Decitabine's mechanism of action in chronic myelomonocytic leukemia remains incompletely understood. We studied the dynamics of neoplastic cell clearance during decitabine treatment (100 mg/m2 per course every 4 weeks) using quantitative monitoring of mutant alleles by pyrosequencing. Patients with chronic myelomonocytic leukemia were first screened for JAK2 and NPM1 mutations, and 3 patients with mutations were identified. Mutant allele percentages in mononuclear cell DNA were followed after treatment, along with methylation of LINE1 and 10 other genes. The clearance of mutant alleles was modest after the first cycle, despite induction of hypomethylation. Delayed substantial clearance was observed after 2 to 4 cycles that correlated with clinical response. Two patients had complete disappearance of mutant alleles and sustained clinical remissions. In another patient, mutant allele was detectable at clinical remission, which lasted for 8 months. Our data suggest a predominantly noncytotoxic mechanism of action for decitabine, leading to altered biology of the neoplastic clone and/or normal cells. This trial was registered at www.ClinicalTrials.gov as #NCT00067808.
Introduction
Decitabine has therapeutic activity in patients with myelodysplastic syndrome and chronic myelomonocytic leukemia (CMML).1,2 Whereas at least part of the mechanism of action is hypomethylation, cytotoxicity is also thought to play a role. The dynamics of neoplastic cells during treatment may be assessed with appropriate monitoring of genetic abnormalities. Wijermans et al have analyzed the dynamics of myelodysplastic syndrome cells after decitabine treatment by following cytogenetic abnormalities.3 However, this method requires a baseline cytogenetic abnormality and multiple bone marrow aspirations. Furthermore, cytogenetic analyses typically examine only 20 cells, which is not suitable for sensitive quantification. Molecular genetic abnormalities can become useful monitoring tools to overcome this issue. Pyrosequencing is a simple method to quantitatively detect nucleotide polymorphisms. The benefits of this method are simplicity and reproducibility. Here we studied the molecular dynamics of CMML in 3 patients with mutations in JAK2 or NPM1 who were treated with decitabine. The percentage of the mutant allele was quantified and stood as a molecular marker of disease response to decitabine.
Methods
Peripheral blood samples were collected after informed consent was obtained in accordance with the Declaration of Helsinki from 16 patients with CMML on entry to a phase 2 decitabine study, where patients were randomized to 1 of 3 decitabine schedules: (1) 20 mg/m2 intravenously daily for 5 days, (2) 20 mg/m2 subcutaneously daily for 5 days, and (3) 10 mg/m2 intravenously daily for 10 days.2 The treatment was planned to be repeated every 4 weeks. Blood collection was scheduled on days 0, 5, 12, and 28 during the first cycle and on day 0 for the following cycles. DNA was isolated from blood samples after density gradient separation of mononuclear cells using standard phenol-chloroform extraction methods. Pretreatment samples were screened for mutations of JAK2 and NPM1 using pyrosequencing as previously reported.4,5 This research was approved by the M. D. Anderson Cancer Center Institutional Review Board.
JAK2 V617F mutation was found in 2 patients, and NPM1 mutation was detected in another patient. In patients with mutations, the proportion of mutant alleles was quantitatively determined by pyrosequencing using samples obtained during and after decitabine therapy. Promoter methylation status of 10 specific genes (C1ORF102, CDH1, CDH13, CDKN2B, ESR1, NPM2, OLIG2, PDLIM4, PGRA, and PGRB)6 that are frequently methylated in myeloid malignancies was also screened in pretreatment samples of these 3 patients using pyrosequencing.7 Methylation status of the LINE1 repetitive element2 and of the genes that showed increased methylation before treatment was followed during and after treatment.
Results and discussion
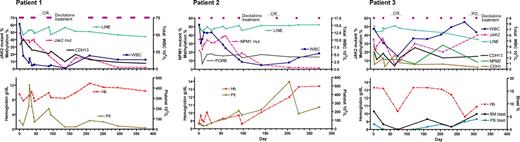
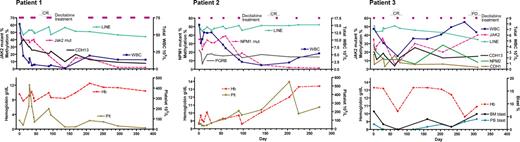
Patient 1 was a 63-year-old woman with CMML with normal karyotype and JAK2 mutation (1849G > T). At diagnosis, white blood count (WBC) was 66 × 109/L with 6% monocytes and 4% peripheral blast cells. JAK2 mutation was detected in 49% of the alleles, suggesting a heterozygous mutation. After the first cycle of decitabine (schedule 2), JAK2 mutant alleles decreased modestly to 40%; LINE1 analysis showed demethylation from 59% at baseline to 49% on day 12, and then remethylation to 56% on day 30. After the second cycle, the patient achieved a complete hematologic remission, whereas 18% of alleles still carried the JAK2 mutation. Three additional courses of decitabine cleared the mutant down to undetectable levels (< 5%). With regards to methylation changes, CDH13 was found methylated (33%) before treatment. At clinical complete response after 2 cycles of decitabine, CDH13 methylation was still 21%. Three additional cycles of treatment decreased methylation down to the level seen in normal controls (10%) (Figure 1). The platelet count deteriorated after 6 cycles of decitabine despite the clearance of JAK2 mutant alleles, and the patient was taken off the study after receiving 8 cycles of treatment. The patient eventually died of pneumonia 1.4 years after initiation of therapy.
Dynamics of DNA methylation, mutant alleles as neoplastic clone markers, and peripheral blood counts. We analyzed methylation of 10 genes at baseline in all 3 patients, and genes with increased baseline methylation were followed during and after the treatment. Patient 1: CMML with JAK2 mutation of 49% alleles before treatment, which decreased to undetectable level after 4 cycles of decitabine. CDH13 methylation was present at initiation of therapy and decreased after decitabine. Patient 2: CMML with NPM1 mutation of 35% alleles before treatment, which decreased to undetectable levels after 4 cycles. PGRB methylation followed the dynamics of LINE1 methylation after decitabine. Patient 3: CMML with JAK2 mutation of 35% alleles before treatment, which decreased to 6% after 2 cycles of decitabine and then increased again. CDH1, CDH13, and NPM2 methylation was present at initiation of therapy and decreased after decitabine. Cycles of decitabine treatment are indicated as purple rectangles above the graphs.
Dynamics of DNA methylation, mutant alleles as neoplastic clone markers, and peripheral blood counts. We analyzed methylation of 10 genes at baseline in all 3 patients, and genes with increased baseline methylation were followed during and after the treatment. Patient 1: CMML with JAK2 mutation of 49% alleles before treatment, which decreased to undetectable level after 4 cycles of decitabine. CDH13 methylation was present at initiation of therapy and decreased after decitabine. Patient 2: CMML with NPM1 mutation of 35% alleles before treatment, which decreased to undetectable levels after 4 cycles. PGRB methylation followed the dynamics of LINE1 methylation after decitabine. Patient 3: CMML with JAK2 mutation of 35% alleles before treatment, which decreased to 6% after 2 cycles of decitabine and then increased again. CDH1, CDH13, and NPM2 methylation was present at initiation of therapy and decreased after decitabine. Cycles of decitabine treatment are indicated as purple rectangles above the graphs.
Patient 2 was a 78-year-old man with CMML with normal karyotype and NPM1 mutation (960-961insTCTG). At diagnosis, WBC count was 15 × 109/L, with 13% monocytes. He had anemia and thrombocytopenia. NPM1 mutation was detected in 35% of the alleles. The first 2 cycles of decitabine (schedule 3) were associated with the typical hypomethylation induction but with a minimal change in the mutant allele percentage (from 35% before treatment to 38% after 2 cycles). This patient achieved a complete hematologic response after 4 courses of decitabine coincident with marked clearance of mutant alleles to barely detectable level (5%). After 2 additional courses of decitabine, the mutant allele was no longer detectable. PGRB was found methylated at baseline (25%). Although transient demethylation of PGRB occurred after the first cycle, remethylation was observed at day 0 of the next cycle. Overall, methylation of PGRB in this 78-year-old patient followed the dynamics of LINE1 methylation, suggesting that PGRB was moderately methylated also in normal cells. The patient remains in complete remission at 2.8 years after initiation of therapy.
Patient 3 was a 55-year-old man with CMML with normal karyotype and JAK2 mutation (1849G > T). At diagnosis, WBC was 7 × 109/L, with 16% monocytes. JAK2 mutation was detected in 35% of the alleles. LINE1 hypomethylation was induced after the first cycle (schedule 2) with transient decrease of JAK2 mutant alleles to 20%. The second cycle of decitabine was associated with marked clearance of the mutant cells (6% mutant alleles), and the patient achieved complete remission evidenced by disappearance of monocytosis and normal platelet count. Subsequently, the mutant JAK2 allele started rising again (Figure 1), although the patient clinically remained in complete remission. The disease overtly progressed after 8 courses; the spleen acutely enlarged and bone marrow blast count increased to 6%. Given otherwise stable condition, this patient received one more course of decitabine and 4 weeks later underwent splenectomy, which confirmed the presence of CMML in the spleen. The patient left our institution after splenectomy to receive supportive care at a local hospital. CDH1, CDH13, and NPM2 were found methylated at baseline. CDH1 and NPM2 showed demethylation after 3 courses (from 17% to 5% and from 28% to 6%, respectively), and the level of methylation remained low until the obvious progression of the disease, except for one point of transiently increased NPM2 methylation. The degree of CDH13 methylation was 20% at baseline. Its methylation dynamics essentially followed changes of JAK2 mutant allele, suggesting the presence of methylation in malignant cells.
In conclusion, we showed that clearance of neoplastic cells after decitabine therapy in CMML was very modest after one cycle despite induction of LINE1 hypomethylation. Subsequent cycles were associated with similar LINE1 hypomethylation dynamics, but a marked delayed clearance of the mutant clones was observed, coincident with clinical remission. Thus, global hypomethylation precedes clonal elimination and clinical responses. The paucity of genes hypermethylated in these cases makes it difficult to comment on the importance of tumor-suppressor gene demethylation. Nevertheless, demethylation dynamics of CDH13 gene temporally coincided with mutant clone elimination. Whole genome analysis of DNA methylation by microarrays or deep sequencing is needed to uncover more gene-specific methylation changes occurring after decitabine treatment. Our data are most consistent with an initial modest cytotoxic effect of decitabine followed by a delayed and complete clearance of the clone, the dynamics of which suggest a noncytotoxic mechanism. Possibilities for this delayed action include altered biology of the neoplastic clone (senescence induction, effects on stem cell renewal), induction of an immune response8 against CMML clone, or effects on normal stem cells. Further investigations should focus on these mechanisms to improve the results seen with decitabine.
Presented in part at the 48th Annual Meeting of the American Society of Hematology, Orlando, FL, December 9-12, 2006.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by Leukemia Specialized Program of Research Excellence (SPORE; grant P50CA100632) from the National Institutes of Health and the Department of Defense (grant W81XWH-05-1-0535). Y.O. is a recipient of an American Society of Clinical Oncology Young Investigator Award 2005–2006. J.-P.J.I. is an American Cancer Society Clinical Research Professor.
National Institutes of Health
Authorship
Contribution: Y.O. and J.J. designed and performed research, analyzed data, and wrote the paper; L.S. performed research; H.M.K. was the principal investigator of the clinical trial and helped analyze the clinical data; J.-P.J.I. supervised all aspects of the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jean-Pierre J. Issa, Department of Leukemia, University of Texas M. D. Anderson Cancer Center, 1515 Holcombe Blvd, Box 428, Houston, TX 77030; e-mail: jpissa@mdanderson.org.
References
Author notes
Y.O. and J.J. contributed equally to this study.