Aberrant methylation of tumor suppressor genes can lead to their silencing in many cancers. TSC-22 is a gene silenced in several solid tumors, but its function and the mechanism(s) responsible for its silencing are largely unknown. Here we demonstrate that the TSC-22 promoter is methylated in primary mouse T or natural killer (NK) large granular lymphocyte (LGL) leukemia and this is associated with down-regulation or silencing of TSC-22 expression. The TSC-22 deregulation was reversed in vivo by a 5-aza-2′-deoxycytidine therapy of T or NK LGL leukemia, which significantly increased survival of the mice bearing this disease. Ectopic expression of TSC-22 in mouse leukemia or lymphoma cell lines resulted in delayed in vivo tumor formation. Targeted disruption of TSC-22 in wild-type mice enhanced proliferation and in vivo repopulation efficiency of hematopoietic precursor cells (HPCs). Collectively, our data suggest that TSC-22 normally contributes to the regulation of HPC function and is a putative tumor suppressor gene that is hypermethylated and silenced in T or NK LGL leukemia.
Introduction
TSC-22 was first isolated from murine osteoblastic cells as a transforming growth factor β (TGF-β)–inducible gene (or TGF-β–stimulated clone).1 The murine homologue (also named Tgfb1i4) encodes a protein of 143 amino acids containing a leucine zipper motif and a N-terminal TSC box that are highly conserved in drosophila, mouse, rat, and human.1,,,–5 Although TSC-22 does not contain a classic DNA binding domain, it can homodimerize or heterodimerize with other transcription factors, repressors, or activators to repress or activate transcription.6,–8 In agreement with this, the TSC-22 protein has been shown to interact with SMAD4 in the U937 cell line.6 TSC-22 is able to inhibit cell growth,9,10 promote apoptosis,11,–13 and stimulate the differentiation of leukemia cell lines into monocytes,14 suggesting that it may act as a putative tumor suppressor gene. TSC-22 has been found to be underexpressed in cancers of the salivary gland, brain, and prostate,15,–17 however, the mechanism(s) by which TSC-22 is down-regulated is largely unknown. Further, genetic disruption of the TSC-22 gene with a functional characterization in an animal model has not been reported.
There is now strong evidence to suggest that aberrant DNA methylation of CpG islands within tumor suppressor genes is important for the development of cancer.18 Unlike genetic control, epigenetic events such as DNA methylation are potentially reversible, making them suitable as targets for both cancer prevention and cancer therapy.19 However, due to the limitation imposed by genetic variability, tumor heterogeneity, availability of genetically identical normal control tissue, and methodologies, the unraveling of methylation and identification of genes targeted by methylation have been slow.20 We previously developed genetically altered mice that constitutively overexpress the proinflammatory cytokine IL-15.21 FVB/NJ IL-15 transgenic (tg) mice have high leukocyte expansion and approximately 30% of them develop an aggressive variant of T or natural killer (NK) large granular lymphocyte (LGL) leukemia compared with wild-type (WT) mice. We previously subjected DNA of cells from WT mice, mice with T-cell and NK cell polyclonal expansion, and mice with T or NK LGL leukemia to a technique called restriction landmark genomic scanning (RLGS) and found that nonrandom aberrant DNA hypermethylation across the mouse genome was associated with T or NK LGL leukemia but not with the T or NK cells in WT mice or in mice with polyclonal expansion.20,22 DNA sequencing indicated that one spot lost in T or NK LGL leukemia with particularly high frequency represented the partial promoter of the TSC-22D1 gene. TSC-22D1 has a genomic sequence of approximately 100 kb but has not been well characterized. Interestingly, TSC-22 is located within TSC-22D1 at the 3′ end and, as noted, has been shown to be down-regulated or silenced in various solid tumors. Our preliminary data also show that TSC-22 is regulated by proinflammatory cytokines. We therefore decided to study this gene in our IL-15tg mouse model of T or NK LGL leukemia.
Here, we show alterations of TSC-22 expression during leukemic transformation. TSC-22 is fully expressed in WT splenocytes and in the expanded, polyclonal T and NK cells, but is underexpressed or silenced in the majority of IL-15tg T or NK LGL leukemia. We provide evidence that this silencing of TSC-22 is associated with methylation of its promoter, and show that this silencing is reversed in vivo with 5-aza-2′-deoxycytidine (5-Aza) therapy in T or NK LGL leukemia resulting in improved survival. Finally, we show that targeted disruption of TSC-22 in mice led to higher proliferation and in vivo repopulation efficiency of hematopoietic precursor cells (HPCs). Collectively, our in vitro and in vivo data support the notion of TSC-22 as a putative tumor suppressor gene with normal regulatory effects on WT HPC proliferation and repopulation. Its underexpression is associated with methylation of its promoter and with the genesis and maintenance of T or NK LGL leukemia.
Methods
Mouse T or NK LGL leukemia samples and cell lines
Mouse T or NK LGL leukemia samples had more than 70% leukemic blasts and were derived from IL-15tg mice. The mouse leukemia (L1210), lymphoma (El-4, TK-1, YAC-1), and plasmacytoma (P1.17) cell lines were purchased from ATCC (Manassas,VA).
Combined bisulfite restriction analysis and bisulfite sequencing
Treatment of genomic DNA with sodium bisulfite has been previously described23 and polymerase chain reaction (PCR) amplifications were performed in 50-μL reaction volumes for 35 cycles. PCR products for combined bisulfite restriction analysis (COBRA)24 of DNA extracted from aforementioned malignant cell lines or fluorescence-activated cell sorting (FACS)–sorted (≥ 95% purity) leukemia cells (DX5+CD3+CD5− for T LGL or DX5+CD3− for NK LGL from leukemic mice) or normal cells (DX5+CD3+, CD5+CD3+, or DX5+CD3− from nonleukemic mice) were purified with QIAquick gel extraction kit (QIAGEN, Valencia, CA), digested with CG-containing restriction enzyme HpyCH4 IV, separated on nondenaturing 8% polyacrylamide gel (PAGE) or 2% agarose gel, stained with ethidium bromide, and visualized under ultraviolet light. For bisulfite sequencing, PCR products were gel-extracted, cloned into the pCR2.1 TOPO vector (Invitrogen, Carlsbad, CA), and sequenced using an ABI Prism 3700 DNA sequencer (Applied Biosystems, Foster City, CA). COBRA primers are available on request.
Treatment of cell lines with 5-Aza in vitro
Cells were treated with 1 to 5 μM 5-Aza, 300 nM trichostatin A, or their combination (Sigma-Aldrich, St Louis, MO) as previously described.20
Regular and real-time RT-PCR
Regular reverse-transcription (RT)–PCR for murine TSC-22 was performed using cDNA template with varied cycles to ensure that the amplification product was in the linear range. Amplification of hypoxanthine phosphoribosyl transferase (HPRT) was included as an internal control. Real-time RT-PCR of murine TSC-22 was performed with primer/probe sets specifically for an exon of the gene. 18S rRNA reactions were included as an internal control. Data were analyzed as previously described.25,26 All primer and probe sequences are available on request.
Plasmid construction, in vitro DNA methylation, and luciferase reporter assays
Plasmid construction and in vitro DNA methylation were detailed in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Luciferase reporter assays in 293T cells were performed as previously described.25
In vivo treatment of T or NK LGL leukemia with 5-Aza
Eight- to 12-week-old C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME). For adoptive transfer of leukemic cells, recipient mice were subjected to a sublethal dose of 6.5 Gy from a gamma source. A total of 106 cells from either freshly isolated or viably frozen aliquots of T or NK LGL leukemia cells was resuspended in 0.5 mL RPMI media without supplements and injected by tail vein. Leukemia in the recipient mice was detected by serial monitoring of total white blood cell count and flow cytometric analysis, and was usually fully established within 4 weeks of adoptive transfer (white blood cell count [WBC] ∼ 10 × 109/L [10 000/μL], with ∼ 25% blasts in blood by flow cytometry). Then, 12 or 32 μg 5-Aza (Sigma-Aldrich) was resuspended in 200 μL PBS and administrated subcutaneously to the mice bearing leukemia, once a day for 4 consecutive days for either one cycle or a second 4-day cycle administered 4 or 8 days after completion of the last dose from cycle 1. For control mice bearing leukemia, 200 μL PBS alone was injected using an identical schedule to experimental animals. After the treatment, leukemic cells (identified as DX5+CD3+CD5− for T LGL or DX5+CD3− for NK LGL) were enriched to 95% purity using a FACSVantage cell sorter (BD Biosciences, San Jose, CA). The enriched leukemia cells from mice treated with 32 μg 5-Aza or PBS were used to make cDNA for real-time RT-PCR analysis. Experiments were approved by Institutional Animal Care and Use Committee of The Ohio State University.
Retroviral infection of cell lines
Retroviral transduction of TSC-22-pMSCV-IRES-GFP-PURO (TSC-22-pMSCV) and the vector alone, followed by cell sorting, was performed following previously published protocols.25,,,–29 Briefly, Phoenix cells were transiently transfected with TSC-22-pMSCV or the vector alone and supernatants were collected 48 hours after transfection and used for 3 cycles of infections. Upon infection, cell lines (eg, YAC-1 and L1210) were sorted by FACSVantage for green fluorescent protein (GFP) expression to at least 99% purity. Overexpression of TSC-22 in sorted populations was confirmed by Western blotting and/or real-time RT-PCR.
Cell growth and tumorigenicity assay
The sorted pMSCV- or TSC-22-pMSCV–infected bulk cells or clones derived from them were used for in vitro cell growth and in vivo tumorigenicity assays. Cells in log-growth phase were plated in triplicate in 6-well plates at the intensity of 104/mL and cultured in RPMI-1640 medium with 1% FBS (YAC-1) or DMEM medium with 5% FBS (L1210) for assessment of in vitro cell growth. Cells were counted 2, 4, 6, and 8 days after plating using trypan blue (Invitrogen) exclusion. The tumorigenicity assay was performed twice with a total of at least 12 mice for each cell line following previously described methods.20
Cell lysis and immunoblotting
Cells lysis and Western blotting were performed as previously described.26 The antibodies used were as follows: rabbit anti–human TSC-22 polyclonal antibody (a kind gift of Drs Hitoshi Kawamata [Dokkyo University School of Medicine, Tochigi, Japan] and Marco G. Cecchini [University of Bern, Bern, Switzerland]); mouse anti–MYC-tag monoclonal antibody (Cell Signaling Technology, Beverly, MA); goat polyclonal anti–β-ACTIN antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
Targeting vector for the generation of TSC-22–deficient mice and genotyping analysis
The genomic clone encoding murine TSC-22 was isolated from SVJ129 genomic library (Stratagene, La Jolla, CA) and was mapped by restriction analyses. A targeting vector was constructed by ligating a 1.7-kb left arm fragment (SacI-XhoI) of TSC-22, a 1.1-kb TK-Neo-cassette (XhoI-BamHI) from a pMC1neo plasmid (Stratagene), an 18-bp adaptor to accommodate incompatible cohesive ends (BamHI-KpnI-XbaI; XbaI is compatible to SpeI), and a 6.3-kb right arm fragment (SpeI-NsiI) of TSC-22. This targeting vector resulted in a replacement of a 1.9-kb XhoI-SpeI fragment containing entire coding region of TSC-22 with the TK-neo cassette through homologous recombination. During ES clone screening, the successful recombination was consecutively confirmed to occur on the left arm and on the right arm, which shows clearer results compared with confirming both arms simultaneously. Genotyping of TSC-22 mice was detailed in Document S2.
In vivo BrdU incorporation assay
Three littermate pairs of TSC-22−/− and WT C57BL/6 strain mice (8-11 weeks old) were given BrdU drinking water changed daily at a concentration of 0.8 mg/mL for 7 consecutive days prior to killing. To determine BrdU incorporation of Lin−CD117+ cells, following killing of the mice, isolated BM cells were stained with CD3-PE, CD11b-PE, CD5-PE, TER119-PE, B220-PE, GR1-PE, NK1.1-PE, CD117-APC, and BrdU-FITC according to the manufacturer's instruction (BD Biosciences).
In vivo repopulation efficiency assay
BM cells from TSC-22+/+ and TSC-22−/− mice aged 8 to 12 weeks (varied cell number, CD45.2) were mixed at 50%:50% ratio and were adoptively transferred into the lethally irradiated (∼ 9 Gy from a gamma source) recipient mice (CD45.1) by tail-vein injection. Three or 6 months after the initial transfer, peripheral blood was collected and the percentage of CD45.2 lineage cells of total lineage cells was analyzed by FACS. Three controlled experiments for the in vivo repopulation efficiency assay are shown in Table 1.
Statistics
The χ2 test was applied to analyze in vivo competitive repopulation assay. Other data were compared using the Student 2-tailed t test. A P value less than .05 was considered statistically significant.
Results
Evidence for methylation of the TSC-22 promoter in murine T or NK LGL
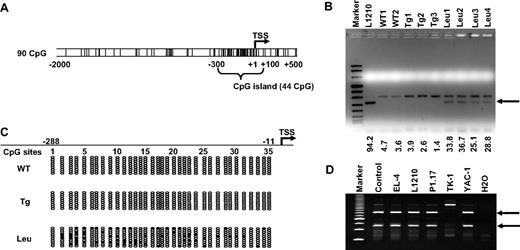
Our previous RLGS analysis20 indicated that a certain spot loss corresponded to the TSC-22D1 proximal promoter, which is located approximately 100-kb upstream of TSC-22 (Figure S1A). The absence of a Not1 methylation-sensitive restriction site in the TSC-22 5′ region limited our access to this gene.20,30 We first examined the TSC-22 promoter sequence using the program EMBOSS31 (Wellcome Trust, Cambridge, United Kingdom) and found a CpG island located between −300 and +100 (Figure 1A). We then performed COBRA analysis within this region on FACS-sorted T or NK LGL leukemia cells from 4 IL-15tg FVB/NJ leukemic mice, and on FACS-sorted normal T or NKT cells from 3 IL-15tg FVB/NJ mice with polyclonal T and NK expansion or from 2 WT FVB/NJ mice. We found that the band indicating TSC-22 promoter methylation appeared in leukemia samples yet was absent in all WT or polyclonal samples (Figure 1B arrow). Our bisulfite sequencing analysis confirmed that methylation in the 5′ regulatory region of TSC-22 preferentially occurred in leukemia samples but not in IL-15tg polyclonal expansion or WT samples (Figure 1C). Finally, we analyzed the methylation status of the TSC-22 promoter by COBRA in EL-4, L1210, P1.17, TK-1, and YAC-1 mouse hematologic malignancy cell lines and found evidence for methylation in all but one of these cell lines (TK-1; Figure 1D). The lack of evidence in the TK-1 cell line might be due to the absence of CpG methylation in this small region detected by COBRA within the TSC-22 promoter or to the absence of methylation on the entire TSC-22 promoter and full expression of the gene.
Methylation of the TSC-22 promoter in mouse T or NK LGL leukemia. (A) Examination of sequences within the murine TSC-22 promoter shows a CpG island located from −300 to +100 (G + C, > 50%; length, > 200; observed/expected ratio, > 0.6). The vertical lines are indicative of CG dinucleotides; TSS indicates transcription start site. (B) Leukemic cells were enriched by flow cytometry (≥ 95%) from spleens of the representative mice with T or NK LGL leukemia. Data of the L1210 T-cell lymphoma cell line and of nonleukemic cells (enriched from WT mice or IL-15tg mice with polyclonal T- and NK-cell expansion) were included for comparison. Combined bisulfite restriction analysis (COBRA) of DNA from these malignant and nonmalignant populations provided evidence of TSC-22 promoter methylation (arrow) in L1210 cell line and in the T or NK LGL leukemia cells (Leu) but not in control WT cells (WT) or in cells from IL-15tg mice with polyclonal T- and NK-cell expansion (Tg). Numbers below the agarose gel represent the intensity of the methylated band, normalized by the upper unmethylated band. (C) Bisulfite sequencing of the TSC-22 promoter region from WT, IL-15tg polyclonal, and T or NK LGL leukemia from mouse spleens. Each row of ovals represents the sequence of an individual clone. Unshaded ovals indicate unmethylated CpG site; shaded ovals, methylated CpG site; TSS, transcription start site, which is designed as +1. (D) COBRA analysis of DNA from the TSC-22 promoter region in 5 murine cell lines derived from hematologic malignancies. The 2 digested fragments (arrows) correspond to methylated DNA separated on a PAGE gel.
Methylation of the TSC-22 promoter in mouse T or NK LGL leukemia. (A) Examination of sequences within the murine TSC-22 promoter shows a CpG island located from −300 to +100 (G + C, > 50%; length, > 200; observed/expected ratio, > 0.6). The vertical lines are indicative of CG dinucleotides; TSS indicates transcription start site. (B) Leukemic cells were enriched by flow cytometry (≥ 95%) from spleens of the representative mice with T or NK LGL leukemia. Data of the L1210 T-cell lymphoma cell line and of nonleukemic cells (enriched from WT mice or IL-15tg mice with polyclonal T- and NK-cell expansion) were included for comparison. Combined bisulfite restriction analysis (COBRA) of DNA from these malignant and nonmalignant populations provided evidence of TSC-22 promoter methylation (arrow) in L1210 cell line and in the T or NK LGL leukemia cells (Leu) but not in control WT cells (WT) or in cells from IL-15tg mice with polyclonal T- and NK-cell expansion (Tg). Numbers below the agarose gel represent the intensity of the methylated band, normalized by the upper unmethylated band. (C) Bisulfite sequencing of the TSC-22 promoter region from WT, IL-15tg polyclonal, and T or NK LGL leukemia from mouse spleens. Each row of ovals represents the sequence of an individual clone. Unshaded ovals indicate unmethylated CpG site; shaded ovals, methylated CpG site; TSS, transcription start site, which is designed as +1. (D) COBRA analysis of DNA from the TSC-22 promoter region in 5 murine cell lines derived from hematologic malignancies. The 2 digested fragments (arrows) correspond to methylated DNA separated on a PAGE gel.
Silent or reduced expression of TSC-22 in lymphoid malignancy and its reversal with 5-Aza in vitro
We used RT-PCR to determine whether TSC-22 promoter methylation altered gene expression in T or NK LGL leukemic mouse samples and in the malignant cell lines. Among the 6 primary T or NK LGL leukemic samples, 5 had absent or very low levels of TSC-22 expression (Figure 2A). Expression of TSC-22 in YAC-1 and EL-4 mouse lymphoma cell lines, which show TSC-22 promoter methylation, was also silenced (Figure 2B). Treatment of these cell lines with the demethylating agent 5-Aza restored TSC-22 expression (Figure 2B).
Reduced or absent TSC-22 expression in hematologic malignant tissue is reversed by the demethylation agent 5-Aza in vitro. (A) Expression of TSC-22 detected by regular RT-PCR in WT, polyclonal IL-15tg (Tg), and primary T or NK LGL leukemia (Leu) mouse splenocytes. (B) RT-PCR of the murine lymphoid tumor cell lines YAC-1 and EL-4 indicated that TSC-22 mRNA was undetectable in untreated cells. Treatment with the demethylation agent 5-Aza restored TSC-22 expression in a dose-dependent fashion. Treatment with trichostatin A (TSA, 300 nM), a histone deacetylase inhibitor, did not have a significant effect on TSC-22 transcription. Amplification of HPRT was included as an internal control.
Reduced or absent TSC-22 expression in hematologic malignant tissue is reversed by the demethylation agent 5-Aza in vitro. (A) Expression of TSC-22 detected by regular RT-PCR in WT, polyclonal IL-15tg (Tg), and primary T or NK LGL leukemia (Leu) mouse splenocytes. (B) RT-PCR of the murine lymphoid tumor cell lines YAC-1 and EL-4 indicated that TSC-22 mRNA was undetectable in untreated cells. Treatment with the demethylation agent 5-Aza restored TSC-22 expression in a dose-dependent fashion. Treatment with trichostatin A (TSA, 300 nM), a histone deacetylase inhibitor, did not have a significant effect on TSC-22 transcription. Amplification of HPRT was included as an internal control.
In vitro methylation of the TSC-22 promoter is associated with lower gene expression
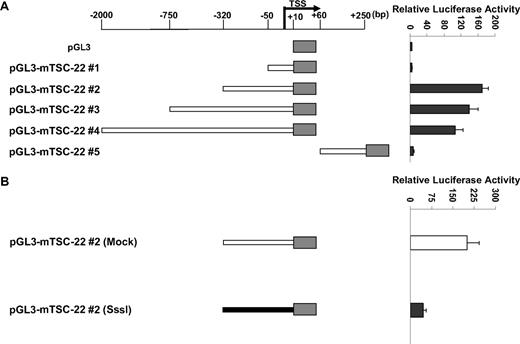
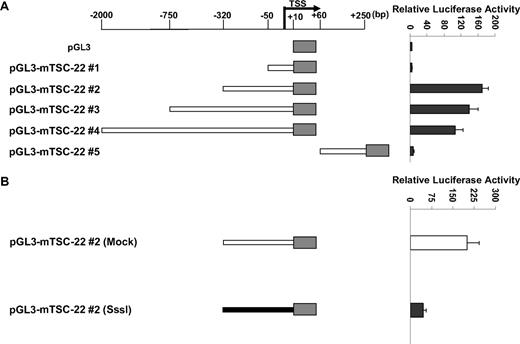
As noted, the TSC-22 promoter is methylated only in primary mouse IL-15tg T or NK LGL leukemia and in some lymphoma and leukemia murine cell lines but not in WT cells or in polyclonal T or NK cells expanded in IL-15tg mice (Figure 1B-D), and this methylation is associated with decreased or absent gene expression (Figure 2). We next assessed the extent and direct effect of TSC-22 promoter methylation on gene expression. Five reporter constructs containing different portions of the unmethylated TSC-22 promoter sequences were cloned (position −2100 bp to +250 bp relative to transcription start site; Figure 3A). Construct pGL3-mTSC-22#2 (−320 bp to +10 bp) was found to contain the maximum promoter activity in 293T cells. To determine the role of DNA methylation in regulation of TSC-22 expression, the promoter sequences from construct pGL3-mTSC-22#2 were excised by restriction digestion, and treated with or without SssI methylase. The methylated and unmethylated DNA fragments were then religated with the unmethylated pGL3 vector. Luciferase assays indicated that promoter activity of the methylated construct was approximately 4.5 times lower than that of the unmethylated construct and showed a significant difference (P < .05; Figure 3B).
TSC-22 promoter hypermethylation attenuates gene expression. (A) Luciferase reporter assays in 293T cells transfected with variable lengths of the mouse (m) TSC-22 promoter indicate that strong promoter activity occurs within the −320-bp proximal region. (B) In vitro methylation of the −320-bp promoter of TSC-22 with Sss1 DNA methyltransferase (shaded bar) indicates that methylation of the −320-bp promoter region significantly inhibits TSC-22 promoter activity, which was detected by luciferase assays, compared with an unmethylated control construct (unshaded bar; P < .05). Error bars indicate standard deviations for triplicates in 1 of 3 representative experiments (A,B).
TSC-22 promoter hypermethylation attenuates gene expression. (A) Luciferase reporter assays in 293T cells transfected with variable lengths of the mouse (m) TSC-22 promoter indicate that strong promoter activity occurs within the −320-bp proximal region. (B) In vitro methylation of the −320-bp promoter of TSC-22 with Sss1 DNA methyltransferase (shaded bar) indicates that methylation of the −320-bp promoter region significantly inhibits TSC-22 promoter activity, which was detected by luciferase assays, compared with an unmethylated control construct (unshaded bar; P < .05). Error bars indicate standard deviations for triplicates in 1 of 3 representative experiments (A,B).
5-Aza therapy results in the reversal of TSC-22 silencing and improves survival of mice bearing T or NK LGL leukemia
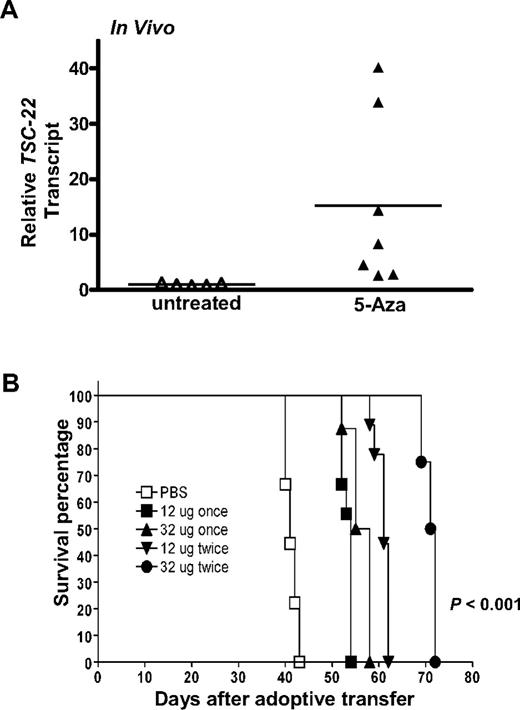
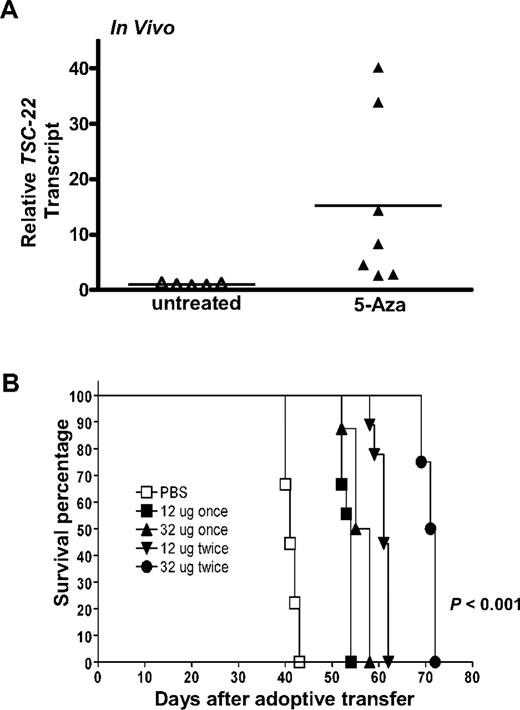
Given that a portion of the TSC-22 promoter is methylated in T or NK LGL leukemia samples and is associated with diminished or absent TSC-22 gene expression, and given that this was reversed in vitro by treatment with 5-Aza, we next tested whether in vivo treatment with 5-Aza could reverse silenced TSC-22 expression and improve survival of the mice. The idea of improved survival came from our previous study of T or NK LGL leukemia that showed a nonrandom, genome-wide pattern of abnormal methylation in the 5′ regulatory regions of 50 to 200 genes.20 To this end, groups of 9 mice received a transplant of 106 primary T or NK LGL leukemia. When symptomatic with tachypnea, decreased activity, and an expanded WBC, each mouse was treated with either 12 or 32 μg 5-Aza daily for 4 days for either one cycle or a second identical cycle 4 or 8 days later. Leukemic cells obtained from the mice treated with 32 μg 5-Aza for 4 days were enriched to 95% purity by cell sorting and showed a significant up-regulation of TSC-22 expression (P < .05; Figure 4A).
In vivo 5-Aza therapy reveres silenced TSC-22 gene expression and prolongs survival in mice bearing T or NK LGL leukemia. (A) In vivo treatment of mice moribund with T or NK LGL leukemia using PBS (“untreated,” ▵) or 5-Aza (▴) for 4 consecutive days shows a significant reversal, with up-regulation of TSC-22 expression as assessed by real-time RT-PCR (P < .05). Horizontal bars indicate mean values. The TSC-22 expression levels of the untreated samples were normalized to 1. (B) One million primary T or NK LGL leukemia cells from IL-15tg mice were adoptively transferred into sublethally irradiated WT FvB mouse recipients. When engrafted mice showed progressive disease, they received daily subcutaneous injections with either 1 or 2 cycles of the indicated dose of 5-Aza or PBS for 4 consecutive days. Mice receiving 2 cycles of therapy had 4 days of rest between cycles at doses of 12 μg or 8 days of rest between cycles at doses of 32 μg. As shown by the Kaplan-Meier curve, mice given increasing doses and/or cycles of 5-Aza displayed prolonged survival compared with control mice treated with PBS (P < .001 among all groups).
In vivo 5-Aza therapy reveres silenced TSC-22 gene expression and prolongs survival in mice bearing T or NK LGL leukemia. (A) In vivo treatment of mice moribund with T or NK LGL leukemia using PBS (“untreated,” ▵) or 5-Aza (▴) for 4 consecutive days shows a significant reversal, with up-regulation of TSC-22 expression as assessed by real-time RT-PCR (P < .05). Horizontal bars indicate mean values. The TSC-22 expression levels of the untreated samples were normalized to 1. (B) One million primary T or NK LGL leukemia cells from IL-15tg mice were adoptively transferred into sublethally irradiated WT FvB mouse recipients. When engrafted mice showed progressive disease, they received daily subcutaneous injections with either 1 or 2 cycles of the indicated dose of 5-Aza or PBS for 4 consecutive days. Mice receiving 2 cycles of therapy had 4 days of rest between cycles at doses of 12 μg or 8 days of rest between cycles at doses of 32 μg. As shown by the Kaplan-Meier curve, mice given increasing doses and/or cycles of 5-Aza displayed prolonged survival compared with control mice treated with PBS (P < .001 among all groups).
Leukemic mice treated with a single cycle of 12 μg 5-Aza survived significantly longer than mice treated with vehicle control (Figure 4B, 56.1 days vs 41.3 days, P < .001), yet increasing the dose of 5-Aza for single cycle of treatment did not further improve survival. Mice treated for 2 cycles at a dose of 12 μg survived significantly longer than mice that received only one cycle at the same dose (60.9 days vs 53.2 days, P < .001), and mice treated for 2 cycles at the higher dose of 32 μg survived significantly longer than all other groups (P < .001 among all groups).
Transduction of TSC-22 in lymphoma and leukemia cells suppresses tumorigenicity
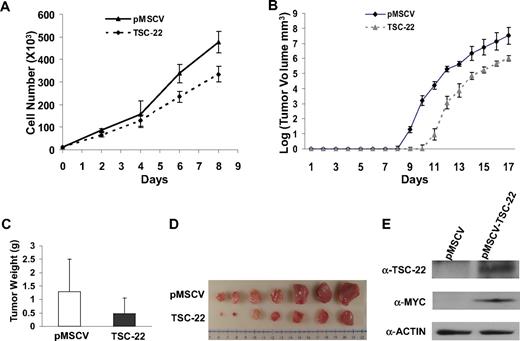
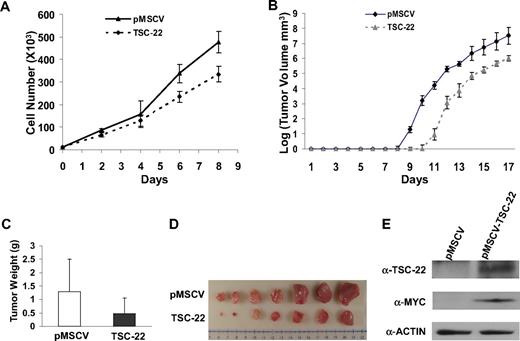
Given the number of genes likely silenced during the genesis of T or NK LGL leukemia in these IL-15tg mice,20 it seems unlikely that the silencing of TSC-22 alone is sufficient to cause leukemia. We therefore investigated whether TSC-22 itself had any effect on growth or survival of leukemic cells. We hypothesized that TSC-22 is a silenced putative tumor suppressor gene whose re-expression or overexpression would therefore have an adverse effect on leukemia or lymphoma cell growth. To test this, we cloned TSC-22 cDNA into a retroviral vector and transfected it or the mock vector into the murine lymphoma cell line YAC-1 and the murine leukemia cell line L1210, each of which has methylation of the TSC-22 promoter. After confirming overexpression (not shown), we performed an in vitro cell growth experiment that showed significant inhibition of tumor cell growth in cells overexpressing TSC-22 compared with the cells transfected with the mock vector in both cell lines (P < .05; Figure 5A and Figure S2A).
Ectopic expression of TSC-22 in malignant cells inhibits cell proliferation and delays tumor formation in vivo. (A) Growth of YAC-1 cells transfected with a pMSCV vector containing TSC-22 exhibited significant inhibition of proliferation in vitro compared with YAC-1 cells transfected with the empty vector alone (P < .05). This experiment was repeated 3 times with similar results. (B) In vivo tumorigenesis resulting from subcutaneous injection of YAC-1 cells ectopically expressing TSC-22 or the pMSCV vector alone into NOD-SCID mice. Tumor size was measured daily. Overexpression of TSC-22 in YAC-1 cells delays the onset of tumor formation in vivo by an average of 2 days (P < .05). (C) The weights of tumors collected from mice killed at the end of the in vivo experiment (day 17). The average weight of tumors formed from YAC-1 cells ectopically expressing TSC-22 was less than that of tumors formed from the YAC-1 cells transfected with the vector alone (P < .05). (D) Photograph of the representative tumor pairs excised from killed mice that had been injected with YAC-1 cells ectopically expressing the pMSCV vector alone (top) or TSC-22 (bottom). (E) Western blot detection of the TSC-22 protein levels in tumors derived from the injection of YAC-1 cells ectopically expressing the pMSCV vector alone or TSC-22 into NOD-SCID mice. Presence of MYC-tag indicates ectopic TSC-22 expression. The tumorigenicity assay in panels B-E was performed twice with a total of 16 mice. Injected transfected YAC-1 tumor cells were first purified to 99% or more purity by cell sorting for GFP. Error bars in panels A-C indicate SDs (n ≥ 3) in one representative experiment.
Ectopic expression of TSC-22 in malignant cells inhibits cell proliferation and delays tumor formation in vivo. (A) Growth of YAC-1 cells transfected with a pMSCV vector containing TSC-22 exhibited significant inhibition of proliferation in vitro compared with YAC-1 cells transfected with the empty vector alone (P < .05). This experiment was repeated 3 times with similar results. (B) In vivo tumorigenesis resulting from subcutaneous injection of YAC-1 cells ectopically expressing TSC-22 or the pMSCV vector alone into NOD-SCID mice. Tumor size was measured daily. Overexpression of TSC-22 in YAC-1 cells delays the onset of tumor formation in vivo by an average of 2 days (P < .05). (C) The weights of tumors collected from mice killed at the end of the in vivo experiment (day 17). The average weight of tumors formed from YAC-1 cells ectopically expressing TSC-22 was less than that of tumors formed from the YAC-1 cells transfected with the vector alone (P < .05). (D) Photograph of the representative tumor pairs excised from killed mice that had been injected with YAC-1 cells ectopically expressing the pMSCV vector alone (top) or TSC-22 (bottom). (E) Western blot detection of the TSC-22 protein levels in tumors derived from the injection of YAC-1 cells ectopically expressing the pMSCV vector alone or TSC-22 into NOD-SCID mice. Presence of MYC-tag indicates ectopic TSC-22 expression. The tumorigenicity assay in panels B-E was performed twice with a total of 16 mice. Injected transfected YAC-1 tumor cells were first purified to 99% or more purity by cell sorting for GFP. Error bars in panels A-C indicate SDs (n ≥ 3) in one representative experiment.
We next compared in vivo tumorigenicity of these transfected cell lines by subcutaneously injecting purified bulk cells into the flanks of nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice and measuring tumor growth. We found that tumor formation was significantly delayed for 2 days in the mice injected with YAC-1 cells expressing exogenous TSC-22 compared with the mice bearing YAC-1 cells transfected with the vector alone (P < .05; Figure 5B). This was confirmed by the weight and volume of tumors collected from killed mice at the end of the 17 days (P < .05; Figure 5C,D). Overexpression of TSC-22 in the TSC-22--transfected cells was confirmed by Western blotting using both an anti–MYC-tag antibody and an anti–TSC-22 antibody (Figure 5E). The in vivo tumorigenicity experiment was repeated using 3 pairs of clones generated from the TSC-22 and mock-transfected YAC-1 cells, and with an additional cell line (L1210), along with a different mouse strain (CB-17 SCID). All results were similar to those shown in Figure 5 (Figures S2B,C and S3, and data not shown).
Generation of TSC-22–deficient mice reveals an increase in proliferation and repopulation activity of HPCs
Our data suggest that TSC-22 is a putative tumor suppressor gene in leukemia. As further evidence, we next investigated whether TSC-22 had any effect on growth of normal HPCs. Targeted disruption of TSC-22 was undertaken (Figure 6). The embryonic stem cells (line ES 14.1) derived from SVJ129 strain mice were electroporated with a TSC-22 targeting construct (Figure 6A) and selected as described previously.32 Five of 401 G418-resistant clones analyzed by Southern blotting carried a TSC-22 allele disrupted by homologous recombination (Figure 6B). TSC-22–targeted embryonic stem cell clones were injected into C57BL6/J blastocysts and transferred into pseudopregnant mice to obtain chimeric mice. Resulting male chimeras were bred with C57BL/6 females and offspring were analyzed for germline transmission of the mutant allele by Southern blot and PCR analyses of genomic DNA. Mice heterozygous for mutation (TSC-22+/−) were intercrossed to yield TSC-22−/− mice. The lack of TSC-22 expression in TSC-22−/− mice was confirmed by Northern hybridization of brain RNA (Figure 6C left panel) and by Western blotting of proteins isolated from BM cells (Figure 6C right panel). TSC-22+/− mice were backcrossed with C57BL/6 mice for at least 5 generations for further experiments.
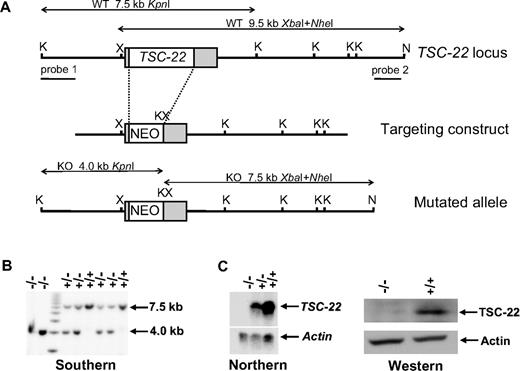
Generation of TSC-22–deficient mice. (A) Schematic representation of the targeting construct and homologous recombination in the TSC-22 region. Restriction enzyme sites of KpnI, NheI, and XbaI at the TSC-22 locus are indicated as K, N, and X, respectively. The probes used in Southern screening of embryonic stem cell and the expected sizes of endogenous and mutated fragments obtained by KpnI single digestion or XbaI and NheI double digestion are shown. (B) Southern blot analysis of KpnI-digested DNA extracted from the littermates derived from a cross between heterozygous TSC-22 mice by using probe 1 (A). The 7.5-kb band indicates the existence of the TSC-22 allele, whereas the 4.0-kb band shows the allele loss. The double digestion of XbaI and NheI followed by hybridization with probe 2 (A) gives rise to a 9.5-kb fragment for the existence of the TSC-22 allele and a 7.5-kb fragment for the allele loss (not shown). (C) Northern blotting (left panel) and Western blotting (right panel) to confirm the lack of TSC-22 expression in the TSC-22−/− mice.
Generation of TSC-22–deficient mice. (A) Schematic representation of the targeting construct and homologous recombination in the TSC-22 region. Restriction enzyme sites of KpnI, NheI, and XbaI at the TSC-22 locus are indicated as K, N, and X, respectively. The probes used in Southern screening of embryonic stem cell and the expected sizes of endogenous and mutated fragments obtained by KpnI single digestion or XbaI and NheI double digestion are shown. (B) Southern blot analysis of KpnI-digested DNA extracted from the littermates derived from a cross between heterozygous TSC-22 mice by using probe 1 (A). The 7.5-kb band indicates the existence of the TSC-22 allele, whereas the 4.0-kb band shows the allele loss. The double digestion of XbaI and NheI followed by hybridization with probe 2 (A) gives rise to a 9.5-kb fragment for the existence of the TSC-22 allele and a 7.5-kb fragment for the allele loss (not shown). (C) Northern blotting (left panel) and Western blotting (right panel) to confirm the lack of TSC-22 expression in the TSC-22−/− mice.
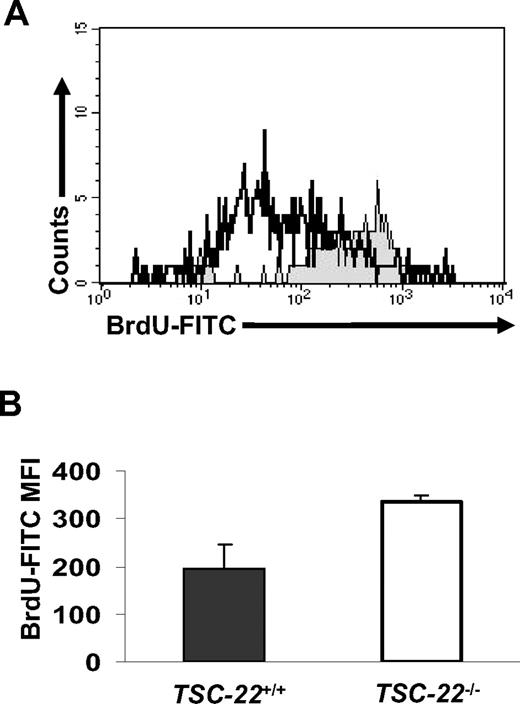
Complete phenotyping of 3 TSC-22−/− mice and their littermate TSC-22+/+ mice revealed an approximately 13% to 15% weight decrease of kidneys and hearts in TSC-22−/− mice measured by absolute weight or the percentage of body weight. To compare the in vivo proliferative ability of TSC-22+/+ and TSC-22−/− Lin−CD117+ BM cells, littermate pairs of TSC-22+/+ and TSC-22−/− mice received BrdU-containing drinking water for 7 days, and fresh Lin−CD117+ BM cells were analyzed for BrdU incorporation by FACS. Figure 7A and B show that Lin−CD117+ BM cells from TSC-22−/− mice had significantly higher BrdU incorporation compared with Lin−CD117+ BM cells from TSC-22+/+ mice (P < .05, n = 3), suggesting a higher rate of proliferation for TSC-22−/− Lin−CD117+ BM cells in vivo. When BM cells from TSC-22−/− and TSC-22+/+ mice were mixed together in a 50%:50% ratio, injected into lethally irradiated TSC-22+/+ recipient mice, and then followed for 3 or 6 months, 70% or 76% of the cells, respectively, were derived from the TSC-22−/− BM (Table 1, P < .01, n ≥ 10). This suggests that the TSC-22−/− HPCs have a higher repopulation efficiency compared with the TSC-22+/+ HPCs. Collectively, these in vivo data support the notion that TSC-22 has a role in regulation of normal HPCs, in that HPCs from TSC-22−/− mice have enhanced in vivo proliferative and repopulation abilities compared with those from TSC-22+/+ mice.
Lin−CD117+ BM cells from TSC-22−/− mice show higher proliferation than those from TSC-22+/+ mice in vivo. (A) Representative example of BM cells obtained from TSC-22+/+ and TSC-22−/− mice after ingesting BrdU-containing drinking water for 7 consecutive days. BM cells were stained for Lin markers, CD117, and BrdU and then were analyzed by flow cytometry. The region shaded in gray represents Lin−CD117+ BM cells from TSC-22−/− mice and shows higher BrdU incorporation than identical BM cells obtained from TSC-22+/+ mice (open area, black line). (B) Summary of data represented in panel A but performed on 3 pairs of littermate mice. There is significantly higher BrdU incorporation in the TSC-22−/− Lin−CD117+ BM cells compared with those of TSC-22+/+ mice (P < .05, n = 3). Error bars indicate standard errors for 3 independent experiments.
Lin−CD117+ BM cells from TSC-22−/− mice show higher proliferation than those from TSC-22+/+ mice in vivo. (A) Representative example of BM cells obtained from TSC-22+/+ and TSC-22−/− mice after ingesting BrdU-containing drinking water for 7 consecutive days. BM cells were stained for Lin markers, CD117, and BrdU and then were analyzed by flow cytometry. The region shaded in gray represents Lin−CD117+ BM cells from TSC-22−/− mice and shows higher BrdU incorporation than identical BM cells obtained from TSC-22+/+ mice (open area, black line). (B) Summary of data represented in panel A but performed on 3 pairs of littermate mice. There is significantly higher BrdU incorporation in the TSC-22−/− Lin−CD117+ BM cells compared with those of TSC-22+/+ mice (P < .05, n = 3). Error bars indicate standard errors for 3 independent experiments.
Discussion
Previous reports have shown that TSC-22 is down-regulated in salivary gland, brain, and prostate tumors,15,–17 but evidence for its down-regulation in leukemia and the mechanism(s) by which it is down-regulated have not been clearly described. Moreover, functional analysis of TSC-22 via the creation of a TSC-22−/− mouse has never been reported. In the current study, using WT, polyclonal IL-15tg, and primary T or NK LGL leukemic splenocytes from genetically identical mice, we discovered that TSC-22 is down-regulated or silenced only in the LGL leukemic cells, and this is associated with abnormal methylation in the TSC-22 promoter. The down-regulated or silenced TSC-22 expression in primary T or NK LGL leukemia and in murine lymphoma or leukemia cell lines was reversed in vivo and in vitro, respectively, by the treatment with the US FDA–approved demethylating drug, 5-Aza. Treatment with 5-Aza also directly correlated with a significantly improved survival in the mice bearing T or NK LGL leukemia. Moreover, ectopic expression of TSC-22 in murine leukemia or lymphoma cell lines delayed tumor growth and reduced tumor size in vivo. Finally, generation and analysis of TSC-22−/− mice described in this report showed higher in vivo proliferation and repopulation efficiency of HPCs compared with their WT littermates.
IL-15 is a proinflammatory cytokine responsible for NK cell and CD8 memory T-cell homeostasis.33 Our IL-15tg mouse model of spontaneous T or NK LGL leukemia supports the notion that proinflammatory cytokines or chronic inflammation resulting from such excessive cytokine production can contribute to tumorigenesis. Our previous study20 suggests that this process may be mediated at least in part by promoter methylation of tumor suppressor genes, and the current study shows that TSC-22 likely plays a role in this process. The genetic disruption of TSC-22 failed to yield any consistent malignancies in mice observed for more than 2 years, suggesting that TSC-22 by itself is not responsible for the genesis of T or NK LGL leukemia in our IL-15tg model.
Interestingly, TSC-22 is up-regulated via signaling induced by the anti-inflammatory cytokine, TGF-β. Our recent data in NK cells imply that some cytokines such as IL-15 promote inflammation not only by their induction of inflammatory mediators such as IFN-γ, but also by down-regulating components of the TGF-β signaling pathway.25 The observation that a proinflammatory cytokine could be capable of functionally inactivating a tumor suppressor or a putative tumor suppressor gene, thereby providing a link between inflammation and tumorigenesis, is not new.34 In agreement with this, we found that costimulation of human NK cells with proinflammatory cytokines (eg, IL-12 and IL-18) not only down-regulates TGF-β signaling25 but also down-regulates TSC-22 transcription (Figure S4). Thus, the current study suggests that in the case of the IL-15tg mouse and the putative tumor suppressor TSC-22, this process of gene inactivation can be accomplished by 2 distinct mechanisms: (1) the methylation of the TSC-22 promoter and (2) a concomitant inhibition of the signaling pathway (eg, TGF-β) that induces TSC-22 expression. The combined consequence of these seemingly distinct actions would be to turn off a gene that we have now shown to regulate cell growth and repopulation using TSC-22−/− mice and to delay tumor formation in vivo, thereby enhancing predisposition to additional genetic alterations while undergoing excessive proliferation.
It is interesting that despite clear in vivo evidence of increased proliferation in the polyclonal T and NK population of the IL-15tg mouse compared with the WT mouse (M.A.C., unpublished observation, July 2007), there is little or no evidence for significant methylation or gene silencing of TSC-22 (or any other gene) in the polyclonal population.20 Therefore, although it is not yet entirely clear if the expansion in T and NK cells seen prior to malignant transformation results only from a physiologic response to genetically induced overproduction of endogenous IL-15, it does seem that this response likely comes before epigenetic silencing of tumor suppressor genes. This further supports the notion that the broad nonrandom epigenetic methylation we observed in the T or NK LGL leukemic mice30 is indeed associated with malignant transformation. Our characterization of TSC-22 in this report suggests that the initiation and maintenance of this transformation require an orchestration of such promoter methylation and associated gene silencing across the genome. Although our studies using TSC-22−/− HPCs showed an increase of proliferation and in vivo repopulation compared with TSC-22+/+ HPCs, it remains to be determined whether the silencing of TSC-22 itself is directly relevant to the process of leukemogenesis.
The TSC-22 protein is highly conserved between different species, such as drosophila, mouse, chicken, and human, with only one amino acid difference between the mouse and human proteins (J.Y., unpublished observation, June 2008). Further, this gene is expressed in many cell types, tissues, and organs, and down-regulated in several types of malignancies that have been examined.1,15,–17 Collectively, the data suggest that it may play a regulatory role in cell development and then in regulating cell homeostasis. In support of this, the drosophila homologue of TSC-22, shortsighted (shs) or bunched (bun), is involved in oogenesis, eye, wing, and peripheral nervous system development,4,35 although the most recent data indicate that the functions of bun in drosophila may be different from those of TSC-22 in humans.36,37 In data not shown, genetic disruption of TSC-22 in mice resulted in a significant alteration of expression of many genes that have previously been reported important for cellular growth and proliferation, cellular development, cell death, or cell cycle.
Many previous mammalian studies of TSC-22 have relied on cell culture experiments, although the genetic structure for this gene is much more complicated in mammals than in drosophila. There are at least 4 genomic loci (TSC22D1 to TSC-22D4) in mammals, each representing different coding sequences and each with different transcripts that can encode different proteins.36 TSC22D1 has genetic sequences of around 100 kb on chromosome 13 that include the entire genomic sequence of the TSC-22 at the 3′end. One of 3 known isoforms for TSC22D3 is GILZ, which has been shown to play an important role in the regulation of lymphocyte proliferation.38 TSC22D.2 and TSC22D.4 are relatively less well understood.
Using our TSC-22−/− mice, we noted that a putative tumor suppressor role of TSC-22 in leukemia can be explained by its ability to regulate HPC proliferation and/or to regulate their survival. HPCs obtained from TSC-22−/− mice showed enhanced ability to repopulate the hematopoietic lineages compared with those from TSC-22+/+ littermate controls. Although we monitored over 100 TSC-22−/− mice and their littermate controls for cancer more than a 2-year period of time, we found only one case of teratoma. We therefore believe that the various TSC-22 paralogues in the genome may have compensated or redundant functions with TSC-22 and therefore may explain the failure of these TSC-22−/− mice to develop leukemia. Our study suggests that a broader genetic approach may need to be considered to fully elucidate the functions of the TSC-22 gene family including TSC-22. Considering that GILZ plays a similar antiproliferative role as TSC-22 and the 2 genes are very homologous to each other, one such future study would be to cross TSC-22−/− mice with GILZ−/− mice.
Certainly, cancers such as leukemia are complex and often consist of multiple genetic changes/mutations that lead to malignant transformation. A single genetic change is rarely sufficient enough for the development of a malignant tumor.39 One of our ongoing studies is to investigate how TSC-22 deficiency cooperates with genetic changes commonly seen in T or NK LGL leukemia in an effort to better replicate this disease in mouse models. Considering the fact that TSC-22 is silenced or has dramatically decreased expression in T or NK LGL leukemia, the findings reported here may aid in providing a new cooperating mutation in instances where a primary mutation does not generate leukemia.40
In summary, we demonstrate that TSC-22 is silenced or down-regulated in IL-15–induced T or NK LGL leukemia and in multiple hematologic cell lines, and that this silencing is associated with methylation in the 5′ regulatory region of the gene. Silenced TSC-22 expression can be reversed in vitro and in mice bearing T or NK LGL leukemia by treatment with the demethylating agent 5-Aza as a single agent, the latter of which is associated with survival improvement of T or NK LGL leukemic mice. Functionally, we demonstrate that, in vivo, TSC-22 delays tumor formation and suppresses proliferation and repopulation of normal HPCs. Our studies substantiate that TSC-22 functions as a putative tumor suppressor gene in T or NK LGL leukemia. Future studies will explore whether deficiency of TSC-22, most likely by cooperating with other genetic alterations, plays a direct role in leukemogenesis, especially in human T or NK LGL leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Hitoshi Kawamata (Dokkyo University School of Medicine, Tochigi, Japan) and Dr Marco G. Cecchini (University of Bern, Bern, Switzerland) for providing TSC-22 antibodies; Zachary Boyd, Michael S. Jaung, Tiffany Hughes, Zhong-yue Wu, Tom Liu, and Dennis Chang for technical assistance; and Dr Baltazar Aguda for data analysis.
This work was supported by the Flow Cytometry, Nucleic Acid, Microarray and Mouse Phenotyping Shared Resources within The Ohio State University Comprehensive Cancer Center, by National Cancer Institute grants (CA95426 and CA68458 to M.A.C.; CA93548 and CA101956 to C.P.), by the Up on the Roof Postdoctoral Fellowship from the Division of Human Cancer Genetics at The Ohio State University (J.Y.), and a scholarship from the Dr Mildred Scheel Foundation for Cancer Research (B.H.).
National Institutes of Health
Authorship
Contribution: J.Y. designed research, performed experiments, analyzed data, and wrote the paper; M.E. and L.Y. designed some of the research and performed some of the experiments; M.W., B.H., A.Y., T.M., C.L., H.M., Z.L., and X.L. performed some of the experiments; S.L., R.T., C.L., J.V., G.M., C.P., and A.V.B. designed some of the research; K.H. analyzed some of the data; and M.A.C. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jianhua Yu or Michael A. Caligiuri, The Ohio State University Comprehensive Cancer Center, 458A Starling Loving Hall, 320 W 10th Ave, Columbus, OH 43210; e-mail: jianhua.yu@osumc.edu or michael.caligiuri@osumc.edu.