-RT-PCR 2 hours, 6 hours, and 16 hours after induction and compared with the value of 0 μg/mL doxycycline sample at each time point; n = 3; *P < .05. (E) Shown are the products of the amplified 5′ cDNA ends from Lin− bone marrow cells for each gene that was sequenced. (F) Using the sequences from panel E, we identified possible binding sites for HOXA10 in the promoter region of Dkk1, Gfi-1, Hlf, and in the intron-1 of Gata-1. (G) EMSA showing the sequences from panel F as competitors to binding of HOXA10 in nuclear extracts from induced transgenic lineage-negative cells to a known HOXA10 probe in 100- and 300-fold excess of unlabeled duplex oligo-nucleotide. (H) Luciferase assay in HeLA cells transfected with 100 or 300 ng HOXA10 expression plasmid reveal induction of luciferase when driven by promoter regions for Dkk1 and Gfi-1. Increasing concentrations of HOXA10 expression results in increasing luciferase activity. The luciferase values were normalized to Renilla, used as an internal transfection control to assess relative intensity (RI) (n = 6). Data represent mean ± SD.
Discussion
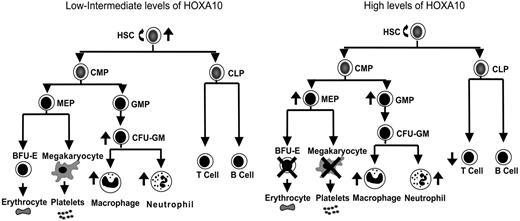
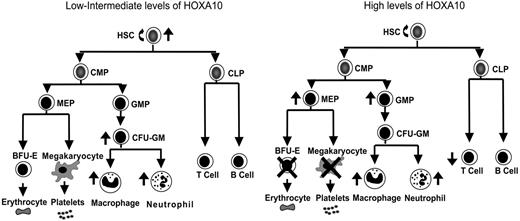
Overexpression of Homeobox transcription factors affects fate options of hematopoietic stem and progenitor cells by altering lineage choice, increasing proliferation, or even causing malignant growth.32,59,60 Because findings have suggested that the expression level of Hox genes within hematopoietic progenitor and stem cells is important for determination of fate outcomes,43,44 we developed a mouse model to ask how graded expression of HOXA10 determines proliferation and differentiation of HSCs and their progeny. We found that HOXA10 governs HSC proliferation and is a critical regulator of erythroid and megakaryocyte development. On the basis of the findings from graded expression of HOXA10 in hematopoietic cells in vitro and in vivo a model is presented in Figure 5 that demonstrates the distinct effects exerted by low or high concentrations of HOXA10 at different levels of the hematopoietic hierarchy. As the model indicates, tight regulation of HOXA10 is essential to allow normal hematopoietic development.
HOXA10 determines the fate of hematopoietic stem and progenitor cells in a concentration-dependent manner. This model explains how distinct hematopoietic cell fates are regulated by HOXA10. Intermediate levels of HOXA10 stimulate proliferation of repopulating HSCs and lead to a 15-fold increase in their repopulation capacity, whereas high concentrations of HOXA10 maintain or have a slight reduction in repopulation ability without affecting viability of HSCs. Low/intermediate concentration of HOXA10 increases the output of CFU-GM progenitors, macrophages, and neutrophils, whereas high concentrations lead to accumulation of GMPs. High concentrations of HOXA10 lead to a block in erythroid and megakaryocytic development and accumulation of MEPs. Similarly, high concentrations of HOXA10 lead to a reduction in T-cell development in vivo without affecting B-cell development. The ↑ indicates increase; ↓, decrease; ×, block.
HOXA10 determines the fate of hematopoietic stem and progenitor cells in a concentration-dependent manner. This model explains how distinct hematopoietic cell fates are regulated by HOXA10. Intermediate levels of HOXA10 stimulate proliferation of repopulating HSCs and lead to a 15-fold increase in their repopulation capacity, whereas high concentrations of HOXA10 maintain or have a slight reduction in repopulation ability without affecting viability of HSCs. Low/intermediate concentration of HOXA10 increases the output of CFU-GM progenitors, macrophages, and neutrophils, whereas high concentrations lead to accumulation of GMPs. High concentrations of HOXA10 lead to a block in erythroid and megakaryocytic development and accumulation of MEPs. Similarly, high concentrations of HOXA10 lead to a reduction in T-cell development in vivo without affecting B-cell development. The ↑ indicates increase; ↓, decrease; ×, block.
We show here for the first time that HOXA10 is an important regulator of HSC proliferation and that the level of HOXA10 is crucial to determine fate options of HSCs. Intermediate levels of HOXA10 increased the repopulation ability of HSCs compared with wt cells after in vitro culture for 13 days. The number of genuine target genes that have been identified for Hox genes is limited, and only a few HOXA10 target genes have been identified in myeloid cell lines (eg, p21 and CYBB).61,62 Here, we identified several new possible downstream target genes that may mechanistically explain how HOXA10 governs HSC proliferation. Hlf, a transcription factor that was recently found to increase the repopulating ability of human HSCs,63 was up-regulated shortly after HOXA10 induction. We found one binding site for HOXA10 close to the transcriptional start site, implicating Hlf as a direct target of HOXA10 in hematopoietic cells. Similarly, Dkk1, a negative Wnt regulator suggested to mediate expansion of adult stem cells in vitro and highly secreted by multiple myeloma cells,64–66 was up-regulated within 16 hours of HOXA10 induction. Remarkably, we found one tandem sequence of 6 TTAT repeats in the promoter region of Dkk1 with very high binding affinity to HOXA10, implicating Dkk1 as an important target gene for 5′ Hox proteins. Furthermore, HOXA10 could efficiently activate the Dkk1 promoter, proposing a direct role for HOXA10 in the regulation of Dkk1. In addition, our gene expression profiling shows that HOXA10 modulates several other genes important for the regulation of stem-cell self-renewal. Gfi-1, a zinc-finger transcriptional repressor, was found to be up-regulated by both low and high levels of HOXA10. However, we were not able to show direct binding of HOXA10 protein to the Gfi-1 promoter sequence by EMSA, whereas luciferase assay revealed a low but clear effect of HOXA10 protein binding to the Gfi-1 promoter, suggesting this gene to be a low-affinity target gene. Pbx1 was down-regulated by HOXA10, which is consistent with a report demonstrating that an antisense RNA toward Pbx1 in combination with overexpression of HOXB4 increases the self-renewal of HSCs in vitro.67
Using graded expression of HOXA10, we demonstrate that proper down-regulation of HOXA10 is essential for normal differentiation of most hematopoietic lineages. High levels of HOXA10 in vitro and in vivo promoted myeloid differentiation and reduced levels of T cells (Figures S1–S2) confirming previously published findings.38,39,41 Crooks et al68 have shown that overexpression of HOXA5 in human hematopoietic progenitors results in increased myeloid differentiation and reduced erythroid differentiation, and, interestingly, HOXA5 was one of the Hox genes found to be up-regulated by the induced HOXA10 expression in our Affymetrix study. It is therefore possible that high expression of several Hox genes could exert similar phenotypes because of highly conserved DNA binding sites. More importantly, high levels of HOXA10 totally blocked the differentiation of erythrocytes as well as megakaryocytes, resulting in an accumulation of their common progenitor (MEP). Several studies have shown that Gata-1 deficiency blocks erythropoiesis as well as megakaryopoiesis.24,27,69 Interestingly, we found that Gata-1 was down-regulated by HOXA10 within 16 hours of induction, indicating a direct effect on this gene, and direct targeting of Gata-1 could explain, at least in part, the findings observed. Furthermore, the erythropoietic block caused by overexpression of HOXA10 or Gata1 deficiency occurs at the same stage of erythropoietic development (proerythroblast).70 However, in contrast to the genes that were positively regulated we were unable to find HOXA10 binding sites in the proximal promoter region of the Gata-1 gene. Instead, we found several high-affinity binding sites in intron 1 of the Gata-1 gene, suggesting that these may mediate repressive functions by HOXA10 on Gata-1.
Because it has been demonstrated that overexpression of HOXA10 in mice can generate AML after a latency period of several months (6-12), we expected that our mice that received transplants of inducible HOXA10 bone marrow would develop AML after long-term induction of HOXA10. However, despite keeping mice on doxycycline for up to 70 weeks (6 wt and 9 inducible rtTA-HOXA10 mice), no leukemia was detected. The absence of leukemia could be due to the lack of MEIS1 expression (a known cofactor of several Hox proteins). In addition, the level of expression in the in vivo setting might not be optimal for leukemic transformation.
Our findings clearly show that different levels of HOXA10 generate different phenotypes in the hematopoietic system; however, the molecular mechanisms governing this have so far not been elucidated. It is known that most Hox monomers recognize the core motif TAAT or TTAT but with low affinity.71 Dimerization with cofactors like Meis and/or Pbx generates heterodimers increasing the DNA binding affinity, giving specificity to different Hox proteins. It has also been reported that phosphorylation of conserved tyrosine residues affects the HOX protein affinity for some DNA bindings sites.72,73 However, because several Hox proteins share the same binding element, it is possible that increasing levels of HOXA10 increases overall binding, resulting in unspecific binding ultimately affecting genes that are not normally activated/repressed. There is also a possibility that Hox proteins have high- and low-affinity binding sites, resulting in different phenotypes depending on the amount of HOXA10 present in the cells, because the probability of low-affinity binding is increased with high levels of HOXA10. In agreement with this hypothesis, we found that high levels of HOXA10 affected more genes than at low levels in the global gene expression analysis (data not shown). From gene expression analysis we selected a number of genes known to be involved in stem-cell self-renewal and lineage choice, and we found possible binding sites in the 5′ region of all selected genes found to be up-regulated (Figure 4). However, HOXA10 did not bind equally to all of them, indicating that, even though the binding sequence is only 4 base pairs long, the specificity to the confirmed Hox target genes is high, further supporting that Hox proteins have low- and high-affinity binding sites.
In summary, we find that HOXA10 governs proliferation of repopulating HSCs and is a critical regulator of erythoid and megakaryocyte development. HOXA10 appears to be a master regulator of hematopoietic stem and progenitor cells by directly regulating genes that control HSC self-renewal and erythroid development. The target genes for HOXA10 identified here are only the beginning of the quest to unravel the complex mechanisms behind the effects of Hox genes. Further advances in the identification of target genes are needed to be able to fully understand how HOXA10 can impact such diverse effects depending on timing and expression levels in hematopoietic cells from the embryonic and fetal stage through postnatal hematopoietic development. Because tight regulation of Hox and other transcription factors is essential for regulation of normal HSC proliferation, our findings may have implications for future applications in stem-cell therapy, including potential stem-cell expansion in vitro.
Authorship
Contribution: M.M., A.C.M.B., J.L., M.S., and S.K. designed the research. M.M., A.C.M.B., N.M., and M.S. performed the research. M.M., A.C.M.B., M.E., and M.S. analyzed the data. J.M.B. and A.W. generated the transgenic mice. M.M., A.C.M.B., and S.K. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
M.M. and A.C.M.B. contributed equally to this work.
Correspondence: Stefan Karlsson, Molecular Medicine and Gene Therapy, Lund, BMC A12, 221 84 Lund, Sweden; e-mail: Stefan.karlsson@med.lu.se.
The online version of this manuscript contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank Eva Gynstam for the expert animal care, Lars Palmqvist and Göran Karlsson for help with statistical calculations and evaluation of microarray data, Silja Dögg Andradottir for excellent technical assistance with the luciferase assay, and Dr R. Jaenisch for making the Rosa26-nls-rtTA mice available to us.
This work was supported by grants from The Swedish Cancer Society (S.K.), The Swedish Children's Cancer Society (S.K.), The Swedish Medical Research Council (S.K.), The European Commission (INHERINET and CONSERT) (S.K.), The Swedish Gene Therapy Program (S.K.), and a Clinical Research Award from Lund University Hospital. The Joint Program on Stem Cell Research Foundation was supported by The Juvenile Diabetes Research Foundation and the Swedish Medical Research Council. The Lund Stem Cell Center is supported by a Center of Excellence grant in life sciences from the Swedish Foundation for Strategic Research.