Abstract
To determine whether aberrantly activated tyrosine kinases other than FLT3 and c-KIT contribute to acute myeloid leukemia (AML) pathogenesis, we used high-throughput (HT) DNA sequence ana-lysis to screen exons encoding the activation loop and juxtamembrane domains of 85 tyrosine kinase genes in 188 AML patients without FLT3 or c-KIT mutations. The screen identified 30 nonsynonymous sequence variations in 22 different kinases not previously reported in single-nucleotide polymorphism (SNP) databases. These included a novel FLT3 activating allele and a previously described activating mutation in MET (METT1010I). The majority of novel sequence variants were stably expressed in factor-dependent Ba/F3 cells. Apart from one FLT3 allele, none of the novel variants showed constitutive phosphorylation by immunoblot analysis and none transformed Ba/F3 cells to factor-independent growth. These findings indicate the majority of these alleles are not potent tyrosine kinase activators in this cellular context and that a significant proportion of nonsynonymous sequence variants identified in HT DNA sequencing screens may not have functional significance. Although some sequence variants may represent SNPs, these data are consistent with recent reports that a significant fraction of such sequence variants are “passenger” rather than “driver” alleles and underscore the importance of functional assessment of candidate disease alleles.
Introduction
Most adult patients with acute myeloid leukemia (AML) are treated with empirically derived cytotoxic chemotherapy, and succumb to complications of their disease or therapy. Tyrosine kinases are key regulators of cellular proliferation, survival, and differentiation. Mutations that result in constitutive kinase activity have been identified in a wide spectrum of human tumors, most commonly in hematologic malignancies.1 Aberrant kinase activation can occur through several mechanisms including point mutations, insertions/deletions, and translocation.2 Mutant tyrosine kinases (TKs) have been validated as therapeutic targets in cancer, first in BCR-ABL–positive chronic myeloid leukemia (CML),3 and subsequently in gastrointestinal stromal cell tumors with mutations in c-KIT,4 and in non–small cell lung cancers associated with mutations in EGFR.5-7 In AML, approximately 30% of patients have activating mutations in FLT3,8 and FLT3 is actively being pursued as a therapeutic target.9
Biochemical and genetic data demonstrate that activation of signal transduction is central to the pathogenesis of AML. For example, the transcription factor STAT5, an important downstream effector of tyrosine kinase signaling, is constitutively phosphorylated in 70% of samples from patients with AML.10,11 In nonmalignant cells, STAT5 is normally phosphorylated upon activation of the JAK family of kinases or upon ligand-mediated activation of receptor tyrosine kinases; however, dysregulated tyrosine kinase activity can lead to constitutive STAT5 phosphorylation. In samples from AML patients in which STAT5 activation can be detected, approximately half have known activating mutations in FLT3.10 The mechanism of STAT5 activation in the remaining cases is not known. Furthermore, there is convincing evidence that the PI3K/AKT and RAS/MAPK pathways are often constitutively activated in AML.12,13 In some cases, this can be attributed to gain-of-function alleles in RAS,14 and less frequently in components of the PI3K/AKT pathway.15 Collectively, these observations suggest the hypothesis that as-yet-unidentified alleles in upstream tyrosine kinases might be responsible for constitutive activation of signal transduction pathways that would confer a proliferative and/or survival advantage to leukemic blasts.
The majority of activating mutations in tyrosine kinases associated with human cancer are gain of function alleles that reside in the activation loop (AL) of receptor or nonreceptor tyrosine kinases, or presumed loss-of-function alleles in autoinhibitory domains, including the juxtamembrane (JM) domain of receptor tyrosine kinases, and the JH2 domain of Janus kinase family members. For example, point mutations and deletions that result in constitutive tyrosine kinase activation have been identified in the ALs of FLT3,16,17 c-KIT,18,19 or EGFR.5,6 In addition, mutations in the JM domain of receptor tyrosine kinases, including internal tandem duplications in the case of FLT3 or KIT,20 or point mutations, as has recently been described for FLT3,21,22 result in constitutive kinase activity and in hematopoietic transformation. These mutant tyrosine kinases have potential as therapeutic targets in treatment of AML, and there are several small molecule FLT3 inhibitors currently being evaluated in clinical trials.23-25 To identify other disease alleles involving tyrosine kinases, we used a HT DNA sequencing strategy.
HT DNA sequence analysis of the tyrosine kinome has been used with some success to screen for novel mutations in different epithelial tumors,5,26 and to identify the JAK2V617F allele in myeloproliferative disorders and in AML.27,28 Despite these advances, the majority of HT DNA sequencing screens thus far result in the compilation of a list of validated nonsynonymous sequence variants that have promise as disease alleles, but that have not been functionally validated. To obtain insight into the specificity and sensitivity of HT DNA sequencing screens to identify functional disease alleles, we prospectively cloned the majority of nonsynonymous sequence variants identified in our screen. We then used a Ba/F3 cell–based screen for constitutive tyrosine kinase activation that has been validated for all receptor and nonreceptor tyrosine kinase alleles identified thus far in human hematologic malignancies, including BCR-ABL, TEL-ABL, TEL-PDGFRB, TEL-JAK2, FLT3-ITD, FLT3 AL mutations, FIP1L1-PDGFRA, a spectrum of KIT alleles, NPM/ALK, and ZNF198-FGFR1.29-40 This functional analysis of putative disease alleles has heightened significance in the context of recent reports providing statistical support for the hypothesis that the majority of nonsynonymous sequence variations may be functionless “passengers” in clonally derived tumors rather than “drivers” of disease phenotype.41,42
Methods
Approval was obtained from the institutional review boards of Oregon Health and Science Cancer Institute (Portland, OR), Harvard Medical School (Boston, MA), and Inserm, Hopital Necker (Paris, France) for research use of deidentified, archived patient samples.
Patient sample collection
Informed consent was obtained in accordance with the Declaration of Helsinki from all patients. Two-hundred sixty-three peripheral blood or marrow samples were obtained from patients with AML. Genomic DNA was prepared from peripheral blood or bone marrow aspirate specimens using the QIAamp DNA Blood Maxi Kit (Qiagen, Hilden, Germany) as previously described.28 KIT and FLT3 mutational analysis was performed as previously described,43,44 and cases with known FLT3-ITD, FLT3-AL, or c-KIT mutations were excluded from tyrosine kinome sequence analysis.
Tyrosine kinase resequencing
External gene-specific primers and internal M13-appended gene-specific primers were designed to amplify and sequence exons encoding the ALs of 85 of the 90 tyrosine kinases (with the exception of AATK, DKFZp434C1418, STYK1, LMTK2, and LMTK3) and the JM regions of receptor tyrosine kinases (with the exception of ERBB4) as previously described or as listed in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).5,27 DNA samples were amplified using whole genome amplification (WGA),45 and 10 ng WGA DNA was used in polymerase chain reaction (PCR) reactions. PCR products were purified (SPRI; Agencourt Bioscience, Beverly MA), followed by sequencing and sequence detection on an ABI Prism 3730 DNA Analyzer (Applied Biosystems, Foster City, CA). Sequence analysis of unidirectional or bidirectional sequence traces was performed using Mutation Surveyor (Softgenetics, State College, PA). Candidate mutations were reamplified and bidirectionally sequenced from the original (non-WGA) DNA sample for independent verification. In some cases, the exons encoding the AL and/or JM domains of tyrosine kinases also encoded residues in adjacent domains, and nonsynonymous variants in these adjacent domains were also validated and subjected to functional assessment.
Generation of kinase mutants
For TEK, FES, and DDR1, mutations were introduced into the parental vector using the QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). Following mutagenesis, the coding regions of each gene were ligated into the EcoRI site of pSRα. All additional kinases were cloned using the Gateway recombination system (Invitrogen, Carlsbad, CA) into the pDONR201 vector, and point mutations were introduced using the QuikChange II XL site-directed mutagenesis kit (Stratagene). The wild-type and mutated cDNA clones were then recombined into a MSCV-Neo Gateway destination clone using an LR Clonase reaction as previously described.46
Cell culture and generation of stable cell lines
293T cells were grown in DMEM with 10% fetal calf serum, and BaF3 cells were maintained in RPMI-1640 supplemented with 10% fetal calf serum and 10% WEHI-conditioned medium as a source of IL-3. Transient transfection of 293T cells and generation of retroviral supernatant were performed as previously described.38 For TEK, FES, and DDR1, 10 μg plasmid DNA representing wild-type and mutant versions of each gene was transfected into 107 Ba/F3 cells via electroporation followed by G418 selection (1 mg/mL) for 4 weeks. For all other tyrosine kinase mutants, Ba/F3 cells were transduced with MSCV-Neo wild-type and mutated kinase retroviral supernatants followed by G418 selection (1 mg/mL) for 4 weeks.
Immunoblot analysis
For each Ba/F3 line expressing the wild-type and mutated kinase of interest, immunoblot analysis of whole-cell lysates was performed to document overexpression of the constructs of interest. For DDR1, TEK, and FES, phosphotyrosine blotting was performed to assess for kinase autophosphorylation when expressed in 293T cells and in Ba/F3 cells. Ba/F3 cells expressing wild-type and mutated kinases were cultured in RPMI containing 0.1% bovine serum albumin in the absence of cytokines for 16 hours, followed by lysis in 3% SDS, 75 mM Tris (pH 6.8), 15% glycerol, and 8% β-mercaptoethanol. Cell lysates were boiled for 5 minutes and separated by polyacrylamide gel electrophoresis. Proteins were transferred to PVDF membranes and immunoblotted for phosphotyrosine (4G10), β-actin, and the kinase of interest.
Assessment of IL-3 independence and hypersensitivity
Following selection with G418 (1 mg/mL) and Western blot analysis to document overexpression of the construct of interest, Ba/F3 cells stably expressing wild-type and mutated kinases were washed 3 times with IL-3–free media and then cultured in the absence of IL-3, and the number of viable cells was assessed daily by trypan blue exclusion. For DDR1, TEK, and FES, Ba/F3 cells stably expressing wild-type and mutated constructs were washed 3 times with IL-3–free media and plated into 96-well plates at 3000 cells per well in media containing log10 serial dilutions of IL-3 ranging from 1 ng/mL to 10−5 ng/mL and in the absence of IL-3. MTS assays were carried out at days 3 and 5 after plating for determination of cellular proliferation in the presence of decreasing IL-3 concentration.
Results
DNA sequence analysis
To assess genetic variability and to identify potentially oncogenic sequence variants in AML, we sequenced the ALs and JM domains of all known tyrosine kinase genes in 188 patients with AML. Two-hundred sixty-three patients were initially screened for c-KIT and FLT3 mutations. Of these, 45 patients contained FLT3-ITD mutations, 8 samples contained FLT3-AL mutations, and 4 samples contained c-KIT-AL mutations, and these were mutually exclusive in consonance with previous reports suggesting functional redundancy of these alleles.47,48 Samples with known FLT3 or c-KIT mutations were excluded from further analysis, and 188 of the remaining 206 samples for which there was adequate quality and quantity of DNA for the HT DNA sequencing platform were analyzed.
Two-hundred seventeen nonsynonymous sequence variations in 58 different genes were identified in the primary screen. Comparison with the NCBI single nucleotide polymorphism (SNP) database49 identified 23 nonsynonymous variants that were not assessed further. The remaining 194 sequence variants in 54 different genes were validated by repeat PCR amplification and sequencing of the amplified and source material that had not been whole genome amplified. Thirty variants in 22 different genes were validated with independent amplification and sequencing. The primary sequence variation was not observed in the 164 remaining candidate variants, suggesting these were false positive results due to WGA errors, PCR amplification errors, sequencing/sequence detection artifacts, or analysis errors. The majority of false-positive mutations likely represented sequencing artifacts or sequence analysis errors. This was especially true where unidirectional sequence screening was used, and the validation rate was approximately 10%. In contrast, the validation rate for mutations identified using first-pass bidirectional screening was approximately 40%, which is consistent with the previous literature.50 These findings illustrate the importance of independent, bidirectional confirmation of candidate mutations.
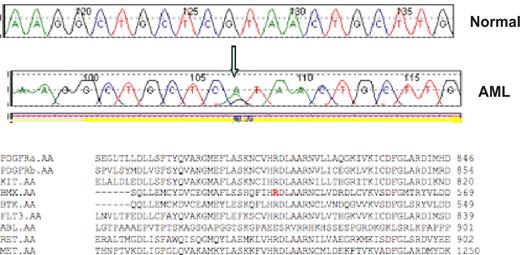
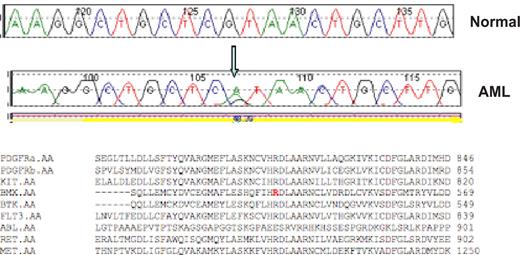
Validated, nonsynonymous candidate mutations that may contribute to leukemogenesis are shown in Table 1, and a representative sequence trace is shown in Figure 1. The majority of these candidate mutations (Table 1) were located in or adjacent to the AL (11 mutations) or in the JM domain (11 mutations). An additional 8 nonsynonymous mutations were in the transmembrane domain or in kinase domain residues outside the AL (Table 2). The only previously reported allele identified in our screen was a point mutation in the JM domain of the MET tyrosine kinase (T1010I) that had previously been identified in small cell lung cancer51 and in gastric cancer.52 None of the novel mutations identified in our screen were present in the COSMIC mutation database.53
Representative sequence trace. A representative reference (normal) and AML sequence trace comparison are shown for the BMX R540H mutation. The arrow notes the guanine to adenine substitution that results in an arginine to histidine substitution at codon 540. Protein alignment demonstrates this residue is highly conserved within the tyrosine kinase family.
Representative sequence trace. A representative reference (normal) and AML sequence trace comparison are shown for the BMX R540H mutation. The arrow notes the guanine to adenine substitution that results in an arginine to histidine substitution at codon 540. Protein alignment demonstrates this residue is highly conserved within the tyrosine kinase family.
Stably expressed nonsynonymous sequence variants do not alter cellular phosphotyrosine content
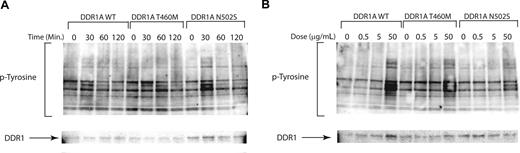
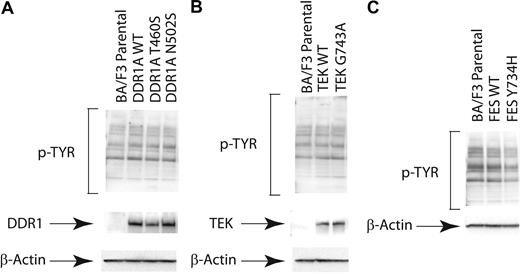
We first assessed the effects of stable expression of selected nonsynonymous sequence variants on global phosphotyrosine content of Ba/F3 cells in the absence of IL-3, with representative examples shown for the DDR1, TEK, and FES wild-type and mutant tyrosine kinases (Figure 2). Expression of each of the respective mutated and wild-type alleles was confirmed, but there was no appreciable alteration in global phosphotyrosine content in stable transduced cell lines. While subtle effects on downstream signaling molecules may have been missed, these results contrast with those of known constitutively activated tyrosine kinases, such as BCR-ABL,54 that result in dramatic increases in phosphotyrosine content. These findings suggest that these alleles are not constitutively activated in this cellular context.
Expression of nonsynonymous sequence variants does not alter cellular phosphotyrosine content. Evaluation of phosphotyrosine levels after expression of mutant DDR1, TEK, and FES. Ba/F3 cells were stably transfected. Cells were serum starved overnight and subjected to immunoblot analysis for phosphotyrosine (4G10) as well as total (A) DDR1, (B) TEK, or (C) FES.
Expression of nonsynonymous sequence variants does not alter cellular phosphotyrosine content. Evaluation of phosphotyrosine levels after expression of mutant DDR1, TEK, and FES. Ba/F3 cells were stably transfected. Cells were serum starved overnight and subjected to immunoblot analysis for phosphotyrosine (4G10) as well as total (A) DDR1, (B) TEK, or (C) FES.
Nonsynonymous sequence variations do not alter sensitivity to cognate ligand for receptor tyrosine kinases
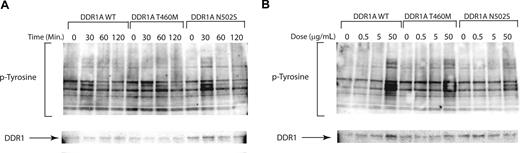
Activating mutations in receptor tyrosine kinases, such as FLT3, may be potentiated by engagement of ligand. We next tested for hypersensitivity of mutated receptor tyrosine kinases for which ligand is known and available, including collagen as a ligand for the DDR1 receptor tyrosine kinase. We tested for prolonged kinase activation following ligand engagement and for activation of tyrosine kinase at lower ligand concentrations by candidate mutant alleles. Ba/F3 cells expressing either wild-type or mutant DDR1 proteins showed the same phosphotyrosine levels at 30 minutes after stimulation with 50 μg collagen/mL. Furthermore, we did not observe prolonged activation of signal transduction in cells expressing mutant DDR1 constructs compared with wild-type DDR1 (Figure 3A), nor did we observe activation of signaling pathways at lower collagen concentrations in DDR1 mutant clones compared with wild type (Figure 3B). Taken together, these data indicate that nonsynonymous sequence variants do not confer hypersensitivity to cognate ligand in this cellular context.
Nonsynonymous sequence variations do not alter sensitivity to cognate ligands. Collagen stimulation of DDR1 mutants for evaluation of ligand hypersensitivity. Ba/F3 cells stably transfected with wild-type and mutant DDR1 as in Figure 2. Cells were serum starved overnight at which time they were collagen stimulated with (A) a time course or (B) a dose response for the indicated times and doses. Cell lysates were subjected to immunoblot analysis for phosphotyrosine (4G10) and total DDR1.
Nonsynonymous sequence variations do not alter sensitivity to cognate ligands. Collagen stimulation of DDR1 mutants for evaluation of ligand hypersensitivity. Ba/F3 cells stably transfected with wild-type and mutant DDR1 as in Figure 2. Cells were serum starved overnight at which time they were collagen stimulated with (A) a time course or (B) a dose response for the indicated times and doses. Cell lysates were subjected to immunoblot analysis for phosphotyrosine (4G10) and total DDR1.
In vitro evaluation of putative oncogenic mutations
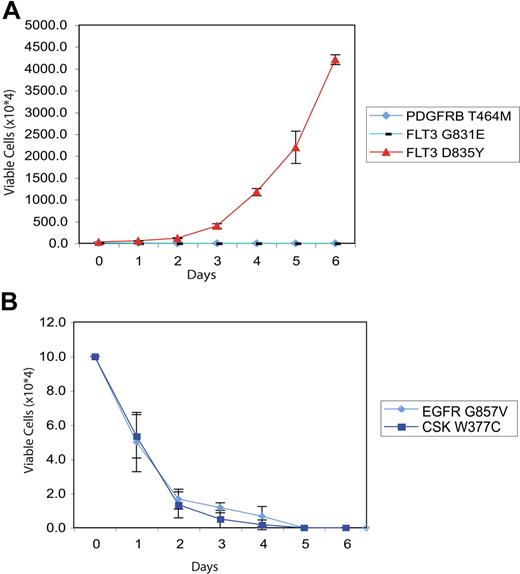
We next examined the ability of the majority of nonsynonymous alleles identified in our HT screen to transform the hematopoietic cell line Ba/F3 to factor-independent growth, a property conferred by a broad spectrum of leukemogenic tyrosine kinase alleles. We generated stable Ba/F3 cells expressing wild-type or mutant alleles for the majority of kinases listed in Tables 1 and 2, and assessed the ability of Ba/F3 lines expressing the different mutated kinases to proliferate in the absence of IL-3. Sequence variants were prioritized for evaluation based on the probability of representing a functional alteration (activation loop, clustering with the juxtamembrane region) and the availability of cDNA constructs. As a control for IL-3 independence and for IL-3 dependence, respectively, Ba/F3 cells expressing a constitutively active FLT3-D835Y mutant and parental Ba/F3 cells were also cultured in the absence of cytokines, and these lines exhibited IL-3–independent and IL-3–dependent growth as previously described.33,36 We did not observe proliferation of Ba/F3 cells expressing the tested mutated kinases in the absence of IL-3 (representative data shown in Figures 4A-C and 5A,B). The only novel allele identified in our screen that demonstrated in vitro transforming ability was FLT3R834Q, which transformed Ba/F3 cells to cytokine-independent growth.55
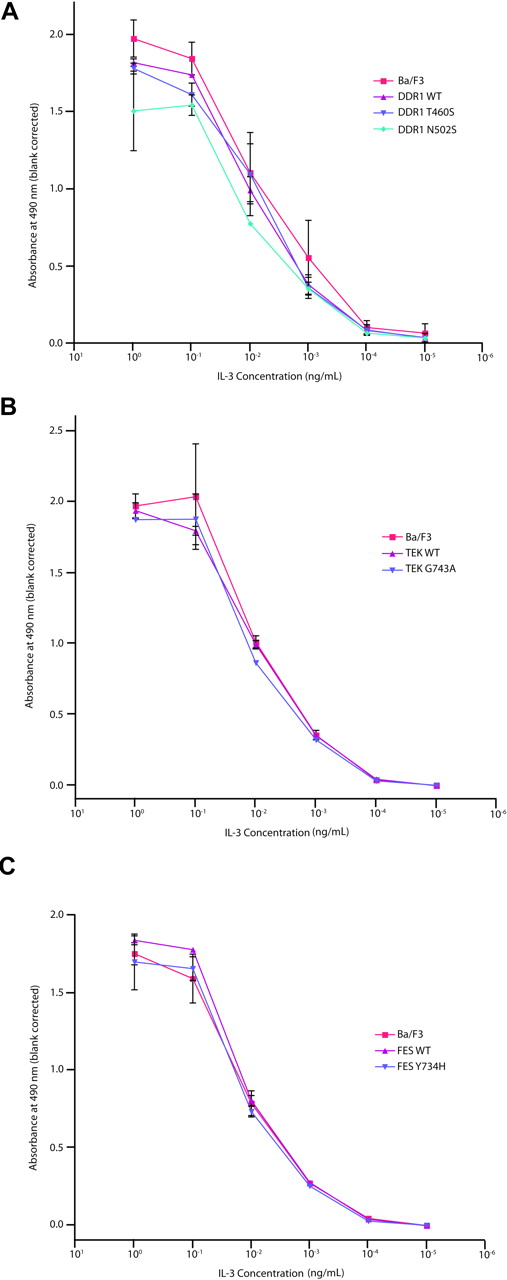
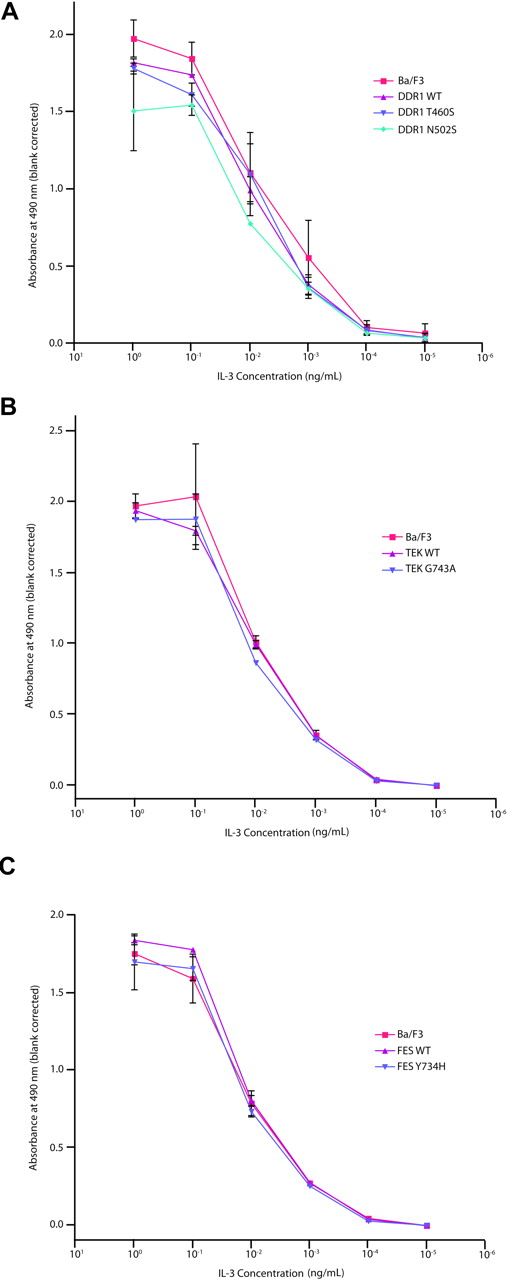
Evaluation of DDR1, TEK, and FES mutations in Ba/F3 WEHI independence assay. Ba/F3 cells were transfected with constructs expressing wild-type or mutant DDR1 (A), TEK (B), or FES (C). Following 4 weeks of neomycin selection, stably transfected cells were washed with WEHI-free media and plated in media containing 10-fold serial dilutions of IL-3, ranging from 1 ng/mL to 10−5 ng/mL. Cells were subjected to an MTS assay at day 3 after plating for determination of total viable cells. Values represent mean plus or minus SEM.
Evaluation of DDR1, TEK, and FES mutations in Ba/F3 WEHI independence assay. Ba/F3 cells were transfected with constructs expressing wild-type or mutant DDR1 (A), TEK (B), or FES (C). Following 4 weeks of neomycin selection, stably transfected cells were washed with WEHI-free media and plated in media containing 10-fold serial dilutions of IL-3, ranging from 1 ng/mL to 10−5 ng/mL. Cells were subjected to an MTS assay at day 3 after plating for determination of total viable cells. Values represent mean plus or minus SEM.
Evaluation of PDGFRB, FLT3, CSK, and EGFR mutations in IL-3 independence assay. Ba/F3 cells were transfected with constructs expressing mutant PDGFRB/FLT3 (A) or CSK/EGFR (B). Following 4 weeks of neomycin selection, stably transfected cells plated in media in the absence of exogenous cytokines. The number of viable cells was assessed by trypan blue exclusion. As a positive control, Ba/F3 cells expressing the constitutively active FLT3-D835Y allele were plated in the absence of IL-3 in panel A to demonstrate IL-3–independent growth. Values represent mean plus or minus SEM.
Evaluation of PDGFRB, FLT3, CSK, and EGFR mutations in IL-3 independence assay. Ba/F3 cells were transfected with constructs expressing mutant PDGFRB/FLT3 (A) or CSK/EGFR (B). Following 4 weeks of neomycin selection, stably transfected cells plated in media in the absence of exogenous cytokines. The number of viable cells was assessed by trypan blue exclusion. As a positive control, Ba/F3 cells expressing the constitutively active FLT3-D835Y allele were plated in the absence of IL-3 in panel A to demonstrate IL-3–independent growth. Values represent mean plus or minus SEM.
Assessment of cytokine hypersensitivity of DDR1, TEK, and FES mutants
We next assessed whether the nonsynonymous alleles might confer hypersensitivity to hematopoietic cytokines. This approach is based in part on the observation that several activating alleles in hematopoietic malignancies confer hypersensitivity to hematopoietic cytokines, including PTPN11 and JAK2V617F.27,56,57 We therefore evaluated a subset of the alleles identified in the primary screen for their ability to confer a cytokine hypersensitivity phenotype. Ba/F3 cells stably expressing wild-type or mutated DDR1, TEK, and FES, as well as parental Ba/F3 cells, were grown at concentrations of IL-3 ranging from 1 ng/mL to 10−5 ng/mL, to determine whether these mutant alleles conferred a cytokine hypersensitivity phenotype. For each DDR1, TEK, or FES mutant assessed, we observed no evidence of cytokine hypersensitivity (Figure 4), further suggesting these alleles do not contribute to the leukemogenic process. Collectively, these data indicate that the nonsynonymous sequence variants identified in our screen are not likely to have a significant functional contribution to the molecular pathogenesis of AML.
Discussion
Although the identification of recurrent cytogenetic abnormalities has allowed for the classification of AML into distinct subgroups,58-60 sequence analysis of selected candidate genes has identified recurrent point mutations in AML that complement the large number of pathogenetic gene rearrangements identified by cloning of chromosomal translocation breakpoints, including FLT3,8 KIT,18 RAS,14 JAK3,61 and others62 that contribute to the leukemogenic process. Because tyrosine kinases are attractive candidates for molecularly targeted therapy with small molecule inhibitors, and only a fraction of AML cases have known mutations in tyrosine kinases, we used HT sequence analysis to screen for novel activating alleles of tyrosine kinases. We identified 30 nonsynonymous sequence variants in 22 different tyrosine kinases. These included the T1010I amino acid substitution in the JM domain of MET in 2 patients that had previously been identified in small cell lung cancer51 and in breast cancer52 as well as a novel FLT3-AL activating mutation.
In addition to these novel and previously described activating alleles, we identified 30 novel nonsynonymous alleles in 21 different tyrosine kinases. Although HT screens of the tyrosine kinome have been performed in a spectrum of human malignancies, including colorectal cancer,26 lung cancer,5 breast cancer,63 and glioblastoma multiforme,64 this study differs from previous reports in 2 important aspects. First, previous reports involved the analysis of the entire coding region of all tyrosine kinases, or of all tyrosine and serine-threonine kinases, in a relatively small (< 50) number of tumors, with secondary screening of a larger set of tumors to assess allele frequency. This approach allows for the identification of recurrent mutations with sufficient frequency to be detected in a small cohort of samples, but does not provide statistical power to enable the identification of rare (< 5%-10%) alleles that can contribute to transformation in a subset of tumors. As AL mutations in FLT3 and in KIT occur in less than 5% to 10% of cases of AML,17,18,65-67 we chose instead to screen a large set of AML samples (188) to identify less frequent disease alleles.
In addition, previous reports of mutational analysis of the tyrosine kinome resulted in the generation of a list of mutations. In these studies, functional assessment either was not performed26,63 or was performed on selected candidate alleles with a likely role in transformation,5 leaving the potential role of the majority of these alleles in malignant transformation unknown. We therefore chose to functionally assess the majority of tyrosine kinase variants identified in our screen, using in vitro assays that are commonly used to assess the transforming potential of activated tyrosine kinases. Although none of the specific alleles identified in our screen was present in the COSMIC database,53 somatic mutations have previously been identified in the majority (20/22) of these genes. We predicted that a subset of these alleles might have transforming properties, based on the observation that many of these alleles result in substitutions at highly conserved AL and JM loop residues. For example, some of the mutations we identified in the JM region of DDR1 are present near or within an isoform-dependent 37–amino acid insertion in exon 11 that encodes a “PRGPGPPTP” SH3-binding site and an “LXNPXY” autophosphorylation site that recruits the docking protein Shc,68,69 suggesting these alleles would result in release of tyrosine kinase autoinhibition and activation of signaling pathways. Similarly, many of the mutations identified in or near the kinase ALs, including BMX, FLT3, and MATK alleles, result in nonsynonymous substitutions at highly conserved residues.
Although we hypothesized that a subset of these alleles would transform hematopoietic cells, with the exception of a novel activating FLT3 allele,55 each of these alleles failed to transform Ba/F3 cells to factor-independent growth. Several explanations may account for these findings. First, transformation of Ba/F3 cells to factor-independent growth may not faithfully assess the functional implications of certain mutations, either because they require coexpression of additional proteins, as we have demonstrated for JAK2V617F,70 or because transformation is lineage specific and would not be observed in the pro-B Ba/F3 cell line. Second, if cooperativity between 2 or more mutations is required for signaling pathway activation, we would not observe hematopoietic transformation in the absence of the second allele. Third, murine systems may be inadequate for mutations identified in human cells, especially when expressed in the context of the human protein. It is also possible a subset of these alleles may contribute to transformation through loss of kinase activity, as has been demonstrated for EphB kinases in colorectal cancer.71 More likely, these nonsynonymous alleles provide further evidence that there are both “driver” and “passenger” allele in human tumors, as has been argued on a statistical basis by Greenman et al.42 Our observations of 2 novel activating alleles of FLT3 and MET in AML, but a large number of apparently nonfunctional alleles, provide functional support for this hypothesis. Greenman et al found that cancer types with high mutation prevalence originate mainly from high turnover tissues. Given the high proliferative rate of leukemia, our data are consistent with the findings of Greenman et al. Specifically, we found a nonsynonymous mutational rate of approximately 2 mutations per megabase (mB) of DNA sequenced (30 mutations in 15.7 mB of sequenced DNA; synonymous mutations were not validated and are not included in the mutational rate).
In addition, our sequencing strategy may have influenced our potential to detect activating mutations through detection bias. We chose to sequence exons encoding tyrosine kinase juxtamembrane regions and activation loops based on the observation that the majority of known activating mutations occur in these regions.5,6,16-22 However, activating mutations outside these regions have been detected in several kinases, including c-KIT,1,72 and would not have been identified in this screen. Recently, high-throughput sequencing of all coding exons in FLT3 have identified a novel activating mutation in the extracellular region.55 We believe future efforts should include sequence analysis of the entire coding region of genes of interest, a strategy that will become more feasible with the development of single-molecule sequencing technologies for analysis of cancer genomes.73 Moreover, kinase overexpression and translocation1 —alterations that are not readily identified by genomic sequencing—can also contribute to the leukemogenic process. Genomic analysis of AML cDNA samples using high-throughput genomic platforms may permit more complete evaluation including expression levels and mutational status, and potentially identify translocations, which should be the subject of future investigation. Although Sanger resequencing has been used to identify oncogenic mutations in a variety of human malignancies, including AML, it is likely this approach will not identify all oncogenic mutations, particularly in AML samples with less than 30% blasts. Mutational analysis using quantitative allele-specific PCR and Sanger resequencing demonstrates that Sanger resequencing is able to detect mutations that are present in at least 20% of a heterogeneous population of clonal and nonclonal cells (Levine et al74 and D.G.G., unpublished data, 2007). Mutational analysis using parallel sequencing technologies75 offers the ability to identify mutations present in a small population of clonal cells, and may lead to the identification of oncogenic mutations in AML that are not detected using Sanger resequencing.
The results of this study have several implications for large-scale cancer sequencing projects, including The Cancer Genome Atlas Project and other similar efforts.41,42 First, these studies rely on discerning somatic mutations from inherited polymorphisms using existing SNP databases and matched normal samples, and use mutational frequency and distribution as well as structural modeling to improve the probability of identifying which alleles contribute to transformation. However, germ-line mutations can contribute to the cancer phenotype, and somatic passenger mutations cannot be distinguished from low-frequency mutations that contribute to tumorigenesis. Our results suggest that the majority of nonsynonymous sequence variants either do not contribute to malignant transformation, or the functional consequences of these mutations may be difficult to assess using standard in vitro assays. Determining the appropriate context to functionally assess the large number of candidate mutations that will be identified in The Cancer Genome Atlas Project and other genomewide screens may prove difficult and time consuming, but our data suggest these efforts will be critical to determine which mutations contribute to transformation and represent true therapeutic targets for the treatment of human malignancies.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to Jeffrey C. Lee and J. Guillermo Paez for assistance with the kinase sequencing platform, and to members of the Gilliland and Druker labs for discussions and suggestions. We are grateful to Alexis Bywater for administrative support.
This work was supported by (NIH) National Institutes of Health grants CA66996 and CA105423 (D.G.G.), HL082677 (R.L.L.), and CA113434 (B.L.), the T. J. Martell Foundation (B.J.D.), VA Merit Review Grant (M.C.H.), and the Leukemia and Lymphoma Society (D.G.G., B.J.D., and M.C.H.). D.G.G. and B.J.D. are Investigators of the Howard Hughes Medical Institute, and D.G.G. and B.J.D. are Distinguished Clinical Scientists of the Doris Duke Charitable Foundation. R.L.L. is a Young Investigator Award recipient of the American Society of Clinical Oncology, a Basic Science Fellow of the American Society of Hematology, a Clinical Scientist Development Award recipient of the Doris Duke Charitable Foundation, and a Howard Hughes Medical Institute Early Career Award Recipient. S.F. and C.S. are supported by grants from the Deutsche Forschungsgemeinschaft. J.W.T. is supported by an NIH Cancer Biology Training Grant.
National Institutes of Health
Howard Hughes Funding
Authorship
Contribution: M.M.L., R.L.L., J.W.T., D.G.G., and B.J.D. designed research and wrote the paper; M.M.L., R.L.L., J.W.T., S.F., C.S., E.P.S., G.W., H.E., C.A.E., R.B., O.A.B., J.D.G., B.L., R.M.S., and M.M. performed experiments, analyzed results, and made figures; M.C.H. supplied some patient samples; and M.W.D. helped with research design.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Brian J. Druker, Oregon Health & Science University, L592, 3181 SW Sam Jackson Park Rd, Portland, OR 97239; e-mail: drukerb@ohsu.edu; or D. Gary Gilliland, Division of Hematology, Brigham and Women's Hospital, Karp Family Research Bldg, Rm 5210, 1 Blackfan Circle, Boston, MA 02115; e-mail: ggilliland@rics.bwh.havard.edu.
References
Author notes
M.M.L. and R.L.L. contributed equally to this work.