Abstract
Glucocorticoids are keystone drugs in the treatment of childhood acute lymphoblastic leukemia (ALL). To get more insight in signal transduction pathways involved in glucocorticoid-induced apoptosis, Affymetrix U133A GeneChips were used to identify transcriptionally regulated genes on 3 and 8 hours of prednisolone exposure in leukemic cells of 13 children as compared with nonexposed cells. Following 3 hours of exposure no significant changes in gene expression could be identified. Following 8 hours of exposure, 51 genes were differentially expressed (P < .001 and false discovery rate < 10%) with 39 genes being up-regulated (median, 2.4-fold) and 12 genes were down-regulated (median, 1.7-fold). Twenty-one of those genes have not been identified before to be transcriptionally regulated by prednisolone. Two of the 3 most highly up-regulated genes were tumor suppressor genes, that is, thioredoxin-interacting protein (TXNIP; 3.7-fold) and zinc finger and BTB domain containing 16 (ZBTB16; 8.8-fold). About 50% of the differentially expressed genes were functionally categorized in 3 major routes, namely MAPK pathways (9 genes), NF-κB signaling (11 genes), and carbohydrate metabolism (5 genes). Biologic characterization of these genes and pathways might elucidate the action of glucocorticoids in ALL cells, possibly suggesting causes of glucocorticoid resistance and new potential targets for therapy.
Introduction
Glucocorticoids such as prednisone and dexamethasone have been used extensively in the treatment of childhood acute lymphoblastic leukemia (ALL) for many years. The in vivo and in vitro prednisone response, as determined with a tetrazolium-based (MTT) toxicity assay, have been shown to correlate with each other and long-term clinical outcome in children with ALL.1–3
The classically proposed way of glucocorticoid action is that glucocorticoids bind to the intracellular glucocorticoid receptor (GR). The glucocorticoid-GR complex then translocates to the nucleus, where it binds to glucocorticoid-responsive elements (GREs), resulting in the transcriptional activation of glucocorticoid-responsive genes.4 Alternatively, the glucocorticoid-GR complex can directly bind to transcription factors such as nuclear factor-κB (NF-κB) or activator protein-1 (AP-1), resulting in so-called transrepression complexes. These complexes disrupt the transcriptional regulation of genes that are normally affected by these transcription factors.5 Depending on the cell type, transcriptional activation and repression result in immunosuppression, stress response, or induction of apoptosis.
Interestingly, glucocorticoids only induce apoptosis in lymphoid cells such as ALL, multiple myelomas, malignant lymphomas, and thymocytes, not in other tissues. Despite the major impact of glucocorticoid resistance on clinical outcome, knowledge about the signal transduction pathways leading to glucocorticoid-induced apoptosis in ALL cells is limited.6,7 Several microarray studies have been performed to determine glucocorticoid-regulated genes in leukemia.8–12 However, the cell lines (ie, immortalized cells) used in these studies do not represent an ideal model to study mechanisms involved in survival and apoptosis of primary ALL cells. Recently, the first microarray study was published in which, besides cell lines, in vivo prednisone-exposed patient ALL samples were analyzed.13 In the present study, we used freshly isolated leukemic cells of pediatric patients with ALL at initial diagnosis to identify which genes are transcriptionally regulated on in vitro prednisolone exposure.
Patients, materials, and methods
The study has been approved by the medical ethical committee of the Erasmus Medical Center Rotterdam, The Netherlands.
Patients
The study population consisted of 13 patients diagnosed with precursor-B ALL or T-ALL. Pretreatment bone marrow or peripheral blood samples were obtained after written informed consent, in accordance with the Declaration of Helsinki, from the patients and/or their legal guardians. The mononuclear cell fraction was separated by Lymphoprep density gradient centrifugation (density, 1.077 g/mL; Nycomed Pharma, Oslo, Norway). When necessary, nonleukemic cells were depleted by immunomagnetic beads to purify the samples to more than 90% of leukemic cells.
Prednisolone exposure
Leukemic cells were incubated in RPMI 1640 medium (Dutch modification without l-glutamine) supplemented with 2 mM l-glutamine, 5 μg/mL insulin, 5 μg/mL transferrin, 5 ng/mL sodium selenite, 20% heat-inactivated fetal calf serum, 100 IU/mL penicillin, 100 μg/mL streptomycin, 0.125 μg/mL fungizone, 200 μg/mL gentamycin with and without 250 μg/mL prednisolone. After 3 and 8 hours of incubation, 20 × 106 cells were removed from the culture for RNA isolation.
RNA extraction, labeling, and hybridization
Total RNA was extracted using the Trizol method (Gibco BRL, Life Technologies, Breda, The Netherlands) according to the protocol provided by the manufacturer with minor modifications.14 RNA integrity was determined using the Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA). RNA (5-15 μg) was used for subsequent production of biotinylated antisense cRNA, as described before.15 Samples with less than 10 μg labeled cRNA were excluded. Labeled cRNA was hybridized to the U133A GeneChip oligonucleotide microarray (Affymetrix, Santa Clara, CA) according to the protocol provided by the manufacturer.
Statistics
Raw gene expression values were calculated using Affymetrix Microarray Suite version 5.0.16 Data were normalized using the variance stabilization procedure (VSN) as proposed by Huber et al.17 Data distribution of unexposed controls suggested a γ- or log-normal distribution. Therefore, a generalized linear model with γ error distribution and identity link function was used to describe the effect of exposure (prednisolone exposed or nonexposed) on gene expression levels. This model was fitted to each gene separately, and the effect of exposure was evaluated via the corresponding ANOVA P values. Differentially expressed genes with respect to exposure were selected by controlling the false discovery rate (FDR; by ANOVA for each gene separately), using the procedure by Benjamini and Hochberg.18 A P value less than .001 and a FDR (adapted for multiple testing) less than 10% was considered statistically significant. Normalization and subsequent analysis were performed using R 1.9.1,19 also making use of the Bioconductor packages VSN and Multtest (www.Bioconductor.org). The fold up- or down-regulation was calculated using the formula: e (vsn value pred sample − vsn value control sample).
Results
Diagnostic samples of 13 patients with ALL were exposed to prednisolone for 3 and 8 hours. Prednisolone exposed and nonexposed (control sample, culture medium only) leukemic cells of the same patient were analyzed pairwise per patient to correct for the effect of culture in time. Paired samples could be successfully analyzed for 9 of 13 patients at 3 hours and for 10 of 13 patients at 8 hours of exposure time.
After 3 hours of prednisolone exposure, no differentially expressed probe sets were identified at P less than .001 and FDR less than 10%. However, 2 genes were differentially expressed with P less than .005 and FDR less than 20%, that is, ZFP36L2 (zinc finger protein 36, C3H type-like 2) and DSIPI (delta sleep-inducing peptide, immunoreactor; alias GILZ). Eight hours of prednisolone exposure revealed differential expression of 57 probe sets (51 unique genes and 2 expressed sequence tags [ESTs]) at P less than .001 and FDR less than 10%. As shown in Table 1, 44 probe sets (39 genes) were up-regulated (median, 2.4-fold; 25th-75th percentile, 1.8- to 3.1-fold) and 13 probe sets (12 genes) were down-regulated (median, 1.7-fold; 25th-75th percentile, 1.5- to 2-fold). Analysis at lower significance level (P < .001 and FDR < 20%) revealed that 144 probe sets were differentially expressed. In general, we observed that the same direction of prednisolone-induced changes in gene expression (down- or up-regulation) was found in the majority of patients, in all 570 observations (57 probe sets, 10 patient samples) only 6 observations (1%) were opposite from the effect seen in the remaining cases (see Table 1).
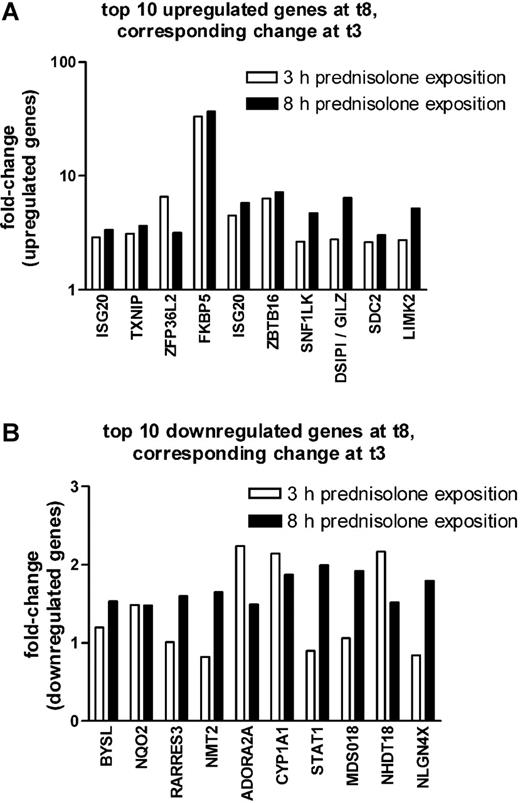
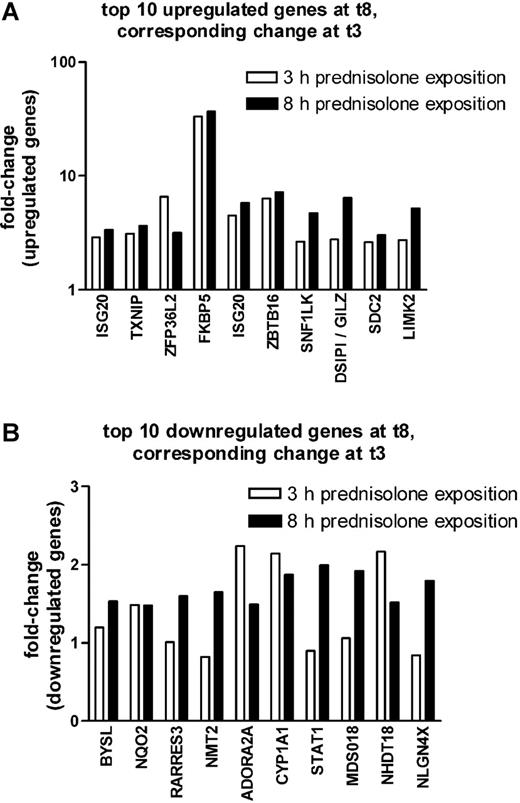
As shown in Figure 1A, the top 10 probe sets that were found to be significantly up-regulated at P less than .005 and FDR less than 10% after 8 hours of prednisolone exposure were also up-regulated after 3 hours of prednisolone exposure, albeit at a lower significance (P < .05). Seven of the 10 most significantly down-regulated genes at 8 hours of prednisolone exposure were also down-regulated after 3 hours of prednisolone exposure but with a much lower significance (P < .35) (Figure 1B).
Comparison between 3 and 8 hours on prednisolone-induced changes in gene expression in pediatric ALL. The fold up-regulation (A) and down-regulation (B) of the top 10 probe sets that were affected after 8 hours of prednisolone exposure are shown at both 3 (□) and 8 (▪) hours of exposure. The median fold change is given.
Comparison between 3 and 8 hours on prednisolone-induced changes in gene expression in pediatric ALL. The fold up-regulation (A) and down-regulation (B) of the top 10 probe sets that were affected after 8 hours of prednisolone exposure are shown at both 3 (□) and 8 (▪) hours of exposure. The median fold change is given.
Table 2 shows a summary of literature on previously identified prednisolone-responsive genes in leukemic cell lines (23 genes) and nonleukemic cells (7 genes) that were also found in the present study to be affected after 8 hours of prednisolone exposure in primary ALL cells. Five genes were found to be up-regulated in our study but down-regulated in array studies that included leukemic cell lines DUSP1 (previously reported to be up-regulated in mast cells and fibroblasts), DUSP10, PGK1, EIF2C2, and PFKP. CYP1A1 was found to be the most prominently down-regulated gene in primary ALL cells. In contrast, this gene was found to be up-regulated in human aorta endothelial cells.
Forty of the 51 genes (57 probe sets) differentially expressed on prednisolone exposure had an annotation in the Gene Ontology database. The representation of these 40 probe sets in each of the functional categories did not statistically differ from the total of U133A Gene Chip probe sets that are annotated by Gene Ontology. Besides analysis through the Gene Ontology database,22 we also analyzed the literature for putative functions of the 51 genes (Table 3). Among the 51 prednisolone responsive genes, 2 of the 3 most highly up-regulated genes are the putative tumor suppressor genes TXNIP (thioredoxin-interacting protein, alias VDUP1; 3.7-fold) and ZBTB16 (zinc finger and BTB domain containing 16, alias PLZF; 8.8-fold). Besides these 2 tumor suppressor genes involved in cell-cycle regulation, approximately 50% of the prednisolone-responsive genes could be assigned to 3 major pathways, the MAPK pathways (9 genes), NF-κB signaling pathways of gene transcription (11 genes), and carbohydrate metabolism (5 genes).
Coordinated up- and down-regulation of multiple genes on prednisolone exposure may depend on the presence of specific transcription factor binding motifs (TFBMs) in the promoter regions of genes. Forty-five of the 51 genes were annotated in the TELiS database,21 of which 35 genes were up-regulated and 10 genes were down-regulated on prednisolone exposure. Seventeen TFBMs were found to be overrepresented or underrepresented in the up-regulated genes compared with nonregulated genes (P < .01 and a FDR < 14%). Of these 17 TFBMs, 11 were overrepresented and 6 were underrepresented (Table 4). cAMP-responsive element-binding protein (CREB) is the most often represented TFBM in the up-regulated genes. We did not find a significant different frequency of GRE motifs in these 45 up-regulated genes as compared with nonregulated genes. The number of 12 down-regulated genes was too small to allow for a meaningful analysis.
Although this study contained both in vitro prednisolone-sensitive and -resistant cases, the sample size in each subgroup was too small for a statistically relevant analysis.
Discussion
Despite the clinical importance of glucocorticoids in the treatment of ALL, the genes which are transcriptionally regulated on glucocorticoid exposure in pediatric ALL and the specific (in)activation of pathways leading to glucocorticoid-induced apoptosis are unknown.
In the present study, 3 hours of prednisolone exposure did not sufficiently alter the level of gene expression to be able to (statistically) discriminate prednisolone-responsive genes in pediatric ALL. In contrast to these primary cells, in leukemic cell lines significant changes in gene expression can be observed already after 3 hours of prednisolone exposure.8,9 The same phenomenon that leukemic cell lines respond faster to a drug than corresponding primary cells was found for L-asparaginase.23 Besides the faster response, different genes were also found to be affected by these drugs in the leukemic cell lines. These studies emphasize that leukemic cell lines behave differently to drugs than primary cells and hence may not be suitable models for pharmacodynamic studies.
Exposure of pediatric ALL cells for 8 hours to prednisolone affected the expression of 57 probe sets (51 genes); 44 probe sets were up-regulated and 13 probe sets were down-regulated. FKBP5 (FK506 binding protein 5) was the most significantly up-regulated gene (35-fold). Its product, FKBP51, functions as co-chaperone molecule of the glucocorticoid receptor that affects the transport of the glucocorticoid receptor into the nucleus. The expression of this gene has been reported before to be highly glucocorticoid inducible in cell lines and primary patient cells.13,24,25 The most significantly down-regulated gene was CYP1A1 (2-fold) which is involved in drug metabolism and detoxification. Although we found only 57 probe sets significantly regulated on 8 hours of prednisolone exposure, more prednisolone-responsive genes can be identified if less-stringent cut-off levels are used for the P value and FDR. For example, at P less than .001 and FDR less than 20%, 144 probe sets are regulated on 8 hours of prednisolone exposure.
Interestingly, a gene expression profiling study has been published recently in which glucocorticoid-responsive genes were studied in both leukemic cell lines and primary cells of pediatric ALL.13 In that study other statistical considerations were made than in our study, such as a minimal change in gene expression of 1.6-fold in at least 6 of 13 studied patients. In our study, 99% of the observations were in correspondence with each other, that is, either up- or down-regulation of gene expression on prednisolone exposure in all tested cases. Despite differences in methodology, 5 of 28 identified responsive genes found in patients that were treated in vivo with prednisone (FKBP5, SOCS1, ZFP36L2, SNF1LK, and ZBTB16) were also found in our study using in vitro–exposed leukemic cells of children with ALL.
Signal transduction pathways possibly involved in glucocorticoid-induced apoptosis
Both TXNIP and ZBTB16 were found to be up-regulated following 8 hours of prednisolone exposure in the present study. TXNIP has been described as a tumor suppressor protein that induces a cell-cycle arrest on formation of a transcriptional repressor complex with ZBTB16.26 TXNIP prevents thioredoxin-mediated apoptosis signal-regulating kinase 1 (ASK1) ubiquitination and degradation27,28 and inhibits the thioredoxin radical scavenging function.29 Thus, TXNIP can act as a proapoptotic regulator. These data support a role for these genes in the induction of apoptosis on prednisolone exposure in childhood ALL, as has been suggested recently by Wang et al.30
MAP kinase pathways.
In the group of 51 prednisolone-regulated genes, 9 genes were associated with the 3 mitogen-activated protein (MAP) kinase pathways (ie, ERK, JNK and p38 MAPK). These MAP kinase pathways are involved in cell survival (ERK and JNK) and cell death (p38 MAPK) and have been reported to play critical roles in the pathogenesis of various hematologic malignancies.31,32 Miller et al33 recently showed that pharmacologic inhibition of ERK and JNK enhanced glucocorticoid-induced apoptosis, whereas inhibition of p38 MAPK activity opposed glucocorticoid-induced apoptosis in lymphoid cells. Four genes (DUSP1, DUSP10, DSIPI [alias GILZ], and SGK) that we found to be induced on prednisolone exposure are negative regulators of the MAP kinase pathways. In the literature, overexpression of DSIPI has been shown to promote apoptosis in thymocytes.34 Overexpression of DUSP1 in a precursor-B ALL cell line did not alter glucocorticoid sensitivity,35 suggesting that DUSP1 may not be essential for mediating the toxic effect of glucocorticoid in ALL cells. SGK is related to the PI3K/AKT pathway (which is linked to the ERK pathway) and has been reported to be a survival molecule.36
Because we found both positive and negative regulators of the MAPK pathways among the prednisolone-regulated genes, the net effect of these genes on cell survival needs to be addressed in functional studies in childhood ALL.
NF-κB.
There are 2 classically proposed ways of an inhibitory effect of glucocorticoids on NF-κB function. First, the glucocorticoid-GR complex may interact with NF-κB directly, thereby opposing its function. Second, glucocorticoids up-regulate IκBα (inhibitor of NF-κB α), which negatively regulates NF-κB. However, we did not find a significant up-regulation of IKBA in our study (P = .002; FDR, 30%). Other postulated mechanisms of glucocorticoid-mediated inhibition of NF-κB are the glucocorticoid-induced up-regulation of DSIPI and TNFAIP3.37,38 Both genes were identified in our study as prednisolone-responsive genes (3.35-fold and 2.12-fold, respectively; P < .001; FDR < 10%). This observation suggests that the last 2 mechanisms might be more relevant for NF-κB inhibition (and hence induction of apoptosis) than the 2 classically proposed models in childhood ALL.
Carbohydrate metabolism.
Five genes directly involved in carbohydrate metabolism are up-regulated on prednisolone exposure: IRS2, INSR, PFKP, GK, and PGK1. Activity of these 5 genes results in higher ATP levels in the cell because of a higher glucose uptake of the cell (IRS2 and INSR), a higher rate of glyconeogenesis (GK), and glycolysis (PFKP and PGK1). Interestingly, in an earlier study looking for the baseline expression of genes determining glucocorticoid resistance in primary, untreated ALL cells, we found GLUT3 and GAPDH, 2 genes involved in carbohydrate metabolism, to be overexpressed in prednisolone-resistant ALL cells.16 Moreover, the glycolytic rate of prednisolone-resistant leukemic cell lines was higher as compared with sensitive cell lines and inhibition of glycolysis by 2-deoxy-D-glucose sensitized resistant cells to prednisolone, whereas no effect on sensitive cells was found (A. Holleman, manuscript in preparation). Taken together, these studies strongly suggest an important role for carbohydrate metabolism in glucocorticoid-induced apoptosis.
Transcription factor binding motifs
The TELiS database20,21 was used to study which TFBMs were overrepresented or underrepresented in the 39 up-regulated genes (Table 4). Four TFBMs representing cAMP-responsive element-binding protein (CREB) were overrepresented in the promoter regions of the 35 up-regulated genes as compared with nonregulated genes. CREB is a ubiquitous transcription factor involved in cell proliferation and survival. Interestingly, the interaction between glucocorticoids and CREB has been reported before.39 Forskolin was shown to increase cellular cAMP levels and to promote the phosphorylation of CREB. In combination with dexamethasone, forskolin synergistically induced apoptosis in the glucocorticoid-resistant CEM-C1 lymphoid cell line, suggesting a role for CREB in glucocorticoid response.
Surprisingly, the glucocorticoid-responsive element (GRE) sequence was not overrepresented in the promoter regions of the 35 up-regulated genes, which is in line with a previous report on glucocorticoid-induced genes in 3 ALL cell lines.40 However, at least 3 of the 51 genes found to be regulated on prednisolone exposure contain GREs in the promoter regions, namely FKBP5,25,41 DSIPI,42 and SGK.43,44 The fact that the GRE TFBMs were not overexpressed in the up-regulated genes in our study might be the result of the delicate positioning of GRE-like sequences in glucocorticoid-responding genes that are not recognized in the TELiS database. Another explanation might be that some genes are regulated “directly” by glucocorticoids binding to their GREs, whereas other genes are regulated more “indirectly” by other transcription factors, which in turn are regulated by glucocorticoids.
In conclusion, we found 51 prednisolone-responsive genes in leukemic cells taken from children at initial diagnosis of ALL. Further functional research may identify which genes/pathways are essential for the glucocorticoid responsiveness of cells. Of the 51 identified glucocorticoid-responsive genes, 50% can be linked to 3 pathways, that is, cell proliferation and survival, NF-κB signaling, and glucose metabolism. Two of the up-regulated genes are tumor suppressor genes: TXNIP and ZBTB16, which is possibly related to the induction of apoptosis by glucocorticoids. Knowledge on the pathways leading to glucocorticoid-induced apoptosis is essential to develop more targeted therapy and ways to modulate glucocorticoid resistance in pediatric ALL.
Authorship
Contribution: W.J.E.T. designed and performed the research, collected and analyzed the data, and wrote the paper; M.L.d.B., J.P.P.M., and R.P. designed the research and analyzed the data; R.X.M., S.S., and P.J.v.d.S. analyzed the data; S.E.S. designed the research; and S.A.A. designed and performed the research and analyzed the data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: M. L. den Boer, Erasmus MC-Sophia Children's Hospital, Department of Pediatric Oncology/Hematology, Dr Molewaterplein 60, 3015 GJ, Rotterdam, The Netherlands; e-mail m.l.denboer@erasmusmc.nl.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This work was supported by the Center of Medical Systems Biology (CMSB) established by The Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research (NGI/NWO) (R.X.M.).