Abstract
Multilevel genetic characterization of Waldenström macroglobulinemia (WM) is required to improve our understanding of the underlying molecular changes that lead to the initiation and progression of this disease. We performed microRNA-expression profiling of bone marrow–derived CD19+ WM cells, compared with their normal cellular counterparts and validated data by quantitative reverse-transcription–polymerase chain reaction (qRT-PCR). We identified a WM-specific microRNA signature characterized by increased expression of microRNA-363*/-206/-494/-155/-184/-542-3p, and decreased expression of microRNA-9* (ANOVA; P < .01). We found that microRNA-155 regulates proliferation and growth of WM cells in vitro and in vivo, by inhibiting MAPK/ERK, PI3/AKT, and NF-κB pathways. Potential microRNA-155 target genes were identified using gene-expression profiling and included genes involved in cell-cycle progression, adhesion, and migration. Importantly, increased expression of the 6 miRNAs significantly correlated with a poorer outcome predicted by the International Prognostic Staging System for WM. We further demonstrated that therapeutic agents commonly used in WM alter the levels of the major miRNAs identified, by inducing downmodulation of 5 increased miRNAs and up-modulation of patient-downexpressed miRNA-9*. These data indicate that microRNAs play a pivotal role in the biology of WM; represent important prognostic marker; and provide the basis for the development of new microRNA-based targeted therapies in WM.
Introduction
MicroRNAs (miRNAs) constitute a class of small, noncoding, 18- to 24-nucleotide RNAs, described for the first time in the nematode Caenorabditis elegans.1 To date, more than 300 miRNAs have been discovered in humans.2 miRNAs act as negative regulators of gene expression by binding to the 3′ untranslated region (UTR) of the target mRNAs with partial sequence complementarity and leading to translational repression.3 By repressing several target mRNAs, mature miRNAs play a pivotal role in regulating development, cell differentiation, apoptosis, and cell proliferation.4 Moreover, miRNAs have been described to play roles in both solid tumors and hematologic malignancies.5-8 Indeed, recently published data have revealed the importance of miRNAs in chronic lymphocytic leukemia and B-cell lymphomas9-11 ; however, the role of miRNAs in Waldenström macroglobulinemia (WM) has not been yet elucidated.
WM is a low-grade lymphoproliferative disorder characterized by the presence of a lymphoplasmacytic infiltrate in the bone marrow and serum monoclonal immunoglobulin M.12-14 Cytogenetic and molecular studies on gene-expression analysis at the mRNA level have demonstrated minimal changes in WM cells.15,16 Therefore, multilevel characterization of this disease at genetic and epigenetic levels is required to improve our understanding of the underlying molecular changes that lead to the initiation and progression of this disease.
In the present study, we evaluated miRNA expression patterns in WM using liquid phase Luminex microbead miRNA profiling (Austin, TX). We showed that selected miRNAs were specifically dysregulated in WM patients compared with healthy controls, and identified a specific miRNA signature that characterizes this disease. We then focused on the functional role of miRNA-155, one of the increased miRNAs in WM that has already been shown to exert a clear oncogenic role in other B-cell malignancies.9-11 We found that miRNA-155 regulates proliferation and growth of WM cells in vitro and in vivo by inhibiting signaling cascades including MAPK/ERK, PI3/AKT, and NF-κB; pathways. Candidate miRNA-155 targets were identified using gene-expression profiling and include genes involved in cell-cycle progression, adhesion, and migration. To assess clinical relevance, we showed that overexpression of the 6 miRNAs significantly correlated with outcome predicted by the International Prognostic Staging System for WM. We further demonstrated that therapeutic agents commonly used in WM (rituximab/perifosine/bortezomib) alter the levels of the 6 signature miRNAs identified, by inducing downmodulation of patient-overexpressed miRNAs (except miR-206) and up-regulation of patient-downexpressed miRNA. These data enhance our understanding of the biologic role of miRNAs in the pathogenesis of WM, and provide the basis for the development of new miRNA-targeted therapeutic strategies.
Methods
Cells and reagents
Primary WM cells were obtained from bone marrow (BM) samples from patients with relapsed/refractory WM (n = 15) or untreated (n = 5), using CD19+ microbead selection (Miltenyi Biotec, Auburn, CA) with more than 90% purity, as confirmed by flow cytometric analysis with monoclonal antibody reactive with human CD20-PE (BD Biosciences, San Jose, CA).17 Similarly, CD19+ cells were isolated from the BM and peripheral blood of 3 healthy donors, and used as controls. Approval for these studies was obtained from the Dana-Farber Cancer Institute Institutional Review Board. Informed consent was obtained from all patients and healthy volunteers in accordance with the Declaration of Helsinki protocol. A recently described WM cell line (BCWM1) was used in this study.18 Cells were cultured at 37°C in RPMI-1640 containing 10% fetal bovine serum (FBS; Sigma Chemical, St Louis, MO), 2 mM l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (GIBCO, Grand Island, NY). GFP-BCWM1 cells were generated using lentivirus infection as previously described.19 Nocodazole has been purchased from Sigma-Aldrich (St Louis, MO).
Therapeutic agents
Perifosine was provided by Keryx Biopharmaceuticals (New York, NY). Bortezomib was obtained from Millennium Pharmaceuticals (Cambridge, MA). Rituximab was provided by Genentech (San Francisco, CA).
DNA synthesis and cytotoxicity assay
Proliferation rate and cytotoxicity on BCWM.1 cells (miR-155 knockdown probe transfected, control probe transfected) and nontransfected BCWM.1 cells were measured by DNA synthesis using [3H]-thymidine uptake (Perkin Elmer, Boston, MA), and by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Chemicon International, Temecula, CA) dye absorbance, respectively, as previously described.17
Cell-cycle analysis
BCWM.1 cells (miR-155 knockdown probe–transfected, control probe–transfected, and nontransfected BCWM.1 cells) were stained with propidium iodine (PI; Sigma-Aldrich) and cell cycle was determined using an Epics (Coulter Immunology, Hialeah, FL) flow cytometer, as previously described.17
Adhesion assay
We used an in vitro adhesion assay (Innocyte ECM Cell Adhesion Assay, Gibbstown, NJ) consisting of 96-well plates coated with fibronectin, a ligand of VLA-4 (EMD Biosciences, San Diego, CA), and WM cells (miR-155 knockdown probe–transfected, control probe–transfected, and nontransfected BCWM.1 cells) were tested. BSA-coated wells served as a negative control. Calcein AM was used to measure adherent cells, and the degree of fluorescence was measured using a spectrophotometer (485-520 nm).17
Transwell migration assay
Migration was determined using the transwell migration assay (Costar, Corning, NY), as previously described. BCWM.1 cells (miR-155 knockdown probe–transfected, control probe–transfected, and nontransfected BCWM.1 cells) were placed in the migration chambers in the presence of SDF-1 (R&D, Minneapolis, MN). After 4 hours of incubation, cells that migrated to the lower chambers were counted, as described.17
Effect of miRNA-155 on paracrine Waldenström macroglobulinemia cell growth in the bone marrow
To evaluate the effect of miRNA-155 on growth of WM cells adherent to bone marrow stromal cells (BMSCs), BMSCs were cultured in the presence or absence of either miRNA-155 knockdown probe–transfected, control probe–transfected, or nontransfected BCWM1 cells. DNA synthesis was measured using the [3H]-thymidine uptake assay, as described.17
NF-κB activity
NF-κB activity was investigated using the Active Motif TransAM kits, a DNA-binding enzyme-linked immunosorbent assay (ELISA)–based assay (Active Motif North America, Carlsbad, CA). Briefly, BCWM.1 cells (miRNA-155 knockdown probe–transfected, control probe–transfected, and nontransfected BCWM.1 cells) were cultured in presence or absence of TNF-α (10 ng/mL) for 20 minutes. NFkBp65 transcription factor binding to its consensus sequence on the plate-bound oligonucleotide was studied from nuclear extracts, following the manufacturer's procedure.
Immunofluorescence
The effect of miRNA-155 on TNF-α–induced nuclear translocation of p-p65 was examined by immunocytochemical method. Briefly, BCWM.1 cells (miR-155 knockdown probe–transfected, control probe–transfected, and nontransfected BCWM.1 cells) were stimulated with TNF-α (10 ng/mL) for 20 minutes. Immunocytochemical analysis was performed using an epifluorescence microscope (Nikon Eclipse E800; Nikon, Avon, MA) and a Photometrics Coolsnap CF color camera (Nikon, Lewisville, TX), as previously described.20
IgM detection
Serum IgM levels were quantified by ELISA (Human IgM ELISA kit; Zeptometrix, Buffalo, NY), according to the manufacturer's instructions.
microRNA expression profiling
RNA was isolated using an RNeasy kit (Qiagen, Valencia, CA), as reported in previous miRNA studies.21-23 Quality control of RNA was done using RNA 6000 Nano assay on the Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA). All RNA samples showed high quality, without RNA degradation or DNA contamination. The expression of 318 miRNAs was investigated using liquid phase Luminex microbead miRNA profiling (Luminex, Austin, TX). Briefly, 500 ng total RNA was labeled with biotin using the FlexmiR MicroRNA labeling kit (Luminex, Austin, TX), which labeled all RNA molecules including small RNAs, by first using calf intestinal phosphatase (CIP) for removal of 5′-phosphatases from the terminal end of the miRNAs. In the second step, a biotin label was then attached enzymatically to the 3′-end of the miRNAs in the total RNA sample. After an enzyme inactivation step, the sample was hybridized with beads containing one of 100 different fluorophores and coated with oligonucleotides complementary to each known miRNA. Addition of streptavidin-phycoerythrin (Molecular Probes, Eugene, OR) then yielded fluorescence with wavelengths and amplitudes characteristic of the identity and quantity of miRNAs, respectively. Normalization of arrays and calculation of median fluorescence intensity was performed according to the manufacturer's instructions.
Quantitative reverse transcription–PCR
Stem-loop quantitative reverse transcription–polymerase chain reaction (qRT-PCR) for mature miRNAs (TaqMan microRNA Assays; Applied Biosystems, Foster City, CA) was performed as described24 on an Applied Biosystems AB7500 Real Time PCR system. All PCR reactions were run in triplicate and miRNA expression, relative to RNU6B, was calculated using the 2−ΔΔCt method.25
miRNA transfection
BCWM1 cells were transfected with either miRNA-155 LNA knockdown probe (Exiqon, Vedbaek, Denmark) or control knockdown probe (Exiqon), at final concentration of 40 nM, using Lipofectamine 2000 following the manufacturer's instructions (Invitrogen, Carlsbad, CA). Culture medium was changed 8 hours after transfection and replaced with RPMI 10% FBS. Cells were then used for functional assays at different time points (24 hours, 48 hours). Both nontransfected or control probe–transfected BCWM1 cells were used as controls. Efficiency of transfection was validated by qRT-PCR and microRNA assay. Fluorescein-labeled miRNA-155 LNA knockdown probe (Exiqon) or fluorescein-labeled control knockdown probe (Exiqon), at final concentration of 40 nM, was also used for transfection, to further confirm the efficiency of the probes as shown by immunofluorescence. Only cells transfected with non–fluorescently labeled probe were used in all the experiments.
Gene expression profile
Total RNA has been isolated from BCWM.1 cells (miR-155 knockdown probe–transfected, control probe–transfected, and nontransfected BCWM.1 cells) using RNeasy kit (Qiagen), as described by the manufacturer, and analyzed with the use of Affymetrix U133 plus 2.0 GeneChip (Affymetrix, Santa Clara, CA). RNA integrity was verified with the Agilent 2100 Bioanalyzer.26
In vivo imaging
In vivo video rate confocal microscopy and 2-photon microscopy WM cell homing to bone marrow vasculature of the skull was analyzed using fluorescence confocal microscopy, as previously described.27 A skin flap was made in the scalp of the mice to expose the underlying dorsal skull surface. Images of the tumors were captured in approximately 1-hour sessions. High-resolution images with cellular detail were obtained through the intact mouse skull at depths of up to 250 μm from the surface of the skull using a 30×/0.9 NA water-immersion objective lens (Lomo, St Petersburg, Russia). Multiple imaging depths were acquired, and a maximum intensity z-projection was performed in Image J (National Institutes of Health, Bethesda, MD) to merge the images. GFP was excited with a 491-nm solid state laser (Dual Calypso; Cobolt AB, Stockholm, Sweden) and detected with a photomultiplier tube through a 528/19-nm band-pass filter (Semrock, Rochester, NY). Blood vessels were imaged using Angiosense-750 (VisEn Medical, Woburn, MA) excited with a 750-nm laser and captured through a 785/25-nm band-pass filter (Chroma Technologies, Rockingham, VT). The frame rate of the confocal microscope was 30 frames per second. Images were captured after averaging 30 frames using a Macintosh computer (Apple, Cupertino, CA) equipped with an Active Silicon snapper card (Active Silicon, Chelmsford, MA). Approval of these studies was obtained by the Dana-Farber Cancer Institute and Massachusetts General Hospital Institutional Animal Care and Use Committees.
Statistical analysis
miRNA expression data were analyzed according to the manufacturer's instructions (Luminex, Houston, TX). The expression patterns of unfiltered data were performed using unsupervised hierarchic clustering of samples, based on centroid linkage and 1-correlation distance metric, using dChip (http://www.dchip.org, Dana-Farber Cancer Institute, Harvard Medical School). To further define those miRNAs differentially expressed between groups (patients vs healthy controls), the data were filtered on significance of differences using ANOVA test (P < .01). To identify specific predicted miRNAs-targeted mRNAs, TargetScan, PicTar, and miRanda algorithms were used (http://microrna.sanger.ac.uk).28,29 To reduce the numbers of false positives, only putative target genes predicted by the 3 algorithms were considered.
To describe the distribution of miRNA levels in WM patients compared with controls, mean values were compared using Mann-Whitney U rank sum test. The expression of miRNA among all WM patients relative to the healthy counterparts has been considered. WM patients have been categorized according to the International Prognostic Staging System (IPSS) for WM,30 and the mean level of miRNA within WM patients (IPSS low vs IPSS intermediate vs IPSS high) has been compared using the Kruskal-Wallis H test. All statistical tests are 2 sided and all analyses were performed with SPSS software (SPSS, Chicago, IL).
Results
Specific miRNA expression signature distinguishes WM lymphocytes from their normal cellular counterparts
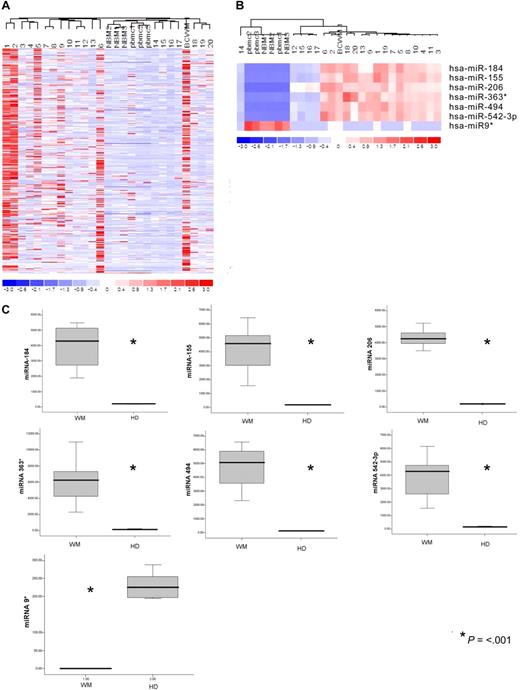
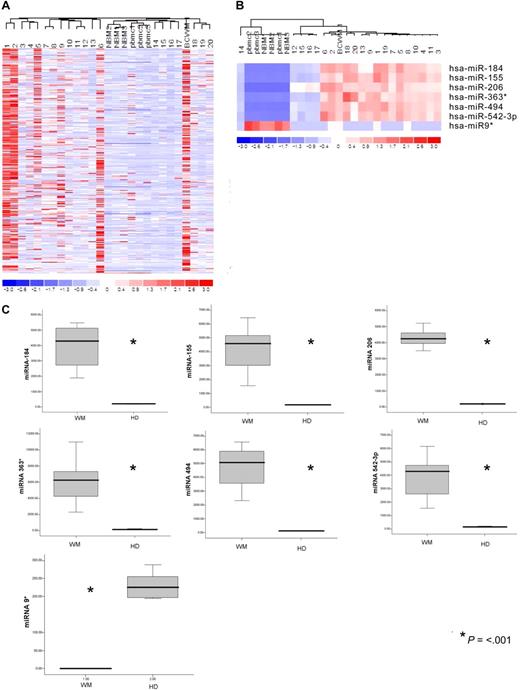
The expression of 318 miRNAs was profiled in BM CD19+ selected WM cells from 20 patients with symptomatic WM in comparison with CD19+ cells isolated from BM and peripheral blood mononuclear cells (PBMCs) of healthy donors, and in WM cell line BCWM.1. The clinical characteristics of the evaluated patients' samples are listed in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Unsupervised hierarchic clustering of samples was performed on all unfiltered data. The generated heat map for the entire population of miRNAs showed differences between miRNA expression profiles of WM samples compared with normal counterparts (Figure 1A).
microRNA expression pattern in WM and healthy donors. (A) miRNA analysis has been performed on total RNA isolated from BM CD19+ WM cells, normal bone marrow (NBM)– and peripheral blood (PBMC)–derived CD19+ counterparts, and WM cell line (BCWM.1). Heat map was generated after unsupervised hierarchic cluster analysis of all unfiltered data. Differential expression of miRNA is shown by the intensity of red (up-regulation) versus blue (down-regulation). (B) Supervised hierarchic clustering analysis was performed using ANOVA test. Differential expression of miRNA patterns is shown by the intensity of red (up-regulation) versus blue (down-regulation). (C) Differential distribution of each indicated miRNA in all WM patients (WM) compared with healthy donors (HDs): mean values were compared using Mann-Whitney U rank sum test; bars indicate standard errors; P value (*) for each miRNA is indicated.
microRNA expression pattern in WM and healthy donors. (A) miRNA analysis has been performed on total RNA isolated from BM CD19+ WM cells, normal bone marrow (NBM)– and peripheral blood (PBMC)–derived CD19+ counterparts, and WM cell line (BCWM.1). Heat map was generated after unsupervised hierarchic cluster analysis of all unfiltered data. Differential expression of miRNA is shown by the intensity of red (up-regulation) versus blue (down-regulation). (B) Supervised hierarchic clustering analysis was performed using ANOVA test. Differential expression of miRNA patterns is shown by the intensity of red (up-regulation) versus blue (down-regulation). (C) Differential distribution of each indicated miRNA in all WM patients (WM) compared with healthy donors (HDs): mean values were compared using Mann-Whitney U rank sum test; bars indicate standard errors; P value (*) for each miRNA is indicated.
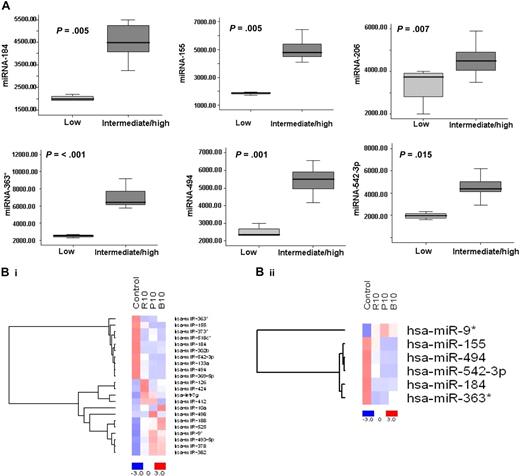
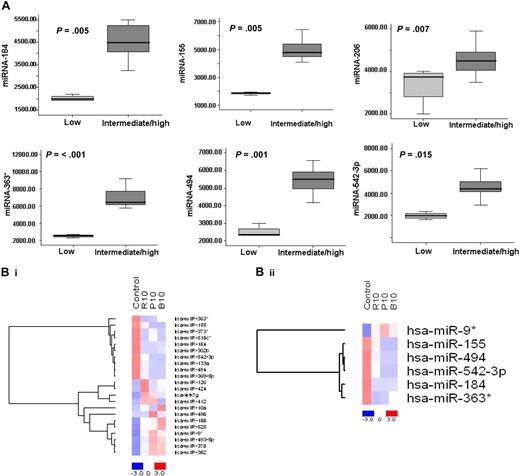
To identify statistically significant differences in miRNA expression profiles between patient and normal donor samples, ANOVA analysis was performed. We identified 7 miRNAs with specific differential expression signatures between WM patients and healthy individuals. Specifically, miRNA analysis showed that miRNA-363*/-206/-494/-155/-184/-542-3p demonstrated increased expression in WM patients; whereas miRNA-9* demonstrated decreased expression in WM patients (P < .01; Figure 1B). Increased miRNAs were significantly higher in the entire cohort of WM patients compared with control samples, whereas miRNA-9* was significantly down-regulated in all patient samples compared with control samples (P ≤ .001; Figure 1C).
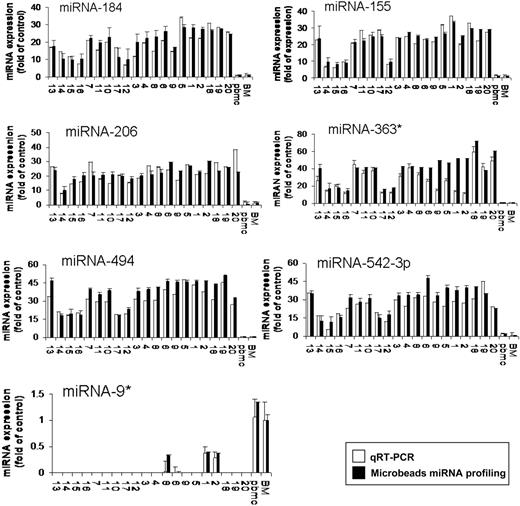
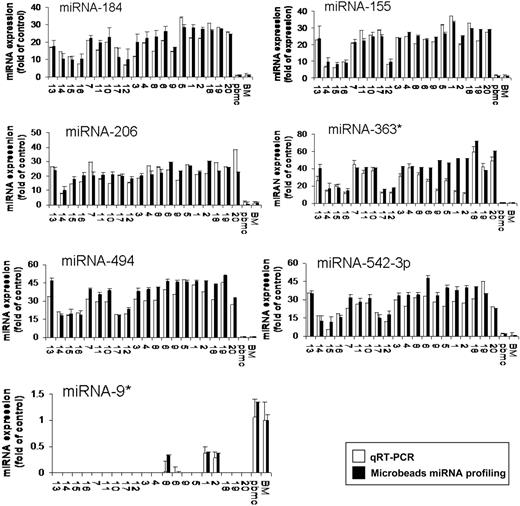
To further validate the results from miRNA expression profiling, qRT-PCR was performed on matched samples. We showed expression patterns similar to those observed in miRNA analysis (Figure 2). We next analyzed the predicted gene targets for each up- or down-regulated miRNA, using algorithms commonly used to predict human miRNA gene targets, miRanda, TargetScan, and PicTar,28,29 and specifically identified genes predicted by all 3 methods. Predicted targets of the increased miRNAs in WM patients included tumor suppressors, cell-cycle inhibitors, cytokine signaling suppressors, and tyrosine phosphatases. Conversely, predicted target genes for the decreased miRNAs in WM included protein kinases, oncogenes, and transcription factors. Moreover, we showed that several protein kinases, transcription factors, and cell cycle regulators were up-regulated in WM patients including protein kinase C, serine/threonine kinase 24, mitogen-activated protein kinase kinase kinase 7; E2F- and pre-B-cell leukemia transcription factors; as well as cyclin D2 and cyclin-dependent kinase inhibitor (p57) (Table S2).
Validation of miRNA expression levels by qRT-PCR. The amount of miRNA-184, -206, -494, -363*, -155, 542-3p, and -9* in WM samples compared with normal controls, evaluated by qRT-PCR (□) and microbead miRNA profiling (■). Results are expressed as fold change of the miRNA expression in bone marrow WM CD19+ cells with respect to average miRNA expression from 3 healthy donors' bone marrow CD19+ cells (BM). qRT-PCR data were obtained using the ΔΔCt method, with normalization to the reference RNU6B microRNA. Bars represent SD.
Validation of miRNA expression levels by qRT-PCR. The amount of miRNA-184, -206, -494, -363*, -155, 542-3p, and -9* in WM samples compared with normal controls, evaluated by qRT-PCR (□) and microbead miRNA profiling (■). Results are expressed as fold change of the miRNA expression in bone marrow WM CD19+ cells with respect to average miRNA expression from 3 healthy donors' bone marrow CD19+ cells (BM). qRT-PCR data were obtained using the ΔΔCt method, with normalization to the reference RNU6B microRNA. Bars represent SD.
miRNA-155 modulates proliferation, adhesion, and migration of WM cells
miRNA-155 plays a pivotal role in B-cell malignancies including diffuse large B-cell lymphomas, primary mediastinal B-cell lymphomas, and Hodgkin lymphomas.9-11 We found that miRNA-155 is increased also in WM and therefore we studied the functional role of miRNA-155 in WM by examining proliferation, adhesion, and migration in miR-155 LNA knockdown WM cells compared with controls (control probe–transfected BCWM1 cells or nontransfected BCWM1 cells). Efficiency of transfection was evaluated by qRT-PCR at 48 hours after transfection (Figure S1A). Fluorescein-labeled miRNA-155 LNA knockdown probe or fluorescein-labeled control knockdown probe (at final concentration of 40 nM) was also used for transfection to further confirm the efficiency of the probes (Figure S1B).
We showed that DNA synthesis was significantly reduced in miRNA-155 knocked-down cells compared with control at 24 and 48 hours (P < .001; Figure 3A), without cytotoxic effects (Figure 3B). We therefore next examined cell cycle profiling using PI staining in control probe– and knockdown probe–transfected cells. Cells were first arrested and synchronized in G2/M phase by growth in 80 nM nocodazole for 16 hours31 ; absence of nocodazole-induced cytotoxicity was observed on treated cells, as assessed by MTT (Figure S1C). miRNA-155 knockdown cells showed increased numbers of cells in G1 phase (76% vs 46% in control) and decreased numbers of cells in S phase (15% vs 42% in control; Figure 3C). Using gene expression profiling, we showed increased expression of cyclin-dependent kinase inhibitors (p18, p19, p21, p27); decreased expression of cyclin-dependent kinases-2, -4, and -6, as well as cyclins D1, D2, D3, and E1 (Figure 3Di); and increased expression of p53 with decreased expression of its negative regulator (Mdm2) in the miR-155 knockdown WM cells compared with controls (Figure 3Dii). Taken together, these data demonstrate changes in cell-cycle regulatory proteins responsible for G1 arrest. We next examined the impact of miRNA-155 knockdown on signaling cascades regulating proliferation. We found that miRNA-155 knockdown strongly inhibited ERK and AKT phosphorylation, as well as p-GSK3α/β and p-S6R, both AKT downstream target proteins. Interestingly, a compensatory increase in the expression of related total protein levels occurred along with the significant inhibition of phosphorylated forms (Figure 3E).
miRNA-155 modulates proliferation and cell cycle of WM cells. (A,B) WM cells (miRNA-155 knockdown probe–transfected, control probe–transfected BCWM.1 cells) were harvested at 24 and 48 hours after transfection; DNA synthesis and cytotoxicity were assessed by thymidine uptake and MTT assays, respectively. Nontransfected BCWM.1 cells were used as controls (ctrl). *P < .001; **P < .001. (C) Cells were first arrested and synchronized in G2/M phase by growth in 80 nM nocodazole for 16 hours. Cells were then washed and regrown using fresh media. After 6 hours, cell-cycle analysis was performed by propidium iodide staining. (D) Purified cRNA (15 μg) isolated from WM cells (control probe–transfected, miRNA-155 knockdown probe–transfected BCWM.1 cells) was hybridized to HG-U133Plus2.0 GeneChip (Affymetrix). (i) Up-regulation of cyclin-dependent kinase inhibitors and down-regulation of cyclins and cyclin-dependent kinases. (ii) Up-regulation of p53 and p53 family members and down-regulation of p53 negative regulators. Fold change is shown by the intensity of induction (red) or suppression (blue). (E) BCWM.1 cells (miRNA-155 knockdown probe transfected, control probe transfected) were harvested at 8 hours after transfection. Whole-cell lysates were subjected to Western blotting using anti–phospho (p)-ERK, –p-AKT, -AKT, –p-GSK3α/β, –p-S6R, and -actin antibodies; nontransfected BCWM.1 cells were used as controls (ctrl). (F) BCWM.1 cells (miRNA-155 knockdown probe transfected, control probe transfected) were harvested at 8 hours after transfection and treated with and without TNF-α (10 ng/mL) for 20 minutes; nontransfected BCWM.1 cells were used as control (ctrl). NF-κBp65 transcription factor binding to its consensus sequence on the plate-bound oligonucleotide was measured in nuclear extracts. Wild-type and mutant are wild-type and mutated consensus competitor oligonucleotides, respectively. All results represent means (±] SD) of triplicate experiments. (G) BCWM.1 cells (miRNA-155 knockdown probe transfected, control probe transfected) were harvested at 8 hours after transfection and treated with and without TNF-α (10 ng/mL) for 20 minutes; nontransfected BCWM.1 cells were used as control (ctrl). Immunocytochemical analysis was assessed using anti–p-NF-κBp65 antibody, with DAPI used to stain nuclei.
miRNA-155 modulates proliferation and cell cycle of WM cells. (A,B) WM cells (miRNA-155 knockdown probe–transfected, control probe–transfected BCWM.1 cells) were harvested at 24 and 48 hours after transfection; DNA synthesis and cytotoxicity were assessed by thymidine uptake and MTT assays, respectively. Nontransfected BCWM.1 cells were used as controls (ctrl). *P < .001; **P < .001. (C) Cells were first arrested and synchronized in G2/M phase by growth in 80 nM nocodazole for 16 hours. Cells were then washed and regrown using fresh media. After 6 hours, cell-cycle analysis was performed by propidium iodide staining. (D) Purified cRNA (15 μg) isolated from WM cells (control probe–transfected, miRNA-155 knockdown probe–transfected BCWM.1 cells) was hybridized to HG-U133Plus2.0 GeneChip (Affymetrix). (i) Up-regulation of cyclin-dependent kinase inhibitors and down-regulation of cyclins and cyclin-dependent kinases. (ii) Up-regulation of p53 and p53 family members and down-regulation of p53 negative regulators. Fold change is shown by the intensity of induction (red) or suppression (blue). (E) BCWM.1 cells (miRNA-155 knockdown probe transfected, control probe transfected) were harvested at 8 hours after transfection. Whole-cell lysates were subjected to Western blotting using anti–phospho (p)-ERK, –p-AKT, -AKT, –p-GSK3α/β, –p-S6R, and -actin antibodies; nontransfected BCWM.1 cells were used as controls (ctrl). (F) BCWM.1 cells (miRNA-155 knockdown probe transfected, control probe transfected) were harvested at 8 hours after transfection and treated with and without TNF-α (10 ng/mL) for 20 minutes; nontransfected BCWM.1 cells were used as control (ctrl). NF-κBp65 transcription factor binding to its consensus sequence on the plate-bound oligonucleotide was measured in nuclear extracts. Wild-type and mutant are wild-type and mutated consensus competitor oligonucleotides, respectively. All results represent means (±] SD) of triplicate experiments. (G) BCWM.1 cells (miRNA-155 knockdown probe transfected, control probe transfected) were harvested at 8 hours after transfection and treated with and without TNF-α (10 ng/mL) for 20 minutes; nontransfected BCWM.1 cells were used as control (ctrl). Immunocytochemical analysis was assessed using anti–p-NF-κBp65 antibody, with DAPI used to stain nuclei.
We have previously shown that WM is characterized by constitutive activation of NF-κB activity, confirming previous reports showing that NF-κB is one of the major pathways implicated in the growth and survival of plasma cell dyscrasias.32,33 We therefore sought to examine the effect of miRNA-155 on NF-κB activity in WM. We first investigated the effect of miRNA-155 knockdown on NF-κBp65 DNA-binding activity, studying nuclear extracts from miRNA-155 knockdown WM cells compared with control nontransfected cells and control probe–transfected cells using Active Motif assay. We showed that TNF-α treatment induced NF-κB recruitment to the nucleus in BCWM.1 control cells, which was inhibited in the knocked-down miR-155 cells (Figure 3F). We further confirmed that phospho-p65 translocation from the cytoplasmic compartment to the nucleus was inhibited in the miRNA-155 knockdown cells, resulting in a significant increase in p-p65 expression in the cytoplasmic compartment as shown by immunofluorescence (Figure 3G).
Inhibition of miRNA-155 targets WM cells in the context of BM milieu in vitro and in vivo
Since the BM microenvironment confers growth advantages to and induces drug resistance in malignant cells,34 we next investigated whether miRNA-155 could affect WM cell growth in the context of the BM milieu. Previous studies have shown that the PI3K/Akt pathway regulates adhesion and migration in B cells35 ; therefore, we examined the effect of miRNA-155 knockdown on adhesion of WM cells using miRNA-155 knockdown probe–transfected, control probe–transfected, and nontransfected BCWM1 cells. A significant inhibition of adhesion to fibronectin was observed in the miRNA-155 knockdown cells compared with controls (P = .02; Figure 4A). We next demonstrated by gene expression analysis that miRNA-155 knockdown probe–transfected cells were characterized by down-regulation of genes such as several Rho GTPase activating proteins, p21(CDKN1A) activating protein, and p21-activated kinase 1 (PAK-1) interacting protein, which are known to be involved in the adhesion process (Figure 4B).36
miRNA-155 knockdown inhibited WM cells in the context of bone marrow microenvironment in vitro. WM cells (miRNA-155 knockdown probe–transfected, control probe–transfected BCWM.1 cells) were harvested at 24 hours after transfection. Nontransfected BCWM.1 cells were used as control (ctrl). (A) Adhesion assay to fibronectin (FN). Control probe–transfected and nontransfected cells showed significant increase in adhesion to FN, compared with BSA, used as control. miRNA-155 knockdown probe–transfected cells showed lower adhesion to FN. (B) Purified cRNA (15 μg) isolated from WM cells (control probe–transfected, miRNA-155 knockdown probe–transfected BCWM.1 cells) was hybridized to HG-U133Plus2.0 GeneChip (Affymetrix). Down-regulation of several genes involved in the adhesion process. Fold change is shown by the intensity of induction (red) or suppression (blue). (C) Transwell migration assay. SDF-1 (30 nM) was placed in the lower chambers and migration was determined after 4 hours. Control probe–transfected and nontransfected cells showed significant migration with SDF-1 30 nM, whereas the miRNA-155 knockdown probe–transfected cells showed minimal migration in response to SDF-1. (D) BCWM.1 cells were cultured for 48 hours in the presence or absence of BMSCs, and cell proliferation was assessed using [3H]-thymidine uptake assay. All data represent mean (±SD) of triplicate experiments.
miRNA-155 knockdown inhibited WM cells in the context of bone marrow microenvironment in vitro. WM cells (miRNA-155 knockdown probe–transfected, control probe–transfected BCWM.1 cells) were harvested at 24 hours after transfection. Nontransfected BCWM.1 cells were used as control (ctrl). (A) Adhesion assay to fibronectin (FN). Control probe–transfected and nontransfected cells showed significant increase in adhesion to FN, compared with BSA, used as control. miRNA-155 knockdown probe–transfected cells showed lower adhesion to FN. (B) Purified cRNA (15 μg) isolated from WM cells (control probe–transfected, miRNA-155 knockdown probe–transfected BCWM.1 cells) was hybridized to HG-U133Plus2.0 GeneChip (Affymetrix). Down-regulation of several genes involved in the adhesion process. Fold change is shown by the intensity of induction (red) or suppression (blue). (C) Transwell migration assay. SDF-1 (30 nM) was placed in the lower chambers and migration was determined after 4 hours. Control probe–transfected and nontransfected cells showed significant migration with SDF-1 30 nM, whereas the miRNA-155 knockdown probe–transfected cells showed minimal migration in response to SDF-1. (D) BCWM.1 cells were cultured for 48 hours in the presence or absence of BMSCs, and cell proliferation was assessed using [3H]-thymidine uptake assay. All data represent mean (±SD) of triplicate experiments.
We next investigated the effect of miRNA-155 knockdown on migration and adhesion of WM cells. We first showed that 30 nM stromal derived factor-1 (SDF-1), an important regulator of migration in B cells, induced migration in BCWM.1 cells (Figure 4C). miRNA-155 knockdown probe–, control probe–transfected, and nontransfected BCWM1 cells were incubated for 2 hours in the presence of SDF-1. miRNA-155 knockdown significantly inhibited BCWM.1 cell migration in response to SDF-1 (P = .04; Figure 4C).
To further confirm that miRNA-155 regulates WM cells proliferation even in the presence of the BM microenvironment, we next examined the effect of miRNA-155 on growth of WM cells in the presence of BM stromal cells (BMSCs). Transfected (either with control probe or miRNA-155 knockdown probe) and nontransfected BCWM1 cells were cultured in the presence or absence of primary BMSCs for 48 hours. Adherence of both nontransfected or control probe–transfected BCWM.1 cells to BMSCs triggered 34% and 33% increase in proliferation, respectively, using [3H]-thymidine uptake. Importantly, we did not observe any significant increase in the proliferation rate of miRNA-155 knocked down BCWM1 cells, indicating that they overcome the growth advantage of bone marrow milieu (Figure 4D).
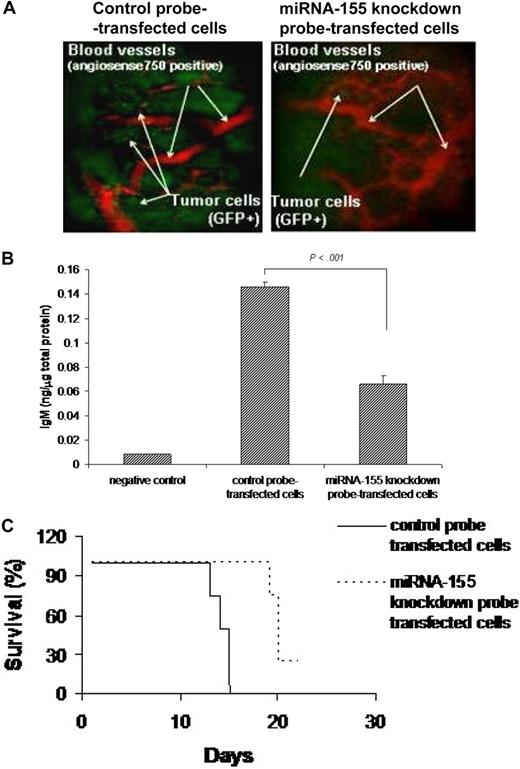
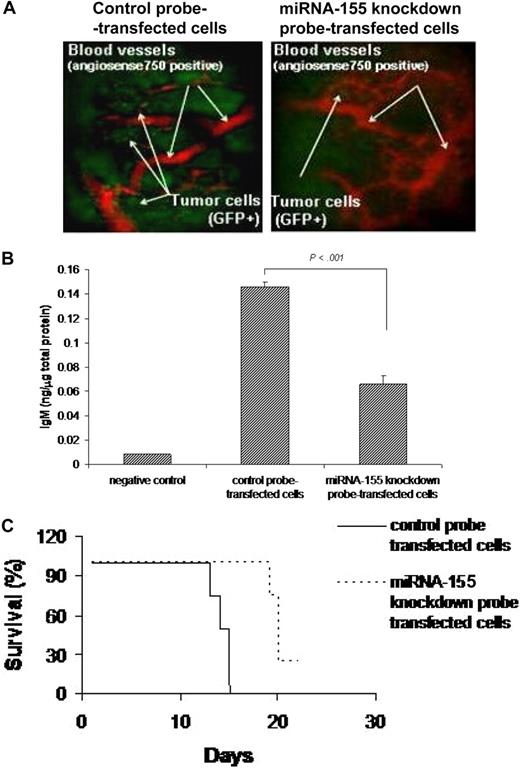
Based on the in vitro data showing inhibition of migration in miRNA-155 knocked-down WM cells, we next investigated the effect of miRNA on migration in vivo, using our previously described in vivo homing model.19 We found that number of miRNA-155 knockdown probe–transfected GFP+ WM cells that homed and proliferated in vivo in bone marrow was lower than the control probe–transfected GFP+ WM cells (Figure 5A). We confirmed that WM cells were efficiently transfected in vivo, as evaluated in cells isolated 21 days after injection from bone marrow of mice injected with either miRNA-155 knockdown probe– or control probe–transfected WM cells, using stem-loop qRT-PCR for human miRNA-155 (Figure S1D). In addition, we determined whether miRNA-155 could affect IgM secretion from WM cells in vivo. Using a human IgM ELISA assay, we showed that IgM secretion was significantly lower in the serum of mice injected with miRNA-155 knockdown probe–transfected cells compared with controls (P < .001; Figure 5B). In addition, we demonstrated that mice injected with miRNA-155 knockdown probe–transfected WM cells had prolonged survival compared with mice injected with control probe–transfected WM cells, with mean survival of 20 days versus 14 days (P < .001) and overall survival of 21 days versus 15 days, respectively (Figure 5C).
miRNA-155 knockdown targets WM cells in the context of bone marrow microenvironment in vivo. (A) In vivo confocal imaging. GFP+ WM cells were transfected using either control probe or miRNA-155 knockdown probe, and then injected in mice 24 hours after transfection. GFP+ WM cells (green) were excited with a 491-nm solid state laser. Blood vessels (red) were imaged using Angiosense-750 (VisEn Medical) excited with a 750-nm laser. Images of parasagittal vasculature and GFP+ tumor cells in the mouse skull bone marrow. Control probe–transfected cells homed and adhered more than the miRNA-155–transfected counterparts. (B) Detection of human IgM from serum of control probe–injected (n = 4) and miRNA-155 knockdown probe–injected (n = 4) mice. Serum of noninjected mice was used as negative control. Average obtained from each group is shown. (C) Kaplan-Meier curve showing survival in each group; mean survival was 14 days versus 20 days (P < .001), and overall survival was 15 days versus 21 days, respectively, in control probe–injected versus miRNA-155 knockdown probe–injected mice.
miRNA-155 knockdown targets WM cells in the context of bone marrow microenvironment in vivo. (A) In vivo confocal imaging. GFP+ WM cells were transfected using either control probe or miRNA-155 knockdown probe, and then injected in mice 24 hours after transfection. GFP+ WM cells (green) were excited with a 491-nm solid state laser. Blood vessels (red) were imaged using Angiosense-750 (VisEn Medical) excited with a 750-nm laser. Images of parasagittal vasculature and GFP+ tumor cells in the mouse skull bone marrow. Control probe–transfected cells homed and adhered more than the miRNA-155–transfected counterparts. (B) Detection of human IgM from serum of control probe–injected (n = 4) and miRNA-155 knockdown probe–injected (n = 4) mice. Serum of noninjected mice was used as negative control. Average obtained from each group is shown. (C) Kaplan-Meier curve showing survival in each group; mean survival was 14 days versus 20 days (P < .001), and overall survival was 15 days versus 21 days, respectively, in control probe–injected versus miRNA-155 knockdown probe–injected mice.
Association between miRNA profiling and WM clinical features
To further investigate the clinical relevance of miRNA in WM in terms of prognosis, we correlated miRNA expression levels with prognostic factors including age, β2microglobulin, hemoglobin, and immunoglobulin M (IgM) levels, and platelet counts, according to the International Prognostic Scoring System (IPSS) for WM which stratifies patients into low-, intermediate-, and high-risk subgroups.30 Twenty-five percent of patients were classified as having IPSS low risk; 35% of patients, as IPSS intermediate risk; and 40% of patients, as IPSS high risk (Table S1). We then compared the mean level of each miRNA within the WM patients IPSS low-risk group with IPSS intermediate- and high-risk groups. There was no statistical difference between IPSS intermediate- and high-risk groups; therefore, these groups were pooled. The expression of each miRNA was significantly different among patients with worse prognosis (high-/intermediate-risk groups) compared with those with better prognostic factors (low-risk group) (Figure 6A); specifically, 6 overexpressed miRNAs were significantly elevated in patients with poor prognostic factors (6 of 6: P < .015). No statistically significant differences were identified for miRNA-9*, since its expression level was low in all the patients studied.
Differentially expressed miRNAs are associated with prognosis in WM patients, and drugs used to treat WM modulate their expression level. (A) Significant differential distribution of each indicated miRNA among subgroups of WM patients with differential clinical-prognostic features (low risk versus intermediate/high risk). Mean values were compared using Mann-Whitney U rank sum test; bars indicate standard errors. P value for each miRNA is indicated. (B) BCWM1 cells were treated with either rituximab (R; 10 μg/mL; 2 hours), perifosine (P; 10 μM; 6 hours), or bortezomib (B; 10 nM; 6 hours), and miRNA analysis was performed on total RNA isolated from treated and untreated cells. Heat map was generated after supervised hierarchic cluster analysis and comparison among the 2 groups (treated vs untreated samples). Differential expression of miRNA is shown by the intensity of red (up-regulation) or blue (down-regulation).
Differentially expressed miRNAs are associated with prognosis in WM patients, and drugs used to treat WM modulate their expression level. (A) Significant differential distribution of each indicated miRNA among subgroups of WM patients with differential clinical-prognostic features (low risk versus intermediate/high risk). Mean values were compared using Mann-Whitney U rank sum test; bars indicate standard errors. P value for each miRNA is indicated. (B) BCWM1 cells were treated with either rituximab (R; 10 μg/mL; 2 hours), perifosine (P; 10 μM; 6 hours), or bortezomib (B; 10 nM; 6 hours), and miRNA analysis was performed on total RNA isolated from treated and untreated cells. Heat map was generated after supervised hierarchic cluster analysis and comparison among the 2 groups (treated vs untreated samples). Differential expression of miRNA is shown by the intensity of red (up-regulation) or blue (down-regulation).
Modulation of miRNA profile in WM cells after treatment with antineoplastic agents
Since the majority of patients with WM used in the miRNA analysis were subsequently treated with rituximab, bortezomib, or the novel Akt inhibitor perifosine on clinical trials, we next examined whether miRNA expression profile was modulated in WM cell line (BCWM1) following treatment with these agents. BCWM.1 cells were treated with either bortezomib (10 nM), perifosine (10 μM) for 6 hours, or rituximab (10 μg/mL) for 2 hours. MTT assay did not demonstrate cytotoxicity at these time points (data not shown). Supervised hierarchic clustering of all unfiltered data, obtained after comparison of treated samples versus untreated cells, was performed; and the generated heat map showed significant differences in 22 miRNAs in treated versus untreated cells (Figure 6Bi). We next investigated whether the expression of miRNA-184, -494, -206, 363*, and -9*, whose expression was altered in patient samples compared with controls, was modulated after treatment of WM cell line with each drug. Interestingly, we found that rituximab, perifosine, and bortezomib reduced the expression of all elevated miRNAs (except miR-206) and increased miRNA-9* whose expression was decreased in WM patients (Figure 6Bii). Moreover, we found that predicted targets of altered miRNAs were reduced after treatment and included oncogenes (RAS oncogene family members and others); protein kinases; transcription factors and positive cell-cycle regulators; as well as antiapoptotic proteins (Table S3). More specifically, we found that bortezomib- and perifosine-treated cells showed miRNA-382–induced downmodulation of 3-phosphoinositide–dependent protein kinase-1 (PIDK1), which has recently been described as an activator of NF-κB and AKT signaling pathways,37 and conversely, miRNA373*-induced up-regulation of the inhibitor of NF-κB (IkB).
Discussion
miRNA expression patterns have been useful in delineating biologic alterations as well prognostic factors in solid and hematologic tumors.5-8 In the present study, we have characterized a specific miRNA signature that distinguishes samples of WM patients from those of control B cells from healthy subjects. The altered WM-related miRNA signature is characterized by increased expression of miRNA-363*, -206, -494, -155, -184, and -542-3p, which was up-regulated in WM samples, and decreased expression of miRNA-9*. The molecular signature of these 7 miRNAs significantly correlated with prognostic factors including age, β2microglobulin, Hb, and IgM levels, and platelet count, according to the IPSS for WM: specifically, the expression of each miRNA was significantly different among patients with worse prognosis compared with those with better prognostic factors, suggesting the importance of miRNA as prognostic markers in this disease. Future studies to prospectively identify the role of this specific miRNA signature as an independent prognostic marker should be conducted to validate our findings.
miRNA-155 is expressed from an exon of the noncoding BIC gene.38 Its role as oncogenic miRNA and its involvement in the initiation and progression of cancers has been reported in several hematologic malignancies, including diffuse large B-cell lymphomas, chronic lymphocytic leukemia, and primary mediastinal B-cell lymphomas, where it has been shown to be highly expressed.9-11 Subsequently, the oncogenic role of miRNA-155 has been confirmed in miRNA-155 transgenic mice, where development of tumors with the characteristics of high-grade lymphoma was observed.39 However, the role of miRNA 155 in WM has not been previously described.
We therefore focused our studies on the biologic role of miRNA-155 in the survival and proliferation of WM cells. We demonstrated that miRNA-155 is a critical regulator of proliferation, but not survival, of WM cells. We therefore hypothesized that miRNA-155 regulates proliferation though cell-cycle transition, and showed using a specific knockdown probe that it blocks G1 to S phase transition, associated with elevated transcripts for p53, p63, and p73, providing a crucial alternate mechanism of cell growth arrest in the absence of p53. Importantly, we found that miRNA-155 plays a key role in targeting both Akt and NF-κB signaling pathways, which have been shown to be constitutively active in primary WM tumor cells.17,33,40 Previous studies have demonstrated a link between NF-κB and PI3K/Akt signaling cascades,41,42 supported by the evidence that after TNFα-induced activation of PI3-kinase and of its downstream target Akt, Akt activation is then responsible for phosphorylation of IKKα and subsequent activation of NF-κB. We may hypothesize that miRNA-155–induced NF-κB modulation could possibly result indirectly from the effect of miRNA-155 on Akt-mediated signaling, and further studies are therefore required.
We next determined the role of miRNA-155 on adhesion and migration of WM cells in vitro and in vivo. We found that knocking down miRNA-155 in WM cells reduced the adhesion and migration in vitro, and significantly decreased proliferation even in the presence of bone marrow stromal cells. In vivo studies confirmed significant inhibition of homing and proliferation of miRNA-155 knockdown WM cells; decreased IgM secretion; and a significant survival benefit in mice. We further confirmed that specific knockdown of miRNA-155 inhibits specific genes regulating cell cycle as well as regulators of adhesion and migration. Together, these studies confirm that miRNA-155 is a critical regulator of proliferation, cell-cycle regulation, migration, and adhesion.
Rituximab, bortezomib, and perifosine represent agents that significantly induce apoptosis and cytotoxicity in WM cells.17 In this study, we demonstrated significant changes in miRNA expression induced by treatment of WM cell line with these agents, suggesting that up- or down-regulation of miRNA expression could mediate, at least in part, effects of drugs on WM tumor cells. Most interestingly, we showed that rituximab, perifosine, and bortezomib induced down-regulation of all increased signature miRNAs (except miRNA-206) and up-regulation of miRNA-9*.
In summary, these data indicate that miRNAs play a pivotal role in the biology of WM; represent important prognostic markers; and provide the basis for the development of new miRNA-targeted therapies for this disease.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by NIH (Washington, DC) grant R21 1R21CA126119-01, International Waldenström Macroglobulinemia Foundation (IWMF), Kirsch Laboratory for Waldenström Macroglobulinemia, Italian Association for Cancer Research (A.M.R.), and Berlucchi Foundation (A.M.R.).
National Institutes of Health
Authorship
Contribution: A.M.R., X.L., K.C.A., B.J.R., K.C.A., I.M.G., and C.D.N. designed the research, performed research, analyzed the data, and wrote the paper; A.M.R., A.S., X.J., H.T.N., and M.R.M. performed research; and A.M.R., J.R., C.C., A.K.A., F.A., N.B., and L.V. analyzed the data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Irene M. Ghobrial, Kirsch Laboratory for Waldenström Macroglobulinemia, Medical Oncology, Dana-Farber Cancer Institute, 44 Binney St, Boston, MA, 02115; e-mail: irene_ghobrial@dfci.harvard.edu.

![Figure 3. miRNA-155 modulates proliferation and cell cycle of WM cells. (A,B) WM cells (miRNA-155 knockdown probe–transfected, control probe–transfected BCWM.1 cells) were harvested at 24 and 48 hours after transfection; DNA synthesis and cytotoxicity were assessed by thymidine uptake and MTT assays, respectively. Nontransfected BCWM.1 cells were used as controls (ctrl). *P < .001; **P < .001. (C) Cells were first arrested and synchronized in G2/M phase by growth in 80 nM nocodazole for 16 hours. Cells were then washed and regrown using fresh media. After 6 hours, cell-cycle analysis was performed by propidium iodide staining. (D) Purified cRNA (15 μg) isolated from WM cells (control probe–transfected, miRNA-155 knockdown probe–transfected BCWM.1 cells) was hybridized to HG-U133Plus2.0 GeneChip (Affymetrix). (i) Up-regulation of cyclin-dependent kinase inhibitors and down-regulation of cyclins and cyclin-dependent kinases. (ii) Up-regulation of p53 and p53 family members and down-regulation of p53 negative regulators. Fold change is shown by the intensity of induction (red) or suppression (blue). (E) BCWM.1 cells (miRNA-155 knockdown probe transfected, control probe transfected) were harvested at 8 hours after transfection. Whole-cell lysates were subjected to Western blotting using anti–phospho (p)-ERK, –p-AKT, -AKT, –p-GSK3α/β, –p-S6R, and -actin antibodies; nontransfected BCWM.1 cells were used as controls (ctrl). (F) BCWM.1 cells (miRNA-155 knockdown probe transfected, control probe transfected) were harvested at 8 hours after transfection and treated with and without TNF-α (10 ng/mL) for 20 minutes; nontransfected BCWM.1 cells were used as control (ctrl). NF-κBp65 transcription factor binding to its consensus sequence on the plate-bound oligonucleotide was measured in nuclear extracts. Wild-type and mutant are wild-type and mutated consensus competitor oligonucleotides, respectively. All results represent means (±] SD) of triplicate experiments. (G) BCWM.1 cells (miRNA-155 knockdown probe transfected, control probe transfected) were harvested at 8 hours after transfection and treated with and without TNF-α (10 ng/mL) for 20 minutes; nontransfected BCWM.1 cells were used as control (ctrl). Immunocytochemical analysis was assessed using anti–p-NF-κBp65 antibody, with DAPI used to stain nuclei.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/18/10.1182_blood-2008-09-178228/6/m_zh80100931820003.jpeg?Expires=1767733575&Signature=4TaA4St0UWEw~pohDCJGPJ18IJeSdkCs1AicsjhCPMZBAcl5fJvG1wIFzi7CWNEWD6oFVyv7JOYdShL3pF8NFyUVMjBRL8nN5VWArI5V01nh66eMVlj2kHYjp~~30Y5gGdIH-ynDiSc730hmMCvriNXiUyC2iP4KoUWcxaS30Z08Hqd6vuz230cIQOrFrjwK8PeNa~PBU2Vne81afpis4ifhOjb7zdRBvhUUuzXhXHdU0TZCTjuD62m9E5~RHM-JZt2755tqGpm1I-bEMSi4iFyAy75uFPdoB5sikT-ZTklruByvw4Q65LOykuGepwUKKWCqAhQ-gzJR7ITiNh9NBg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. miRNA-155 knockdown inhibited WM cells in the context of bone marrow microenvironment in vitro. WM cells (miRNA-155 knockdown probe–transfected, control probe–transfected BCWM.1 cells) were harvested at 24 hours after transfection. Nontransfected BCWM.1 cells were used as control (ctrl). (A) Adhesion assay to fibronectin (FN). Control probe–transfected and nontransfected cells showed significant increase in adhesion to FN, compared with BSA, used as control. miRNA-155 knockdown probe–transfected cells showed lower adhesion to FN. (B) Purified cRNA (15 μg) isolated from WM cells (control probe–transfected, miRNA-155 knockdown probe–transfected BCWM.1 cells) was hybridized to HG-U133Plus2.0 GeneChip (Affymetrix). Down-regulation of several genes involved in the adhesion process. Fold change is shown by the intensity of induction (red) or suppression (blue). (C) Transwell migration assay. SDF-1 (30 nM) was placed in the lower chambers and migration was determined after 4 hours. Control probe–transfected and nontransfected cells showed significant migration with SDF-1 30 nM, whereas the miRNA-155 knockdown probe–transfected cells showed minimal migration in response to SDF-1. (D) BCWM.1 cells were cultured for 48 hours in the presence or absence of BMSCs, and cell proliferation was assessed using [3H]-thymidine uptake assay. All data represent mean (±SD) of triplicate experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/18/10.1182_blood-2008-09-178228/6/m_zh80100931820004.jpeg?Expires=1767733575&Signature=x86pJvwxEHkRD9ymWWtlZgYC7Icb0gumh2KfQB8d8dre8u7z1OzOJFLb8TZK7zuwZrLqYp4V34EsV1kX9o-PvHTI~xDhkd3UMtLqbY0QOxvvuPrGl6aM2fktKCTVyt~BXMPpzFLz-zrpWSW~AdxdsVfEmPNDO0M0MDkEQf1xW84BUqpc2o83st5UBB9Yy7oFEGAcx4PMAWGsjwoY5Tjlg7DKcK49g0JpnZIOyOprwd4GWXXCe3UiXGQSyOILbQpywMr5dV6lzZtxuZNVRpU11a~zJpg0UyyTaep8x89l-44XTJpKPUyX1wK1OCNTV8WCN9waZ54TJ54vVFxSGJ9W6g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 3. miRNA-155 modulates proliferation and cell cycle of WM cells. (A,B) WM cells (miRNA-155 knockdown probe–transfected, control probe–transfected BCWM.1 cells) were harvested at 24 and 48 hours after transfection; DNA synthesis and cytotoxicity were assessed by thymidine uptake and MTT assays, respectively. Nontransfected BCWM.1 cells were used as controls (ctrl). *P < .001; **P < .001. (C) Cells were first arrested and synchronized in G2/M phase by growth in 80 nM nocodazole for 16 hours. Cells were then washed and regrown using fresh media. After 6 hours, cell-cycle analysis was performed by propidium iodide staining. (D) Purified cRNA (15 μg) isolated from WM cells (control probe–transfected, miRNA-155 knockdown probe–transfected BCWM.1 cells) was hybridized to HG-U133Plus2.0 GeneChip (Affymetrix). (i) Up-regulation of cyclin-dependent kinase inhibitors and down-regulation of cyclins and cyclin-dependent kinases. (ii) Up-regulation of p53 and p53 family members and down-regulation of p53 negative regulators. Fold change is shown by the intensity of induction (red) or suppression (blue). (E) BCWM.1 cells (miRNA-155 knockdown probe transfected, control probe transfected) were harvested at 8 hours after transfection. Whole-cell lysates were subjected to Western blotting using anti–phospho (p)-ERK, –p-AKT, -AKT, –p-GSK3α/β, –p-S6R, and -actin antibodies; nontransfected BCWM.1 cells were used as controls (ctrl). (F) BCWM.1 cells (miRNA-155 knockdown probe transfected, control probe transfected) were harvested at 8 hours after transfection and treated with and without TNF-α (10 ng/mL) for 20 minutes; nontransfected BCWM.1 cells were used as control (ctrl). NF-κBp65 transcription factor binding to its consensus sequence on the plate-bound oligonucleotide was measured in nuclear extracts. Wild-type and mutant are wild-type and mutated consensus competitor oligonucleotides, respectively. All results represent means (±] SD) of triplicate experiments. (G) BCWM.1 cells (miRNA-155 knockdown probe transfected, control probe transfected) were harvested at 8 hours after transfection and treated with and without TNF-α (10 ng/mL) for 20 minutes; nontransfected BCWM.1 cells were used as control (ctrl). Immunocytochemical analysis was assessed using anti–p-NF-κBp65 antibody, with DAPI used to stain nuclei.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/18/10.1182_blood-2008-09-178228/6/m_zh80100931820003.jpeg?Expires=1767775333&Signature=owMyM9UMym8Y4TflADNZC5szwhE5UkeqsWdgUWgsYiSkvXHJsOsi6n~LXOxwtCJBpsWgekLrIE9y2hDK94GIs52-ds9rpScbx9J29aOPzYtJXdc-P9iELWVkwyYC6WbbXaabytQuMZrsHGE8WLh2EcImZ10v~F1Wu14TOWZiAIk456JLpqm8prCEh0VIeKqjvSu1U-drKMP153DZq5VSYcoW8JIAwxiKDF-cqIH7PC8Ve-IAKMiBB3iuU0igkWB8W7VKojDMCpFQ5SrnYoHPVxg4n7pI2hxa-vzURgOJrVvT8XjjIEGE9H-BwZNooP-2XB9qTjVn-imkdT83CAsxtw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. miRNA-155 knockdown inhibited WM cells in the context of bone marrow microenvironment in vitro. WM cells (miRNA-155 knockdown probe–transfected, control probe–transfected BCWM.1 cells) were harvested at 24 hours after transfection. Nontransfected BCWM.1 cells were used as control (ctrl). (A) Adhesion assay to fibronectin (FN). Control probe–transfected and nontransfected cells showed significant increase in adhesion to FN, compared with BSA, used as control. miRNA-155 knockdown probe–transfected cells showed lower adhesion to FN. (B) Purified cRNA (15 μg) isolated from WM cells (control probe–transfected, miRNA-155 knockdown probe–transfected BCWM.1 cells) was hybridized to HG-U133Plus2.0 GeneChip (Affymetrix). Down-regulation of several genes involved in the adhesion process. Fold change is shown by the intensity of induction (red) or suppression (blue). (C) Transwell migration assay. SDF-1 (30 nM) was placed in the lower chambers and migration was determined after 4 hours. Control probe–transfected and nontransfected cells showed significant migration with SDF-1 30 nM, whereas the miRNA-155 knockdown probe–transfected cells showed minimal migration in response to SDF-1. (D) BCWM.1 cells were cultured for 48 hours in the presence or absence of BMSCs, and cell proliferation was assessed using [3H]-thymidine uptake assay. All data represent mean (±SD) of triplicate experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/18/10.1182_blood-2008-09-178228/6/m_zh80100931820004.jpeg?Expires=1767775333&Signature=UAP2gwwUNUUpDmIOGtOKl~Movg6sqZmeQ6x2tXr4WiutNF6lLS8g37v~6KEbQG3E6am2~bL0xwnPrzgmduAw6IMfuwhUJvRpGh08YcxdvyxjLSwEesCyQ-CLysFoQ6aNpYJ7bV8wo9m16GISWBmHOYXjgNrChL-DKKC8u0ZQTH~PlVBJJymV8S9mvgFfp5q3O5rfuHpEp~giOGuxf0iVqx4Yx3JB64hV3Sak~tZ-3BrKNJQV7YodGxD5aTMutAudjqIau5haAVR0zc2hEH1vZikf10QaeEH5afjco4gXeLrhkEi5v~n2h5~jeGp654CrsWo7fk4VZeFFnvt5sGSKsw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)