Abstract
Defining the genetic pathways essential for hematopoietic stem cell (HSC) development remains a fundamental goal impacting stem cell biology and regenerative medicine. To genetically dissect HSC emergence in the aorta-gonad-mesonephros (AGM) region, we screened a collection of insertional zebrafish mutant lines for expression of the HSC marker, c-myb. Nine essential genes were identified, which were subsequently binned into categories representing their proximity to HSC induction. Using overexpression and loss-of-function studies in zebrafish, we ordered these signaling pathways with respect to each other and to the Vegf, Notch, and Runx programs. Overexpression of vegf and notch is sufficient to induce HSCs in the tbx16 mutant, despite a lack of axial vascular organization. Although embryos deficient for artery specification, such as the phospholipase C gamma-1 (plcγ1) mutant, fail to specify HSCs, overexpression of notch or runx1 can rescue their hematopoietic defect. The most proximal HSC mutants, such as hdac1, were found to have no defect in vessel or artery formation. Further analysis demonstrated that hdac1 acts downstream of Notch signaling but upstream or in parallel to runx1 to promote AGM hematopoiesis. Together, our results establish a hierarchy of signaling programs required and sufficient for HSC emergence in the AGM.
Introduction
Specification of definitive hematopoietic stem cells (HSCs) capable of generating the blood cell lineages is a vertebrate-specific process that occurs in the aorta-gonad-mesonephros (AGM) region of the developing embryo.1 In the mouse, HSCs are located near the ventral endothelium of the dorsal aorta on embryonic day (E) 10, approximately the same time that HSC activity is present in the AGM region.2-4 The proto-oncogene c-myb and the transcription factor runx1 are both excellent markers of these emerging HSCs and are essential for mammalian HSC development.5
Runx1 is thought to operate very early in HSC specification as the mouse knockout lacks intraaortic hematopoietic clusters.4 Based on analysis of a point mutant, c-Myb is thought to act in concert with p300 to regulate the proliferation and differentiation of HSCs.6 Similarly, zebrafish have an AGM-like region in the ventral wall of the dorsal aorta also marked by c-myb and runx1 expression.5,7-11 Lineage tracing experiments have shown cells exiting this region and ultimately seeding the kidney and thymus, the sites of definitive hematopoiesis in the zebrafish.12-14 Consistent with the mouse knockout data, morpholino knockdown of runx1 translation in zebrafish results in the loss of c-myb cells in the aortic region as well as a loss of definitive marker expression in the thymus.8
Although many studies have focused on the molecular events important for HSC emergence from the AGM, very little is understood about the signals essential for specifying these cells.15 Some progress has been made through the analysis of several zebrafish mutants. In zebrafish, definitive HSCs are located between and in the axial vasculature,14 which is developmentally derived from intermediate mesoderm. Mutants with defects in vascular formation and patterning show deficiencies in HSC specification. For example, the cloche (clo) mutant lacks endothelium and thus a vasculature, has no AGM HSCs and does not undergo definitive hematopoiesis.11 Loss of vasculature organization, such as that in the spadetail (spt) mutant harboring a mutation in the tbx16 locus, also impacts formation of c-myb–expressing HSCs.11 Another zebrafish mutant, phospholipase C gamma-1 (plcγ1), which lacks Vegf signaling, fails to establish aortic identity and induce HSCs.9 Therefore, the vasculature must be properly formed and patterned to have normal HSC emergence during embryogenesis, yet the stages of AGM HSC formation in relation to vasculogenesis have not been described.
Mutants have been isolated that show defects in definitive hematopoiesis; however, where these genes lie in proximity to HSC specification remains unclear. Work on both mouse and zebrafish notch pathway mutants have demonstrated a Notch signaling requirement for AGM HSC specification.8,9,16,17 The absence of HSCs in these mutants may be an indirect result of defects in arterial specification. Whereas some studies suggest notch signaling is required for establishing proper artery identity,18-20 experiments analyzing the effect of overexpression of an activated form of notch (NICD) demonstrate that these 2 pathways can be uncoupled. In these studies, activation of NICD in zebrafish can expand c-myb+ and runx1+ progenitors in the vein independently of arterial marker expansion.8 Moreover, runx1 overexpression can rescue AGM HSCs in the mindbomb mutant, which is defective for an E3 ubiquitin ligase essential for Notch signaling.21 These conclusions were confirmed in the notch1 knockout mouse, in which overexpression of runx1 rescued AGM cluster formation.22 Furthermore, Gata-2 in AGM-derived HSCs is regulated by RBPjκ-dependent Notch function.16 In addition, recent characterization of the notch ligand Jagged1 mutant revealed that Jagged1 function is not involved in establishing artery identity but is essential for AGM hematopoiesis in mouse.23 Taken together, these studies suggest that Notch signaling may be specifically required for HSC formation separate from a function in vascular patterning.
Herein, we performed a screen on a collection of insertional mutant zebrafish lines to identify novel pathways essential for AGM HSC formation. From this screen, we were able to establish a hierarchy of programs required for HSC emergence, describe distinct steps essential for HSC induction, and present new mutants in AGM hematopoiesis. We find that overexpression of vegf, notch, or runx1 can rescue HSC induction in tbx16, a class of the HSC mutants lacking vascular organization. Activated notch and runx1 rescues HSCs in mutants lacking plcγ1, an obligate effecter of the Vegf signaling program and necessary for proper artery identity. In addition, we show that histone deacetylase 1 (hdac1) represents a novel locus required for definitive HSC induction, yet unlike the previously described mutants, hdac1−/− embryos undergo normal vessel development and artery establishment. Finally, activated notch fails to rescue HSCs, whereas runx1 suppresses the hematopoietic defect in hdac1−/− animals. These studies define the genetic relationship between critical developmental programs required for HSC emergence during embryogenesis.
Methods
Zebrafish maintenance, lines, and insertional shelf screen
Embryos and adult fish were raised and maintained under standard laboratory conditions. Ethical approval was obtained from the Institutional Animal Care and Use Committees of Children's Hospital at Harvard Medical School. We used the following lines: insertional mutant collection for screening24,25 (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) and Tg(hsp70:gal4); Tg(uas:notch1a-intra).26 Heterozygous pairs from the mutant lines were crossed and their progeny blindly screened by in situ hybridization. After in situ hybridization, only a subset of the lines could be critically evaluated for HSC defects as a portion exhibited a bloodless (bls)–like phenotype (Excel spreadsheet for lines unable to be evaluated), suggesting the presence of a second mutation in the background.27 To exclude contamination by this second mutation, we performed an exhaustive genotyping analysis after the initial in situ hybridization. Stained embryos were arrayed in a 96-well plate based on c-myb expression and individually genotyped for the viral insertion to confirm proper segregation of the genotype (wt, heterozygote, or mutant) with the phenotype (normal or loss of c-myb expression). Lines with unclear genotype and/or phenotype analysis were unable to be evaluated and were discarded from our screen numbers. Expression analyses of HSC markers were repeated multiple times on the mutants identified by the screen and always verified by genotyping (n > 50 on at least 3 sets of clutches from different heterozygous matings). Genotyping conditions and primer sequences for all 9 lines are available on request.
In situ hybridization, morpholinos, and mRNAs
Whole-mount in situ hybridization was performed as described.28 Digoxigenin-labeled antisense RNA probes were synthesized using a DIG RNA Labeling Kit (SP6/T7; Roche Diagnostics, Indianapolis, IN). Antisense morpholinos were synthesized as follows: tbx16 (www.openbiosystems.com; 5′AAGACAAGTACTCACCTCTGATAGC3′), targets the exon 1 donor spicing site; plcγ129 (www.gene-tools.com), 5′ATTAGCATAGGGAACTTACTTTCG3′), targets the exon1/intron1 boundary sequence. tbx16 MO and plcγ1 MO were injected at a concentration of 0.1 to 0.2 mmol and approximately 15 ng, respectively, in 1× Danieau Medium/1× Phenol Red8 into 1 to 4 cell embryos generated by Tg(hsp70:gal4) and Tg(uas:nicd) matings. runx1 and vegf121 RNAs were synthesized using mMessage Machine (Ambion, Austin, TX) and 20 to 30 pg and 35 to 40 pg were injected into one or 2 cell embryos, respectively. After in situ hybridization, embryos were photographed and arrayed in a 96-well plate format and subsequently genotyped.
Embryo heat-shock experiments
Tg(hsp70:gal4) adults were mated to Tg(uas:nicd) fish and their embryos harvested. For the epistasis experiments, the resulting clutches were injected with the gene-specific morpholino between the 1- to 2-cell stage. Between the 6- and 12-somite stage, embryos were collected in 50-mL BD Biosciences Discovery Labware tubes (Bedford, MA) containing approximately 5 mL of E3 and submerged in a 39°C water bath for 30 minutes. Subsequently, embryos were placed in Petri dishes, allowed to develop until 38 to 40 hpf, collected in 4% paraformaldehyde, and processed by in situ hybridization. Individual embryos were photographed in glycerol using a Nikon Coolpix camera (Nikon, Tokyo, Japan) mounted on a Nikon E600 compound microscope. After photography, embryos were individually arrayed and genotyped.
Results
A forward genetic screen identifies genes crucial for AGM HSC specification
To uncover essential genes for HSC induction during embryogenesis, we screened 194 independent insertional mutant zebrafish lines in which the defective genes have been isolated24,25 (“Methods” and Document S1: a complete list of genes screened). Heterozygous adults were mated, their embryos collected, and processed by in situ hybridization for alterations in c-myb expression (Figure 1A). Although most mutations had no effect on HSC induction (n = 185), 9 loci were found to be essential for AGM hematopoiesis (Figure 1B). One insertion identified was in the mind bomb (mib) locus,8 which encodes an E3 ligase essential for Notch signaling.21 As the HSC phenotype of an ENU-induced mib allele was previously characterized,8 we did not include insertional allele data as they were similar.
A genetic screen in zebrafish defines 9 essential genes for HSC emergence. (A) Screening strategy. Heterozygous adults from each insertional (ins.) mutant line were crossed, embryos raised until 36 to 40 hpf of development, fixed in paraformaldehye, and processed by in situ hybridization for c-myb expression in the AGM. (B) Table listing the hi alleles uncovered in the screen, the gene disrupted in each line, and previously isolated alleles. (C) Whole-mount in situ hybridization for c-myb, runx1, and rag1 transcripts in wild-type, hdac, plcg1, and tbx16 mutant animals. (Columns 1 and 2) Lateral views, anterior left; 36 to 40 hpf; low magnification (original magnification ×10) and higher magnification (original magnification ×40) of the trunk region. HSC gene expression in the AGM is indicated by ➤ in subpanels i and ii. (Column 3) Dorsal view, anterior up; 4 dpf; bilateral rag1 expression in thymi is indicated by ➤ in subpanel iii.
A genetic screen in zebrafish defines 9 essential genes for HSC emergence. (A) Screening strategy. Heterozygous adults from each insertional (ins.) mutant line were crossed, embryos raised until 36 to 40 hpf of development, fixed in paraformaldehye, and processed by in situ hybridization for c-myb expression in the AGM. (B) Table listing the hi alleles uncovered in the screen, the gene disrupted in each line, and previously isolated alleles. (C) Whole-mount in situ hybridization for c-myb, runx1, and rag1 transcripts in wild-type, hdac, plcg1, and tbx16 mutant animals. (Columns 1 and 2) Lateral views, anterior left; 36 to 40 hpf; low magnification (original magnification ×10) and higher magnification (original magnification ×40) of the trunk region. HSC gene expression in the AGM is indicated by ➤ in subpanels i and ii. (Column 3) Dorsal view, anterior up; 4 dpf; bilateral rag1 expression in thymi is indicated by ➤ in subpanel iii.
Mutants from each line showed loss of AGM c-myb and runx1 expression compared with wild-type siblings (Figures 1C, S1). Differentiated thymic T cells that express rag1 and are exclusively derived from definitive HSCs were also absent from each mutant examined at 4 days after fertilization (Figures 1C, S1). Thus, our screen defined a set of genes required for specification of the definitive HSC fate, and analysis of these mutants will provide a better understanding of stem cell emergence during vertebrate embryogenesis.
Mutants define critical stages of HSC emergence
Several early embryonic events, including endothelial cell fate specification, vasculogenesis, and the establishment of artery identity, must occur for the mesoderm to generate definitive HSCs. To assess the stage at which each mutant is defective, we performed in situ hybridization on embryos generated by heterozygous matings to examine markers of vessel formation (flk1; Figure 2A) and artery identity (efrinb2a; Figure 2B). One of the hallmarks of proper arterial fate specification is the presence of branching intersomitic vessels from the dorsal aorta.29 Artery identity is disrupted in tbx16−/− animals as indicated by the loss of intersomitic flk1-expressing vessels and efrinb2a transcripts (Figure 2G,H). Similar to the previously described spadetail/tbx16b104 deficiency,30 tbx16 mutants induce endothelial cells, as shown by flk1 expression, but fail to organize them into a proper vasculature (Figure 2G), positioning this locus most distal to HSC specification (class I; Figure 2I). plcγ1−/−, 40s ribosomal protein s29−/−, and wdr43-like−/− mutants undergo vasculogenesis normally (Figures 2E, S2) but fail to establish artery identity as shown by lack of flk1-expressing intersomitic vessels and efrinb2a expression (class II; Figures 2E,F, S2). Thus, analysis of these mutants demonstrates that proper vasculogenesis and establishment of artery identity are events crucial for normal AGM hematopoiesis. Interestingly, our screen identified a set of genes, including hdac1, splicosome associated factor 61, nop14-like, and U1 small ribonucleoprotein, which are completely dispensable for flk1 and efrinb2a aortic expression (Figures 2C,D, S2). These mutants define a new class of loci that function most proximally to HSC induction and at a time point after which endothelial specification and artery identity are evident (class III; Figure 2I). Thus, we classified the stages required for HSC specification as determined by the mutants isolated in our screen (Figure 2I).
Classification of the stages required for HSC emergence. Whole-mount in situ hybridization for flk1 and efrinb2a transcripts in wild-type, hdac, plcg1, and tbx16 mutant animals. (A-H) Lateral views, anterior left; 24 to 28 hpf; low magnification (original magnification ×10) and higher magnification (original magnification ×40) of the trunk region. (Column 1) flk1 gene expression in the dorsal aorta is indicated by black arrowheads, and intersomitic vessels are marked with green arrowheads. (Column 2) efrinb2a transcripts are highlighted with red arrowheads. (I) Stages required for HSC induction with the genes listed under each arrow that are needed for the transition to occur.
Classification of the stages required for HSC emergence. Whole-mount in situ hybridization for flk1 and efrinb2a transcripts in wild-type, hdac, plcg1, and tbx16 mutant animals. (A-H) Lateral views, anterior left; 24 to 28 hpf; low magnification (original magnification ×10) and higher magnification (original magnification ×40) of the trunk region. (Column 1) flk1 gene expression in the dorsal aorta is indicated by black arrowheads, and intersomitic vessels are marked with green arrowheads. (Column 2) efrinb2a transcripts are highlighted with red arrowheads. (I) Stages required for HSC induction with the genes listed under each arrow that are needed for the transition to occur.
Vegf acts downstream of tbx16-directed specification of paraxial mesoderm
Recent coexpression9 and fate-mapping31 studies have highlighted the shared ancestry between the endothelial and hematopoietic lineages. The Vegf signaling pathway, which is mediated by plcγ1, is one example of a program that controls both aorta identity and HSC induction9,29 Because ectopic Vegf signaling is known to promote arterial fate by up-regulating artery-specific transcripts in the cardinal vein,29 we tested whether Vegf is also sufficient to induce ectopic HSCs. Synthetic vegf121 RNA was injected at the 1- to 2-cell stage, and embryos were raised until 40 hpf. Compared with uninjected sibling controls that show ventrally restricted dorsal aorta c-myb expression (Figure 3A), vegf121 mRNA expanded c-myb-expressing HSCs throughout the aortic roof and vein (Figure 3B). To order the Vegf-plcγ1 program relative to tbx16, we next analyzed vegf expression in tbx16 mutants and found that transcripts were relatively lower and more disorganized within the trunk somites compared with wild-type siblings (Figure 3C,D). Next, we tested whether vegf121 mRNA could rescue the definitive hematopoietic defect in tbx16 mutants (Figure 3E,F). Injection of vegf121 mRNA rescued c-myb+ HSCs in the trunk of tbx16−/− animals (the area most affected by loss of tbx16; Figure 3E,F), further indicating that Vegf-Plcγ1 pathway functions downstream of tbx16. Moreover, overexpression of vegf also partially rescued vascular organization and artery identity in the trunk of tbx16 mutants (Figure 3G-J), further supporting the stages required for HSC induction put forth by the mutants isolated (Figure 2I). Together, these data show that the Vegf pathway, like the Notch program,8 is sufficient to promote HSC specification in wild-type animals and positions plcγ1 genetically downstream of tbx16 in HSC induction.
The Vegf pathway functions downstream of tbx16 in HSC specification. Whole-mount in situ hybridization of embryos between 36 and 40 hpf (c-myb expression) or approximately 28 hpf (vegf, flk1, or efrinb2a). Red arrowheads represent arterial expression of HSC markers; green arrowheads, intersomitic vessels. Lateral views, anterior left; low magnification (original magnification ×10) and higher magnification (original magnification ×40) of the trunk region. (A,B) Injection of approximately 40 pg vegf121 RNA into wild-type embryos expands c-myb–positive HSCs. (C,D) vegf expression in wild-type (C) and tbx16−/− mutants (D). (E-J) Uninjected (E,G,I) or vegf injected (F,H,J) tbx16 mutant embryos showing substantial rescue of c-myb, flk1, and efrinb2a in the trunk.
The Vegf pathway functions downstream of tbx16 in HSC specification. Whole-mount in situ hybridization of embryos between 36 and 40 hpf (c-myb expression) or approximately 28 hpf (vegf, flk1, or efrinb2a). Red arrowheads represent arterial expression of HSC markers; green arrowheads, intersomitic vessels. Lateral views, anterior left; low magnification (original magnification ×10) and higher magnification (original magnification ×40) of the trunk region. (A,B) Injection of approximately 40 pg vegf121 RNA into wild-type embryos expands c-myb–positive HSCs. (C,D) vegf expression in wild-type (C) and tbx16−/− mutants (D). (E-J) Uninjected (E,G,I) or vegf injected (F,H,J) tbx16 mutant embryos showing substantial rescue of c-myb, flk1, and efrinb2a in the trunk.
Notch acts downstream of tbx16 and vegf signaling for HSC specification
As previous reports established the Notch-Runx pathway as critical and sufficient for HSC specification during embryogenesis, we next focused on one mutant from each class and analyzed its genetic relationship relative to Notch and Runx in HSC development. tbx16hi3714 represents the class I mutants and generates a weaker morphologic phenotype than the spadetail/tbx16b104 deficiency.30 Both genetic lines, however, maintain trunk endothelial cell induction,32 display a disorganized vasculature,11 and show loss of definitive HSCs.10,11 plcγ1hi2335, the class II mutant, is phenotypically indistinguishable from the reported plcγ1y10 allele29 in that the dorsal aorta is well organized but lacks artery identity and HSC specification is disrupted.9 Because tbx16 and plcγ1 both function in earlier embryologic processes relative to HSC specification, constitutive Notch activation may rescue their definitive hematopoietic phenotype.
To position these genes in a genetic hierarchy relative to each other and the Notch-Runx pathway, we performed classic epistasis experiments. Heat induction of the constitutively active zebrafish Notch1 receptor intracellular domain (NICD) via a Gal4/UAS transgenic system26 expands HSCs in the AGM as shown by expansion of c-myb and runx1 transcripts (Figure 4A,B).8 To knock down tbx16 or plcγ1 translation, gene-specific morpholinos (MO)33 were injected into embryo clutches derived from Tg(hsp70:gal4) and Tg(uas:NICD) matings. After injection, the entire clutch was exposed to high heat (“Methods”). Whereas 75% of the resulting progeny were control siblings, 25% carried both transgenes and therefore activated the Notch signaling pathway. Control morphant embryos showed loss of both c-myb (Figure 4C,G) and runx1 (Figure 4D,H) transcripts in the AGM, thereby phenocopying the genetic mutants. tbx16MO- or plcγ1MO-injected Tg(hsp70:gal4);Tg(uas:NICD) embryos, however, showed the Notch gain-of-function phenotype in that both c-myb (Figure 4E,I) and runx1 (Figure 4F,J) transcripts were expanded in the AGM. These data suggest that Notch signaling acts downstream of tbx16 and plcγ1 in HSC fate determination.
Genetic interaction of the tbx16, plcγ1, and notch signaling pathways for the induction of HSCs. Whole-mount in situ hybridization of embryos between 36 and 40 hpf. Embryo manipulation, genotype, and probes used are described for each panel. Red arrowheads denote arterial expression of HSC markers. Lateral views, anterior left; low magnification (original magnification ×10) and higher magnification (original magnification ×40) of the trunk region. After heat shock (“Methods”), NICD expands c-myb and runx1 expression in wild-type embryos (A,B). NICD rescues c-myb and runx1 expression in tbx16 morphants (C-F) and plcγ1 morphants (G-J).
Genetic interaction of the tbx16, plcγ1, and notch signaling pathways for the induction of HSCs. Whole-mount in situ hybridization of embryos between 36 and 40 hpf. Embryo manipulation, genotype, and probes used are described for each panel. Red arrowheads denote arterial expression of HSC markers. Lateral views, anterior left; low magnification (original magnification ×10) and higher magnification (original magnification ×40) of the trunk region. After heat shock (“Methods”), NICD expands c-myb and runx1 expression in wild-type embryos (A,B). NICD rescues c-myb and runx1 expression in tbx16 morphants (C-F) and plcγ1 morphants (G-J).
Hdac1 regulates HSC formation after artery specification
hdac1, which encodes an enzyme responsible for catalyzing the removal of acetyl groups from core histones and transcription factors,34 represents the class III mutants. Biochemical evidence suggests that HDAC1 normally functions as a transcriptional repressor and attenuates Notch signaling by sequestering a Notch pathway activator, RBPjκ, to a repressor complex.35 In the nervous system, absence of HDAC1 results in high levels of Notch signaling as RBPjκ target genes are continuously activated.8,9,36,37 Surprisingly, our studies demonstrate that hdac1 mutants fail to specify HSCs similar to the Notch pathway loss-of-function mutant, mindbomb.8,9 Thus, the relationship between notch and hdac1 may differ in the hematopoietic system compared with the nervous system.
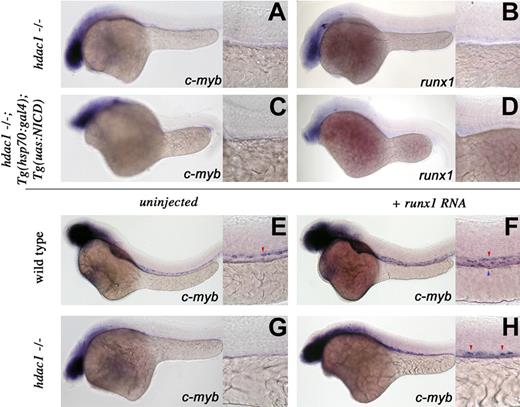
To investigate potential interactions, we crossed the hdac1 mutation onto the Tg(hsp70:gal4);Tg(uas:NICD) background and analyzed HSC specification in their progeny after heat induction. Both control hdac1−/− siblings (Figure 5A,B) and Tg(hsp70:gal4);Tg(uas:NICD);hdac1−/− animals (Figure 5C,D) showed loss of c-myb and runx1 expression, suggesting that hdac1 is required downstream of Notch signaling for HSC emergence.
hdac1 acts between notch and runx1 in HSC specification. Whole-mount in situ hybridization of embryos between 36 and 40 hpf. Embryo manipulation, genotype, and probes used are described for each panel. Red arrowheads represent arterial expression of HSC markers; blue arrowhead, venous expression of HSC markers. Lateral views, anterior left; low magnification (original magnification ×10) and higher magnification (original magnification ×40) of the trunk region. Overexpression of NICD does not rescue the hdac−/− HSC phenotype (A-D). After heat shock (“Methods”), control hdac1 mutant siblings and hdac1−/−;Tg(hsp70:gal4);Tg(uas:nicd) embryos fail to induce c-myb (A) or runx1 (B) expression, whereas Tg(hsp70:gal4);Tg(uas:nicd) embryos show expanded c-myb and runx1 expression (Figure 3A,B). Overexpression of runx1 RNA expands c-myb–expressing cells in wild-type (compare panel F with panel E) and partially suppresses the hdac−/− HSC phenotype (G,H).
hdac1 acts between notch and runx1 in HSC specification. Whole-mount in situ hybridization of embryos between 36 and 40 hpf. Embryo manipulation, genotype, and probes used are described for each panel. Red arrowheads represent arterial expression of HSC markers; blue arrowhead, venous expression of HSC markers. Lateral views, anterior left; low magnification (original magnification ×10) and higher magnification (original magnification ×40) of the trunk region. Overexpression of NICD does not rescue the hdac−/− HSC phenotype (A-D). After heat shock (“Methods”), control hdac1 mutant siblings and hdac1−/−;Tg(hsp70:gal4);Tg(uas:nicd) embryos fail to induce c-myb (A) or runx1 (B) expression, whereas Tg(hsp70:gal4);Tg(uas:nicd) embryos show expanded c-myb and runx1 expression (Figure 3A,B). Overexpression of runx1 RNA expands c-myb–expressing cells in wild-type (compare panel F with panel E) and partially suppresses the hdac−/− HSC phenotype (G,H).
Runx1 function is required for Notch to induce and expand HSC numbers and runx1 overexpression can rescue HSCs in the absence of Notch signaling.8,22 As runx1 transcripts are lost in hdac1 mutants, we tested whether synthetic runx1 RNA could rescue the hdac1−/− HSC phenotype. Injection of runx1 RNA into 1- to 4-cell wild-type embryos expanded c-myb expression throughout the aortic wall and vein (Figure 5E,F). Uninjected hdac1−/− animals failed to specify HSCs (Figure 5G); however, overexpression of runx1 in the mutant background partially rescued c-myb transcripts in the AGM (Figure 5H). This reemergence of myb+ cells indicates that hdac1 acts genetically upsteam of or in parallel to runx1 in HSC induction. Our work demonstrates that hdac1, a novel HSC regulator, acts mostly proximally to HSC induction.
Discussion
HSC emergence in the vertebrate AGM may require genes that are more broadly critical for stem cell specification and self-renewal. Our studies uncovered a new set of pathways required for HSC formation during vertebrate development. In total, our screen identified 9 genes required for AGM HSCs in zebrafish. By focusing on 3 of these genes, we established distinct stages required for HSC formation and developed a genetic paradigm by which HSCs are induced during development. The remaining genes await further study to determine their specific role in HSC formation.
Whereas 3 loci involved in ribosomal subunit biogenesis were identified in our screen (rps29, wdr43-like, possible homolog to yeast ribosome biogenesis factor UTP5, and nop-1438 ), 26 other ribosomal subunit/biogenesis factors (supplemental data) were excluded, strongly arguing that only certain subunits are required for definitive hematopoiesis. Lending credence to this hypothesis, mutations in rps1939 and other specific ribosomal proteins40 lead to Diamond-Blackfan anemia, and knockdown of rps14 has been associated with the erythroid differentiation defect found in 5q- syndrome.41
tbx16/spadetail represents the most distal pathway to AGM hematopoiesis as it has been shown to directly regulate paraxial mesoderm convergence during early gastrulation.30 Loss of proper convergence movements results in a broadened fli1-expressing endothelial progenitor domain during early somitogenesis32 and subsequent disorganization of the trunk vasculature.11,30 Whether tbx16 exerts a cell-autonomous and/or nonautonomous effect on definitive HSC specification is not known. During primitive hematopoiesis, tbx16 plays both an autonomous and nonautonomous role in specifying the red blood cell lineage within intermediate mesoderm.32 During embryogenesis, mesoderm from the AGM becomes regionalized into specific fates. Many studies show that a “hemogenic endothelial cell” acts as a bipotential precursor to the HSC and vascular lineages. Other studies show that a subaortic mesenchymal cell population independently migrates though endothelial cells to become hematopoietic.13,42 We hypothesize that tbx16 plays an indirect role in HSC induction by providing a proper vascular environment in which definitive hematopoiesis can initiate.
Our genetic studies demonstrate that loss of HSCs in tbx16 mutants can be rescued by Vegf signaling. Although the vegf-plcg1 pathway has been shown to regulate aortic identity,19,29 vegf has also been identified as up-regulated in human AGM HSC intraaortic clusters.43 It is unclear whether Vegf signaling is only required for establishment of arterial fate, which in turn is necessary for AGM HSCs formation, or whether it may have a more direct function in specifying AGM HSCs. Mice deficient in the Vegf receptor, flk-1, die very early during embryogenesis from severe vascular and hematopoietic defects.44 Accordingly, analysis of chimeric mice revealed that flk1−/− cells fail to contribute to primitive yolk sac, definitive fetal liver, or adult hematopoiesis.44 Although this finding initially suggested a direct role for Vegf signaling in hematopoiesis, further analysis found that flk1−/− ES cells were capable of differentiating into primitive red blood cells in vitro. Furthermore, as the flk1−/− cells were also found to be absent from the vasculature in chimeras, it is not possible to distinguish whether the definitive hematopoietic defect is primary (cell-autonomous) or secondary to an endothelial deficiency.44 Similarly, in zebrafish loss of the vegf-plcg1 pathway has no effect on primitive hematopoiesis but is required for artery identity and definitive hematopoiesis.9,19,29 Because overexpression of Vegf has also been found to expand arterial fates29 and c-myb expression (shown here) in wild-type embryos, it appears that the function of Vegf in artery fate decisions is linked to AGM HSC emergence.
Unlike Vegf signaling, the role of Notch signaling in artery identity can be uncoupled from AGM HSC formation. Overexpression of an activated form of Notch expands AGM HSCs, but not arterial marker expression8 and loss of the Notch ligand, Jagged1, results in impaired AGM hematopoiesis without concomitant defects in aortic identity.23 Our work has shown that Notch signaling can rescue AGM HSCs in the absence of vegf-plcg1 signaling, demonstrating that Notch signaling can bypass the need for the Vegf-plcg1 pathway in AGM hematopoiesis.
A novel HSC regulator, hdac1, is required for runx1 expression downstream of the Notch pathway. Previous studies have shown that HDAC factors biochemically interact with Runx proteins to actively repress Runx-target gene transcription.45,46 Specifically, Runx1 was found to strongly associate with HDAC1 via immunoprecipitation,47 suggesting that Runx factors recruit distinct HDACs to mediate repression. Based on these studies, loss of HDAC1 would seemingly result in an increase in Runx1-target gene activation and stem cell emergence, yet our results suggest the opposite. We find that runx1 expression fails to initiate in the hdac1 mutant and HSCs do not form. Moreover, injection of runx1 RNA rescues some c-myb+ cells in the AGM of hdac1 mutants but does not fully suppress the HSC deficiency. There are several plausible mechanisms that can be envisioned. One possibility is that HDAC1 represses transcription of a factor that normally silences runx1 transcription. In this case, loss of hdac1 would lead to increased levels of a runx1 transcriptional repressor. Alternatively, HDACs have been found to directly regulate whether a transcription factor will act to promote or repress target gene activity by altering acetylation.48 Because HDAC1 and Runx1 proteins have been found to physically interact,45,46 it is plausible that HDAC1 is required to convert Runx1 from a repressor to an activator (or vice versa). In this case, loss of hdac1 would lead to constitutive Runx1 repressor activity such that no target genes are transcribed. Although future biochemical studies would be required to address the precise HDAC1/RUNX1 interplay within HSCs, our genetic results show that HDAC1 activity is extremely proximal to and required for HSC emergence.
There has been considerable effort aimed at understanding the developmental origins of the AGM cells using many vertebrate model organisms. Our work using forward genetic screening in zebrafish demonstrates that 2 processes, the proper formation of the vasculature, which is dependent on the tbx16 pathway, as well as the proper patterning and specification of the hemogenic endothelium, which requires the VEGF/Notch programs, are absolutely essential for the emergence of AGM HSCs. The output of these pathways is the induction of runx1 expression, which is critical for HSC formation. Our work now places hdac1 between these 2 pathways and runx1 induction and identifies it as an essential regulator of HSC formation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Xiaoying Bai and Trista North for critical reading of the manuscript and C. Belair, B. Barut, and A. McCollum for fish care and laboratory management.
This work was supported by the National Heart, Lung, and Blood Institute (grant 5 R01 HL48801). C.E.B. is supported by the National Institute of Diabetes and Digestive and Kidney Diseases (Research Career Award 1 K01 DK067179). A.H.A. is supported by a National Institutes of Health grant from the National Center for Research Resources (2 R01 RR012589). L.I.Z. is supported by Howard Hughes Medical Institute.
National Institutes of Health
Authorship
Contribution: C.E.B., J.L.G., A.C.H.S., M.D.K., E.A.M., E.J.P., and A.H.A. performed the screen; C.E.B., J.L.G., A.C.H.S., and T.J.C. performed overexpression studies; C.E.B., J.L.G., and A.C.H.S. performed the mutant analyses; and C.E.B., J.L.G., and L.I.Z. conceived of experiments, analyzed experiments, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Leonard I. Zon, Stem Cell Program and Division of Hematology/Oncology, Children's Hospital and Dana-Farber Cancer Institute, 320 Longwood Ave, Enders Bldg Rm 761, Harvard Medical School, Boston, MA 02115; e-mail: zon@enders.tch.harvard.edu.
References
Author notes
*C.E.B. and J.L.G. contributed equally to this study.