Abstract
RUNX1 (AML1) encodes the core binding factor α subunit of a heterodimeric transcription factor complex which plays critical roles in normal hematopoiesis. Translocations or down-regulation of RUNX1 have been linked to favorable clinical outcomes in acute leukemias, suggesting that RUNX1 may also play critical roles in chemotherapy responses in acute leukemias; however, the molecular mechanisms remain unclear. The median level of RUNX1b transcripts in Down syndrome (DS) children with acute megakaryocytic leukemia (AMkL) were 4.4-fold (P < .001) lower than that in non-DS AMkL cases. Short hairpin RNA knockdown of RUNX1 in a non-DS AMkL cell line, Meg-01, resulted in significantly increased sensitivity to cytosine arabinoside, accompanied by significantly decreased expression of PIK3CD, which encodes the δ catalytic subunit of the survival kinase, phosphoinositide 3 (PI3)–kinase. Transcriptional regulation of PIK3CD by RUNX1 was further confirmed by chromatin immunoprecipitation and promoter reporter gene assays. Further, a PI3-kinase inhibitor, LY294002, and cytosine arabinoside synergized in antileukemia effects on Meg-01 and primary pediatric AMkL cells. Our results suggest that RUNX1 may play a critical role in chemotherapy response in AMkL by regulating the PI3-kinase/Akt pathway. Thus, the treatment of AMkL may be improved by integrating PI3-kinase or Akt inhibitors into the chemotherapy of this disease.
Introduction
Acute myeloid leukemia (AML) is the second most common acute leukemia in children, and treatment outcomes have been consistently inferior to childhood acute lymphoblastic leukemia (ALL). Improvements in clinical outcomes for childhood AML are probably to come from better understanding of both the molecular alterations responsible for development and proliferation of leukemia cells and the molecular mechanisms underlying drug resistance.1
One of the most frequently translocated genes in adult and childhood leukemias is RUNX1 (also known as AML1) localized to 21q. RUNX1 is a member of a family of DNA-binding proteins that includes RUNX2 and RUNX3.2-4 In association with a second subunit, CBFβ (core binding factor β), RUNX1 binds to the core enhancer DNA sequence TGT/cGGT.2-4 Both DNA binding and heterodimerization are mediated through a central domain in the RUNX1 family of proteins termed the runt-homology domain.2-4 RUNX1 plays a critical role in the regulation of expression of many genes involved in normal hematopoiesis. Homozygous knockout of either RUNX1 or CBFβ results in embryonic lethality, with mouse embryos dying at embryonic day 12.5 from a lack of definitive hematopoiesis and from central nervous system hemorrhages.5-8 The biologic functions of RUNX1 appear to be exquisitely sensitive to its intracellular levels, as reflected in the observation that germline AML1 deletions or mutations in patients with a familial platelet disorder are associated with a high predisposition to develop AML.9,10 RUNX1 is targeted either directly or indirectly in t(8;21), t(3;21), and inv (16) AML, and in t(12;21) ALL.2,4
Translocations involving RUNX1 have also been linked to clinical outcomes in acute leukemias. Patients with t(8;21) or inv(16) AML (CBF AML) are considered to be favorable subgroups compared with AML cases overall,11,12 although the presence of mutations in additional oncogenes (eg, KIT) may have a negative effect on prognosis.13-16 t(12;21) B precursor–ALL is considered one of the most favorable prognostic subgroups of childhood ALL with cure rates greater than 90%.17 Interestingly, gene amplification of RUNX1 is associated with a poor prognosis of ALL.18 Despite its localization to chromosome 21, RUNX1 expression was reported to be lower in megakaryoblasts from children with Down syndrome (DS) with acute megakaryocytic leukemia (AMkL) compared with children without DS with AMkL19 and thus may contribute to differences in event-free survival (EFS) rates between these groups (80%-100% vs < 25%, respectively) when treated with cytosine arabinoside (ara-C)/anthracycline-based protocols.20-26 These clinical observations suggest that RUNX1 may also play critical roles in chemotherapy response/resistance in acute leukemias, in addition to its vital role in leukemogenesis; however, the molecular mechanisms remain largely unestablished.
In the present study, we used real-time reverse transcription–polymerase chain reaction (RT-PCR) to quantitate transcript levels for RUNX1a (the shorter RUNX1 transcript isoform) and RUNX1b and RUNX1c (the 2 larger RUNX1 transcript isoforms) in blast cells from children with and without DS with AMkL. Our results show that RUNX1b is the major transcript isoform expressed and that its transcript levels are significantly higher in non-DS AMkL blasts compared with DS AMkL blasts. Short hairpin (shRNA) knockdown of RUNX1 in a non-DS AMkL cell line, Meg-01, resulted in significantly increased ara-C sensitivity. This was accompanied by significantly decreased expression of phosphoinositide-3-kinase catalytic delta polypeptide (PIK3CD; encoding the delta catalytic subunit of PI3-kinase [p110δ]), determined by oligonucleotide microarray and real-time RT-PCR analyses. Down-regulation of RUNX1 by shRNA in the Meg-01 cells also resulted in decreased expression of p110δ and decreased phosphorylation of Akt, the downstream target of p110δ. Our results suggest that RUNX1 may play critical roles in chemotherapy response in AMkL by regulating genes related to cell survival and apoptosis. Better understanding of the role of RUNX1 in chemotherapy response may lead to improvements in the therapy of AMkL in particular, and potentially AML overall.
Methods
Clinical samples
Diagnostic blasts from DS (n = 16; 66% median blasts) and non-DS (n = 12; 81.5% median blasts) children with AMkL were obtained from the Children's Hospital of Michigan leukemia cell bank and from the Pediatric Oncology Group 9421 study, as previously described.27,28 The diagnosis of AMkL was confirmed by flow cytometry detection of the megakaryocytic lineage antigens, CD41 and CD61. Nonspecific adherence of platelets to monocyte lineage cells was identified by simultaneous staining with CD14. Mononuclear cells were purified by standard Ficoll-Hypaque density centrifugation. Informed consent was provided according to the Declaration of Helsinki. Sample handling and data analysis protocols were approved by the Human Investigation Committee of the Wayne State University School of Medicine.
Cell culture
The K562 AML and Meg-01 AMkL cell lines were obtained from the ATCC. The parental and the engineered sublines were cultured in RPMI 1640 with 10% fetal bovine serum (FBS; Hyclone) and 2 mM l-glutamine plus 100 U/mL penicillin and 100 μg/mL streptomycin in a 37°C humidified atmosphere containing 5% CO2/95% air.
Expression of an inducible RUNX1b construct in K562 cells
A commercial “Tet-on” (Clontech) system was used to express RUNX1b in K562 cells under control of a tetracycline/doxycycline (Dox)–inducible promoter.29 The K562pTet-on cell line was developed from wild-type K562 cells by transfection with the regulator pTet-on plasmid with the use of Lipofectin (Invitrogen), as previously reported.30 To construct the pTRE2hyg/RUNX1b plasmid, wild-type RUNX1b cDNA from pcDNA3-AML1b31 and the pTRE2hyg vector were digested with KpnI and BamHI, respectively, and the resulting overhangs were filled in using T4 DNA polymerase. The blunt-ended pcDNA3-RUNX1b and the pTRE2hyg were then digested with XhoI and SalI, respectively. The resulting RUNX1b coding cDNA fragment was subcloned into the pTRE2hyg vector, prepared as described above. After confirmation by automated DNA sequencing, the plasmid was then transfected into the K562pTet-on cell line by electroporation (200 V, 950 μF) and screened by selection with hygromycin (200 μg/mL). Colonies were isolated in soft agar, expanded in the presence of 200 μg/mL hygromycin, and tested for RUNX1b expression in the presence or absence of Dox (2 μg/mL) by Western blotting, as described in “Western blot analysis.” A “double-stable” clone K562pTet-on/AML1b (designated KA1b), which exhibited low background and a high level of Dox-induced RUNX1b expression, was identified and used for further analyses.
Western blot analysis
Soluble protein extracts were prepared by sonication in hypotonic buffer (10 mM Tris-Cl, pH 7.0), containing 1% SDS and proteolytic inhibitors, and subjected to SDS–polyacrylamide gel electrophoresis. Separated proteins were electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes (DuPont) and immunoblotted with anti-RUNX1 antibody, anti-p110δ (Santa Cruz Biotechnology Inc), anti–phospho-Akt or anti-Akt antibody (Cell Signaling Technology), as described previously.32 Immunoreactive proteins were visualized with the Odyssey Infrared Imaging System (Li-Cor), as described by the manufacturer.
In vitro ara-C cytotoxicity assays
For cytotoxicity determinations, cell lines were cultured in complete medium with dialyzed FBS in 96-well plates at a density of 4 × 104 cells/mL. Cells were cultured for 96 hours with a range of concentrations of ara-C at 37°C, and cell numbers were determined with the Cell Titer–blue reagent (Promega) and a fluorescence microplate reader. Fifty percent inhibitory concentration (IC50) values were calculated as the concentrations of drug necessary to inhibit 50% growth compared with control cells cultured in the absence of drug. Data are presented as mean values plus or minus SEs from at least 3 independent experiments.
The levels of interaction between ara-C and LY294002 manifest as drug sensitivities were evaluated as described previously.33 Briefly, synergism, additivity, or antagonism was quantified by determining the combination index (CI), where CI less than 1, CI equal to 1, and CI greater than 1 indicate synergistic, additive, or antagonistic effects, respectively. On the basis of the classic isobologram, the CI was calculated with the following equation: CI = [(D)1/(Dx)1] + [(D)2/(Dx)2]. At the 50% inhibition level, (Dx)1 and (Dx)2 are the concentrations of ara-C and LY294002, respectively, which induce a 50% inhibition in cell growth. (D)1 and (D)2 are the concentrations of ara-C and LY294002, which also inhibit cell growth by 50% (compared with single agents alone).
MTT cytotoxicity assay
In vitro ara-C and LY294002 sensitivities of primary pediatric AMkL patient samples were measured by MTT (3-[4,5-dimethyl-thiazol-2-yl]-2,5-diphenyltetrazolium-bromide) assays, as previously described.27 Briefly, blasts from children newly diagnosed with de novo AMkL were cultured in 50 μL RPMI 1640/20% dialyzed FBS in 96-well plates at a density of 106 cells/mL. Cells were incubated at 37°C in the presence of various concentrations of ara-C and LY294002. After 72 hours, MTT was added to a final concentration of 1 mM. After 4.5 hours, formazan crystals were dissolved by the addition of 50 μL 10% SDS in 10 mM HCl. The optical densities were measured by a microplate reader at 590 nm. IC50 values were calculated as the drug concentrations necessary to inhibit 50% growth compared with untreated control cells. The level of interaction between ara-C and LY294002 was evaluated by calculating the CI, as described in “In vitro ara-C cytotoxicity assays.”
Oligonucleotide microarray sample preparation and hybridization
Microarray samples were prepared essentially as described.28 On each microarray, a labeled RUNX1-positive (Meg-01 negative) sample was cohybridized with an oppositely labeled RUNX1-negative (Meg-01 RUNX1 1-1) sample. Two arrays were completed for each of 2 independent pairs of biologic samples, for a total of 4 microarrays. RNA samples from 2 independent experiments were prepared from Meg-01 negative and Meg-01 RUNX1 1-1 cells. A dye-swapped pair of arrays was completed for each pair of biologic samples. The microarray data associated with this study have been deposited to Gene Expression Omnibus under accession no. GSE17311.
Quantification of gene expression by real-time RT-PCR
cDNAs were prepared from 1 μg total RNA with the use of random hexamer primers and a RT-PCR kit (Perkin Elmer Life Sciences) and were purified with the QIAquick PCR Purification Kit (QIAGEN). Transcript levels for RUNX1a, total of RUNX1b and RUNX1c (designated RUNX1b/1c), RUNX1c, and PIK3CD were quantitated with the use of a LightCycler real-time PCR machine (Roche), as previously described.31,32 PCRs contained 2 μL purified cDNA or standard plasmid, 4 mM MgCl2, 0.5 μM each of sense and antisense primers, and 2 μL FastStart DNA Master SYBR Green I enzyme–SYBR reaction mix (Roche). Primers and PCR conditions are described in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Real-time PCR results were expressed as mean values from 3 independent experiments with the same cDNA preparation and normalized to 18S rRNAs. Transcript levels for RUNX1b were calculated by subtracting RUNX1c transcript levels from those for RUNX1b/1c.
Chromatin immunoprecipitation assays
The chromatin immunoprecipitation (ChIP) assay was performed as described previously.27,34 Purified chromatin fragments from Meg-01 cells were incubated with anti-RUNX1 antibody or normal IgG. Standard PCR (qualitative) for the RUNX1 binding region in the PIK3CD promoter was performed with the use of forward (5′-GAATAGGGCTTGGGTCAGGT-3′) and reverse (5′-ATTTTGAGGAATCTTGTTCTGTTG-3′) primers spanning the 2 putative RUNX1 binding sites. An upstream unrelated (ie, to RUNX1 binding) region in the PIK3CD gene was amplified by PCR with the use of upper (5′-GCTTTGTTTGAATCAGTTGG-3′) and lower (5′-TCTTTCTATCATCTCCCCTTTT-3′) primers to further validate the specificity of the ChIP assays. The same PIK3CD promoter region was also amplified by real-time PCR (quantitative), using the same primers, on a Roche LightCycler 480 real-time PCR machine.
Construction of PIK3CD promoter construct, transient transfections, and luciferase assays
The 5′-upstream sequence of PIK3CD gene (positions −2005 to +92, relative to the transcription start site based on PIK3CD mRNA sequence NM_005026) was PCR amplified with the use of forward (5′-ATTGACTGGCTCACCCTTCACCCTATCC-3′) and reverse (5′-TCCTCTTCCCACCTCCCCTGACG-3′) primers. The amplicons were cloned into a TA cloning vector, pCRIITOPO. After confirming identity of the gene sequence by automated DNA sequencing, a sense clone was then digested with BamHI and XhoI. The resulting smaller DNA fragment was subcloned into pGL3Basic with the use of the same restriction sites to generate pGL3Basic-PIK3CDpro reporter gene construct.
To determine whether RUNX1b transactivates the PIK3CD promoter, KA1b cells (5 × 106) were transfected by electroporation (200 V, 950 μF) with the pGL3Basic-PIK3CDpro (10 μg) and pRLSV40 (2 μg). Cells were split into 2 aliquots and resuspended into 5 mL complete media in a 6-well plate. Dox was added to 1 of the 2 cell aliquots to a final concentration of 2 μg/mL. Cells were harvested, and lysates were prepared 24 hours after transfection. Firefly luciferase activities were assayed with a Dual-Luciferase Reporter Assay System (Promega) in a Turner TD2420 luminometer, and results were normalized to Renilla luciferase activities. The PIK3CD promoter activities in Dox-induced KA1b cells are presented as mean fold changes relative to uninduced control cells from at least 3 independent experiments.
shRNA knockdown of AML1 in Meg-01 cells
AML1 shRNA lentivirus clones were purchased from the RNAi Consortium (Sigma-Aldrich). Meg-01 cells were infected by shRNA lentivirus clones. After selection with puromycin, infected Meg-01 cells were plated in soft agar. Colonies were isolated, expanded, and tested for RUNX1 expression by Western blotting and real-time RT-PCR. Two colonies (designated Meg-01 RUNX1 1-1 and RUNX1 1-2, respectively) were selected for loss of RUNX1 gene expression and used for further study. A pool of cells from the negative control transduction was used as the control (designated Meg-01 negative).
Assessment of baseline apoptosis
The Meg-01 RUNX1 shRNA stable clones (negative, RUNX1 1-1, and RUNX1 1-2) in logarithmic growth phase in RPMI 1640 plus 10% FBS were vigorously pipetted, and triplicate samples were taken for baseline apoptosis assay with the use of the Apoptosis Annexin-V FITC/PI Kit (Beckman Coulter). Briefly, 50 μL resuspended cells were transferred to clean 12 × 75 culture tubes containing 50 μL Annexin-V FITC/PI reagent in 1× binding buffer and incubated in the dark for 15 minutes. At the end of incubation, an additional 0.4 mL of 1× binding buffer was added to each tube, vortexed, and analyzed with the use of a Coulter XL Flow Cytometer (Coulter) equipped with an Argon laser. Cells were gated to include the main viable cell population based on forward scatter (FS)/side scatter (SS) characteristics. Cellular events with low FS/high SS character were not included in the gate because these represent a mixture of overtly necrotic cells and apoptotic bodies that can artifactually elevate apoptosis levels. Apoptotic events from this viable cell gate were recorded as a combination of the Annexin-V+/PI− (early apoptotic) and Annexin-V+/PI+ (late apoptotic/dead) events.
Statistical analysis
Differences in transcript levels between different AML patient groups were compared with the nonparametric Mann-Whitney 2-sample test. Differences in basal apoptosis between the RUNX1 shRNA clones and the negative control, and differences in the ara-C IC50 values between LY294002 treated and untreated cells or between Meg-01–negative and RUNX1 1-1 or RUNX1 1-2 stable clones were compared with paired t tests. The nonparametric Spearman rank correlation coefficient was used to analyze the relation between RUNX1b and PIK3CD transcript levels. Statistical analyses were performed with GraphPad Prism 4.0 (GraphPad Software Inc).
Results
Decreased expression of RUNX1b in DS AMkL compared with non-DS AMkL cases
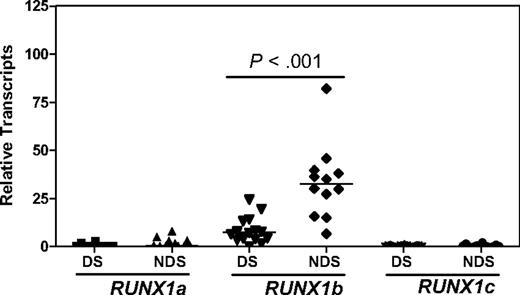
It is predicted that RUNX1 transcript levels in DS AMkLs would be 1.5-fold higher than those in non-DS AMkLs because of the gene dosage effect of constitutional trisomy 21 in DS cells. However, transcript levels for RUNX1b and RUNX1c, the 2 larger transcript isoforms, were found to be significantly lower in DS AMkL cases than that in non-DS AMkL cases.19 In our cohort of DS and non-DS AMkLs, RUNX1b transcripts quantitated by real-time RT-PCR represented the major transcript isoform (> 90% of the total RUNX1 transcripts, including RUNX1a, 1b, and 1c) and were significantly lower in 16 DS AMkL cases than those in 12 non-DS AMkL cases (4.4-fold; P < .001; Figure 1). These results suggest that the differential expression of RUNX1 between non-DS and DS AMkLs could also contribute to the marked differences in EFS rates between these groups,20-26 in addition to its critical role in leukemogenesis.9,10
Real-time RT-PCR quantification of RUNX1 transcripts in megakaryoblasts from children with and children without DS newly diagnosed with AMkL. Transcript levels for RUNX1a, RUNX1b/1c, and RUNX1c were measured by real-time RT-PCR and normalized to 18S rRNA levels. RUNX1b transcript levels were calculated by subtracting RUNX1c transcript levels from the total transcript levels for RUNX1b/1c. The horizontal lines indicate median RUNX1 transcript levels in each group of patient samples. The P value was determined by the nonparametric Mann-Whitney U test.
Real-time RT-PCR quantification of RUNX1 transcripts in megakaryoblasts from children with and children without DS newly diagnosed with AMkL. Transcript levels for RUNX1a, RUNX1b/1c, and RUNX1c were measured by real-time RT-PCR and normalized to 18S rRNA levels. RUNX1b transcript levels were calculated by subtracting RUNX1c transcript levels from the total transcript levels for RUNX1b/1c. The horizontal lines indicate median RUNX1 transcript levels in each group of patient samples. The P value was determined by the nonparametric Mann-Whitney U test.
shRNA knockdown of RUNX1 in a non-DS AMkL cell line Meg-01 increased sensitivity to ara-C
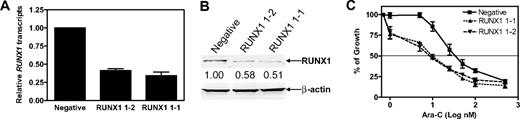
To investigate the role of RUNX1 in chemotherapy responses in AMkL, shRNA knockdown of RUNX1 by lentivirus AML1 shRNA clones was performed in an AMkL cell line, Meg-01 (Figure 2A-B). Down-regulation of RUNX1 by shRNA knockdown in RUNX1 1-1 and RUNX1 1-2 clones was consistently accompanied by significantly increased ara-C sensitivities (Figure 2C). Thus, shRNA silencing of RUNX1 resulted in 3.4- and 3.8-fold increased ara-C sensitivities in RUNX1 1-1 (mean IC50, 8.4 ± 0.3 nM) and RUNX1 1-2 (mean IC50, 7.4 ± 1.4 nM), respectively, compared with that in negative cells (mean IC50, 28.4 ± 3.3 nM; P < .03).
shRNA knockdown of RUNX1 in Meg-01 cells resulted in increased sensitivity to ara-C. (A-B) Meg-01 cells were infected by RUNX1 shRNA lentivirus clones. After selection with puromycin, infected Meg-01 cells were plated in soft agar. Colonies were isolated, expanded, and tested for RUNX1 expression by real-time RT-PCR (A) and Western blotting (B). Two colonies (designated RUNX1 1-1 and RUNX1 1-2) with decreased RUNX1 gene expression were selected as candidates for further study. A pool of cells from the negative control transduction was used as the control (designated negative). (C) The RUNX1 1-1, RUNX1 1-2, and negative cells were cultured in complete medium with dialyzed FBS in 96-well plates at a density of 4 × 104 cells/mL. Cells were cultured continuously with a range of concentrations of ara-C at 37°C, and cell numbers were determined with the Cell Titer–blue reagent and a fluorescence microplate reader. The data represent mean values ± SEs from at least 3 independent experiments.
shRNA knockdown of RUNX1 in Meg-01 cells resulted in increased sensitivity to ara-C. (A-B) Meg-01 cells were infected by RUNX1 shRNA lentivirus clones. After selection with puromycin, infected Meg-01 cells were plated in soft agar. Colonies were isolated, expanded, and tested for RUNX1 expression by real-time RT-PCR (A) and Western blotting (B). Two colonies (designated RUNX1 1-1 and RUNX1 1-2) with decreased RUNX1 gene expression were selected as candidates for further study. A pool of cells from the negative control transduction was used as the control (designated negative). (C) The RUNX1 1-1, RUNX1 1-2, and negative cells were cultured in complete medium with dialyzed FBS in 96-well plates at a density of 4 × 104 cells/mL. Cells were cultured continuously with a range of concentrations of ara-C at 37°C, and cell numbers were determined with the Cell Titer–blue reagent and a fluorescence microplate reader. The data represent mean values ± SEs from at least 3 independent experiments.
Identification of RUNX1 responsive genes in Meg-01–negative and RUNX1 1-1 cells by oligonucleotide microarray analysis
To determine the mechanisms that contribute to increased ara-C sensitivity accompanying RUNX1 down-regulation in Meg-01 cells, oligonucleotide microarray analysis was performed on 2 independent pairs of total RNAs prepared from Meg-01–negative and RUNX1 1-1 cells. Average log ratios representing the difference in expression were derived for each array feature by combining the replicate array data, using the error-weighted algorithm of Rosetta Resolver.35 Differentially expressed genes were identified by their P values, calculated with the Resolver error-model and the replicate data. At a P value of .005 or less, 7374 differentially expressed features (probes) were identified by combining all 4 arrays with a false discovery rate of 2.78% (supplemental Table 2). Among the 7374 features, genes related to apoptosis (supplemental Table 3), cell proliferation (supplemental Table 4), cell differentiation (supplemental Table 5), and immune response (supplemental Table 6) were identified. When the 7374 differentially expressed probes were cross referenced with the 551 genes differentially expressed between primary DS and non-DS AMkL cases from our previous study,27 351 probes, including PIK3CD (3.4-fold higher in non-DS AMkL cases than that in DS AMkL cases), were identified (Figure 3A; supplemental Table 7).
Overlapping genes between the oligonucleotide microarray dataset derived from Meg-01 shRNA stable clones and the Affymetrix microarray dataset derived from primary DS and non-DS AMkL patient blasts. (A) Differentially expressed genes identified by oligonucleotide microarray from Meg-01–negative and RUNX1 1-1 cells (left circle) were cross referenced, by comparing GenBank accession numbers, with genes differentially expressed between primary DS and non-DS AMkL patient specimens identified by Affymetrix microarray (right circle) in our previous study.27 A total of 351 probes (supplemental Table 7) were identified in both microarray datasets (overlapping area), including PIK3CD. (B) Transcript levels for PIK3CD were quantitated by real-time RT-PCR in the Meg-01 shRNA stable clones to validate the oligonucleotide microarray data. Real-time PCR results were expressed as mean values from 3 independent experiments with the same cDNA preparation and normalized to 18S rRNA. **Statistically significant differences (P < .005). Error bars indicate SEs. (C-D) PIK3CD transcript levels in primary DS and non-DS AMkL blasts were quantitated by real-time RT-PCR. Median PIK3CD transcript levels were compared between the 2 patient groups with the use of the nonparametric Mann-Whitney U test (C), and their relation to RUNX1b transcript levels was determined by the nonparametric Spearman rank correlation coefficient (D).
Overlapping genes between the oligonucleotide microarray dataset derived from Meg-01 shRNA stable clones and the Affymetrix microarray dataset derived from primary DS and non-DS AMkL patient blasts. (A) Differentially expressed genes identified by oligonucleotide microarray from Meg-01–negative and RUNX1 1-1 cells (left circle) were cross referenced, by comparing GenBank accession numbers, with genes differentially expressed between primary DS and non-DS AMkL patient specimens identified by Affymetrix microarray (right circle) in our previous study.27 A total of 351 probes (supplemental Table 7) were identified in both microarray datasets (overlapping area), including PIK3CD. (B) Transcript levels for PIK3CD were quantitated by real-time RT-PCR in the Meg-01 shRNA stable clones to validate the oligonucleotide microarray data. Real-time PCR results were expressed as mean values from 3 independent experiments with the same cDNA preparation and normalized to 18S rRNA. **Statistically significant differences (P < .005). Error bars indicate SEs. (C-D) PIK3CD transcript levels in primary DS and non-DS AMkL blasts were quantitated by real-time RT-PCR. Median PIK3CD transcript levels were compared between the 2 patient groups with the use of the nonparametric Mann-Whitney U test (C), and their relation to RUNX1b transcript levels was determined by the nonparametric Spearman rank correlation coefficient (D).
PIK3CD encodes the δ catalytic subunit (p110δ) of PI3-kinase, one of the class IA PI3-kinase catalytic subunits (p110α, p110β, and p110δ).36 A previous report showed that the p110δ isoform of PI3-kinase is consistently expressed at a high level in blast cells from AML.37 However, its regulation in myeloblasts has not been established. Our microarray studies with the use of both engineered cell models and primary megakaryoblasts suggest that PIK3CD could be a RUNX1 target in AMkL. To validate the oligonucleotide microarray data derived from the Meg-01 shRNA stable clones, PIK3CD transcript levels were quantitated by real-time RT-PCR in the clones. As shown in Figure 3B, PIK3CD transcript levels were 36% and 75%, respectively, decreased in the RUNX1 1-2 and RUNX1 1-1 cells compared with that in the negative cells (P < .005). Further, expression of PIK3CD generally paralleled changes in RUNX1 transcript and protein levels shown in Figure 2 (Figure 3B). These results confirm the PIK3CD gene expression pattern determined by oligonucleotide microarray and suggest that PIK3CD could be a RUNX1 target.
To assess the relation between RUNX1 and PIK3CD in AMkL, transcript levels for PIK3CD were quantitated by real-time RT-PCR in primary AMkL specimens from both children with DS and children without DS children and correlated with that for RUNX1b, the major RUNX1 transcript isoform in AMkL cells. As shown in Figure 3C, PIK3CD transcript levels were significantly higher in non-DS AMkL than that in DS AMkL (P = .02) cells. Further, transcript levels for PIK3CD significantly correlated with that for RUNX1b over a 328-fold range (r = 0.49, P = .008; Figure 3D).
PIK3CD is a direct AML1 target in Meg-01 cells
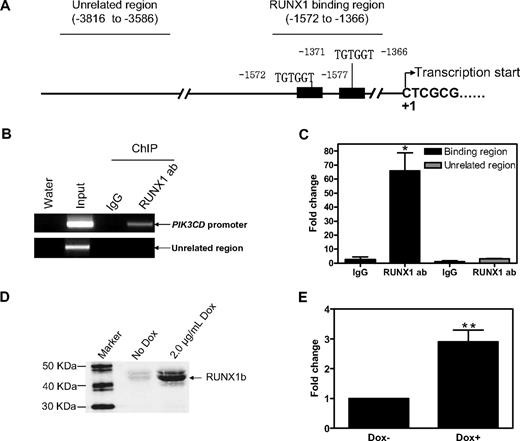
It was important to establish whether PIK3CD is a direct RUNX1 target. There are 2 putative RUNX1 binding sites (TGTGGT) located at positions −1572 to −1577 and −1371 to −1366 (relative to the transcription start site, based on PIK3CD mRNA sequence NM_005026; Figure 4A). In Meg-01 cells, RUNX1 protein directly binds to the RUNX1 binding sites of PIK3CD in vivo when assayed by ChIP with either regular PCR (Figure 4B) or real-time PCR (Figure 4C). To assess transactivation of the PIK3CD promoter by RUNX1b, the pGL3Basic-PIK3CDpro reporter gene construct was transfected into KA1b cells along with pRLSV40 by electroporation. Induction of RUNX1b by Dox in KA1b cells (Figure 4D) transfected with pGL3Basic-PIK3CDpro resulted in 3-fold increased PIK3CD promoter activity compared with uninduced cells (Figure 4E). Thus, our results from oligonucleotide microarray, real-time RT-PCR, ChIP assays, and reporter gene assays show that PIK3CD is indeed a direct RUNX1 target in Meg-01 cells.
PIK3CD is a bona fide RUNX1 target gene in AMkL. (A-C) In vivo binding of RUNX1 to the putative RUNX1 binding sites located in the upstream region of the PIK3CD gene (A) in Meg-01 cells was determined by ChIP assays with the use of regular PCR (B) and real-time PCR (C), as described in “Methods.” (D-E) KA1b cells were transfected with pGL3Basic-PIK3CDpro along with pRLSV40 by electroporation. The transfected cells were split into 2 equal aliquots with 1 induced by 2 μg/mL Dox. Cells were harvested 24 hours after transfection, and induction of RUNX1b was determined by Western blotting (D) and luciferase activities were assayed (E) as described in “Methods.” **Statistically significant differences (P < .005). (C and E) Error bars indicate SEs.
PIK3CD is a bona fide RUNX1 target gene in AMkL. (A-C) In vivo binding of RUNX1 to the putative RUNX1 binding sites located in the upstream region of the PIK3CD gene (A) in Meg-01 cells was determined by ChIP assays with the use of regular PCR (B) and real-time PCR (C), as described in “Methods.” (D-E) KA1b cells were transfected with pGL3Basic-PIK3CDpro along with pRLSV40 by electroporation. The transfected cells were split into 2 equal aliquots with 1 induced by 2 μg/mL Dox. Cells were harvested 24 hours after transfection, and induction of RUNX1b was determined by Western blotting (D) and luciferase activities were assayed (E) as described in “Methods.” **Statistically significant differences (P < .005). (C and E) Error bars indicate SEs.
Effect of RUNX1 on Akt phosphorylation and basal apoptosis in AMkL
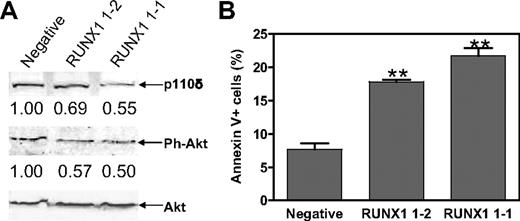
Akt is a downstream target of PI3-kinase. It seems likely that RUNX1 could modulate Akt phosphorylation by regulating PIK3CD. To test this hypothesis, phosphorylation status of Akt in the Meg-01 RUNX1 shRNA stable clones was determined with an antibody against phosphoSer473 of Akt. As predicted from the loss of PIK3CD transcripts, shRNA silencing of RUNX1 resulted in 45% and 31% decreased p110δ protein levels in RUNX1 1-1 and RUNX1 1-2 stable clones, respectively, compared with that in the Meg-01–negative stable clone (Figure 5A top). This was accompanied by decreased phosphorylation of Akt in the RUNX1 1-1 and RUNX1 1-2 shRNA stable clones (50% and 43%, respectively) (Figure 5A middle). However, shRNA knockdown of RUNX1 did not affect total Akt protein levels (Figure 5A bottom).
Functional impact of RUNX1 on PI3-kinase/Akt signaling and cell apoptosis in Meg-01 cells. (A) The RUNX1 1-1, RUNX1 1-2, and negative stable clones were lysed in PBS buffer containing both protease and phosphatase inhibitors. Soluble proteins were analyzed by Western blots with anti-p110δ, anti–ph-Akt, and anti-Akt antibodies. Intensity of each band was quantified with the use of the Odyssey software. Protein levels for p110δ and Ph-Akt in RUNX1 1-1 and RUNX1 1-2 cells are presented relative to that in negative cells after normalization to levels of total Akt. (B) Baseline apoptosis in the RUNX1 1-1, RUNX1 1-2, and negative stable clones was determined by flow cytometry with annexin V-fluorescein isothiocyanate/propidium iodide staining. **Statistically significant differences (P < .005). Error bars indicate SEs.
Functional impact of RUNX1 on PI3-kinase/Akt signaling and cell apoptosis in Meg-01 cells. (A) The RUNX1 1-1, RUNX1 1-2, and negative stable clones were lysed in PBS buffer containing both protease and phosphatase inhibitors. Soluble proteins were analyzed by Western blots with anti-p110δ, anti–ph-Akt, and anti-Akt antibodies. Intensity of each band was quantified with the use of the Odyssey software. Protein levels for p110δ and Ph-Akt in RUNX1 1-1 and RUNX1 1-2 cells are presented relative to that in negative cells after normalization to levels of total Akt. (B) Baseline apoptosis in the RUNX1 1-1, RUNX1 1-2, and negative stable clones was determined by flow cytometry with annexin V-fluorescein isothiocyanate/propidium iodide staining. **Statistically significant differences (P < .005). Error bars indicate SEs.
Activation of the PI3-kinase/Akt pathway has an antiapoptotic effect.36 Thus, shRNA silencing of RUNX1 would be expected to result in increased basal apoptosis in the RUNX1 shRNA clones compared with the shRNA-negative control cells. As shown in Figure 5B, shRNA knockdown of RUNX1 in Meg-01 cells resulted in significantly increased (> 2-fold; P < .005) basal apoptosis, determined by flow cytometric analyses, compared with the negative control cells (Figure 5B).
Inhibition of PI3-kinase enhances the cytotoxic effect of ara-C in Meg-01 and primary pediatric AMkL cells
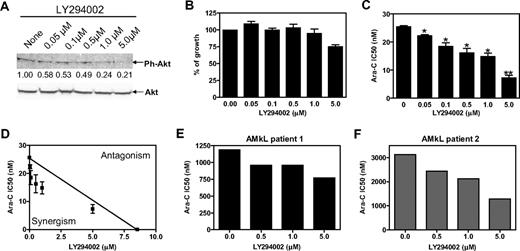
PI3-kinase/Akt signaling contributes to AML cell survival, and its constitutive activation can reduce the effectiveness of cytotoxic agents.36 Conversely, blockade of PI3-kinase/Akt pathway may enhance the effectiveness of cytotoxic agents. Incubation of Meg-01 cells with various concentrations (0-5 μM) of LY294002, a generic PI3-kinase inhibitor, resulted in decreased phosphorylation of Akt. This was especially obvious with higher doses of LY294002 (1.0 and 5.0 μM; Figure 6A). However, these latter doses of LY294002 showed modest cytotoxic effects in the absence of ara-C (Figure 6B). Interestingly, when LY294002 was combined with ara-C, it significantly enhanced ara-C cytotoxicities in a dose-dependent manner (Figure 6C). The interaction between LY294002 and ara-C resulting in antileukemia effects was clearly synergistic when determined by standard isobologram analysis (Figure 6D). Synergism was further assessed by CI values, for which a CI less than 1 (indicative of synergism) was found in all the combinations of LY294002 and ara-C tested. The greatest synergism was seen for 0.5 μM LY294002 (CI = 0.69; Table 1).
Synergistic antileukemia activity between the PI3-kinase inhibitor, LY294002, and ara-C in Meg-01 cells and primary blasts from children without DS with AMkL. (A) Meg-01 cells were harvested and lysed after incubation with a range of concentrations of the nonselective PI3-kinase inhibitor, LY294002, for 48 hours. Soluble proteins were subjected to Western blots probed with anti-Akt and anti–ph-Akt antibodies. (B) Meg-01 cells were continuously cultured in 96-well plates for 96 hours in dialyzed FBS in the presence of various concentrations of LY294002. Cell numbers were determined by the Cell Titer–blue reagents, as described in “Methods.” (C) Ara-C IC50 values were determined in Meg-01 cells in the presence of different concentrations of LY294002. *Statistically significant differences (P < .05); **statistically significant differences (P < .005). (B-C) Error bars indicate SEs. (D) Standard isobologram analysis of Meg-01 cell growth inhibition by LY294002 and ara-C in combination. The IC50 values of each drug are plotted; the solid line represents the additive effect, whereas the points represent the concentrations of each drug resulting in 50% growth inhibition. (E-F) Ara-C IC50 values were determined in the presence of a range of concentrations of LY294002 in 2 primary AMkL blasts from 2 children newly diagnosed with de novo AMkL by MTT assay as described in “Methods.”
Synergistic antileukemia activity between the PI3-kinase inhibitor, LY294002, and ara-C in Meg-01 cells and primary blasts from children without DS with AMkL. (A) Meg-01 cells were harvested and lysed after incubation with a range of concentrations of the nonselective PI3-kinase inhibitor, LY294002, for 48 hours. Soluble proteins were subjected to Western blots probed with anti-Akt and anti–ph-Akt antibodies. (B) Meg-01 cells were continuously cultured in 96-well plates for 96 hours in dialyzed FBS in the presence of various concentrations of LY294002. Cell numbers were determined by the Cell Titer–blue reagents, as described in “Methods.” (C) Ara-C IC50 values were determined in Meg-01 cells in the presence of different concentrations of LY294002. *Statistically significant differences (P < .05); **statistically significant differences (P < .005). (B-C) Error bars indicate SEs. (D) Standard isobologram analysis of Meg-01 cell growth inhibition by LY294002 and ara-C in combination. The IC50 values of each drug are plotted; the solid line represents the additive effect, whereas the points represent the concentrations of each drug resulting in 50% growth inhibition. (E-F) Ara-C IC50 values were determined in the presence of a range of concentrations of LY294002 in 2 primary AMkL blasts from 2 children newly diagnosed with de novo AMkL by MTT assay as described in “Methods.”
Similar results were obtained when AMkL blasts collected at diagnosis from 2 children with de novo AMkL were evaluated after cotreatment of ara-C with a range of concentrations of LY294002 (0.5-5.0 μM). In both primary AMkL specimens LY294002 enhanced ara-C cytotoxicity in a dose-dependent manner (Figure 6E-F). Interactions between ara-C and LY294002 were synergistic to additive in both primary AMkL specimens by isobologram analyses (data not shown) and CI values (Table 1).
Discussion
AML accounts for one-fourth of the acute leukemias in children, but it is responsible for more than one-half of the leukemic deaths in this patient population.38 In contrast to the significant success in the treatment of pediatric ALL in the past 3 decades,39 improvements in AML therapy have been limited, such that only approximately 50% of children with AML are cured.38,40 The development of new treatment strategies for childhood AML must be based on an improved understanding of the molecular mechanisms underlying chemotherapy drug resistance.
AMkL (M7) is a biologically heterogeneous form of AML, representing approximately 10% of pediatric AML cases and 1% to 2% of adult AML cases.41-45 AMkL is the most common AML subtype in children with DS.46 Children with DS with AMkL have an excellent prognosis with EFS rates of 80% to 100% when treated with ara-C/anthracycline–based protocols,20-26 in contrast to the less than 30% EFS rates of patients without DS with AMkL.45,47 This also contrasts to the approximately 50% EFS rates of children without DS with AML overall38,40 and indicates that AMkL is an extremely poor risk group among patients without DS with AML despite the use of intensive chemotherapy-based protocols.
Previous studies from our group and others have shown that DS AMkL blasts were significantly more sensitive to a panel of chemotherapy drugs with different mechanisms of action than were non-DS AML cells (including AMkL blasts).27,48,49 This suggests that a more global mechanism of chemotherapy sensitivity and/or pathway (eg, apoptosis) may account for the differences between the DS and non-DS groups. Indeed, microarray studies from our own group and others have shown significantly decreased expression of several antiapoptotic genes (such as Bcl2, Hsp70, PIK3CD) in DS AMkL compared with non-DS AMkL.19,27 However, the mechanisms responsible for the differential expression of these genes between the DS and non-DS AMkL groups have not been established.
An interesting finding was that expression of the chromosome 21–encoded transcription factor RUNX1 was decreased in DS AMkL compared with non-DS AMkL, despite the predicted gene dosage effect of trisomy 21 in DS blasts.19 We hypothesized that elevated expression of RUNX1 may contribute to the increased chemotherapy resistance in non-DS AMkL by up-regulating antiapoptotic genes, such as Bcl-2.50 In the non-DS AMkL cell line, Meg-01, shRNA knockdown of RUNX1 was accompanied by significantly increased sensitivity to ara-C and significantly decreased expression of PIK3CD, which encodes the δ catalytic subunit of PI3-kinase (p110δ),36 determined by oligonucleotide microarray and real-time RT-PCR analyses. Higher expression of PIK3CD in non-DS AMkL patient specimens than that in DS AMkL patient blasts was also detected by microarray analysis in our previous study.27 These results strongly support our hypothesis that RUNX1 may directly regulate PIK3CD in AMkL. In vivo binding of RUNX1 to the 2 putative RUNX1 binding sites located in the upstream region of PIK3CD gene and direct transactivation of PIK3CD promoter by RUNX1b were established by ChIP and reporter gene assays, respectively. Finally, PIK3CD transcript levels were significantly higher in non-DS AMkL patient specimens than those in DS AMkL patient specimens and significantly correlated with RUNX1b transcript levels. Collectively, these results confirm that PIK3CD is a bona fide RUNX1 target in AMkL and that the differential expression of RUNX1 between DS and non-DS AMkL is responsible, at least partly, for the differential expression of PIK3CD between these groups of patients.
The PI3-kinase/Akt signaling network is critical to widely divergent physiologic processes, including cell-cycle progression, differentiation, transcription, translation, and apoptosis.36 It is clear now that up-regulation of PI3-kinase will result in activation of its downstream target, Akt (also known as protein kinase B), leading to increased cell survival and proliferation.36 We found that shRNA knockdown of RUNX1 in Meg-01 cells resulted in decreased p110δ protein level and a corresponding decrease in Akt phosphorylation. This was accompanied by significantly increased basal level apoptosis and sensitivity to ara-C in the cells. Alternately, inhibition of PI3-kinase by compound LY294002 resulted in significantly increased ara-C sensitivity in Meg-01 cells. Analogous results were also obtained in 2 primary AMkL specimens from children newly diagnosed with de novo AMkL.
Several reports have highlighted that constitutive action of PI3-kinase/Akt signaling is a common feature of AML.36 From 50% to 70% of patients with AML display phosphorylation of both Thr308 and Ser473 Akt, and the overall survival time for patients harboring Akt activation was significantly shorter compared with patients with no Akt activation. The mechanisms underlying the up-regulation of PI3-kinase/Akt signaling in AML cells remain unclear, although point mutations of N-Ras or K-Ras, internal tandem duplication of the juxtamembrane domain of FLT3, and activating mutations in c-Kit have all been implicated to play important roles in the activation of PI3-kinase/Akt pathways.51 Our results have shown, for the first time, that RUNX1 probably contributes to chemotherapy resistance in non-DS AMkL, and it is responsible, at least partly, for the activation of PI3-kinase/Akt signaling in this group of patients. It would be difficult to identify specific inhibitors to target RUNX1 in AMkL because RUNX1 also plays very important roles in normal hematopoiesis. Therefore, targeting its downstream targets, such as PI3-kinase/Akt pathways, with specific inhibitors could be a good strategy to improve the treatment outcome of children without DS with AMkL.
It is important to note that Malinge et al52 recently reported a frequency of approximately 30% (2 of 7) of FLT3 D835Y mutations in DS AMkL but none in non-DS AMkL. It is possible that the PI3-kinase/Akt pathway is active in some DS AMkL cases because of FLT3 D835Y mutations. However, our results suggest that the decreased expression of PIK3CD in DS AMkL, at least partly because of decreased expression of RUNX1, may minimize the effects of FLT3 D835Y mutation on the activation of the PI3K-Akt pathway. Further studies are warranted to determine the potential effect of FLT3 D835Y mutation on chemotherapy response in DS AMkL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the Children's Oncology Group AML Biology Subcommittee for authorizing and providing the clinical specimens that made this study possible.
This work was supported by grants from the Karmanos Cancer Institute, the Children's Research Center of Michigan, Leukemia Research Life, the Children's Leukemia Foundation of Michigan, the National Cancer Institute (CA120772), the Leukemia & Lymphoma Society, BPCT Golf Charity, the Sehn Family Foundation, and the Dale Meyer Memorial Endowment for Leukemia Research and the Ring Screw Textron Endowed Chair for Pediatric Cancer Research (J.W.T.). Microarray data analysis was supported by the Microarray and Bioinformatics Facility Core, Wayne State University (National Institute of Environmental Health Science [NIEHS] Center grant P30 ES06639).
National Institutes of Health
Authorship
Contribution: H.E. performed the molecular biology study and wrote the paper; C.X. and K.M.L. performed the molecular biology study; A.A.D. performed the microarray analyses; S.A.B. performed the flow cytometric study; J.L.B. assisted Y.G. to design the study and performed some of the molecular biology study; J.W.T. assisted Y.G. to design the study and arrange clinical samples; L.H.M. assisted Y.G. to design the study, provided intellectual and technical support, and wrote the paper; and Y.G. designed the study and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yubin Ge, Developmental Therapeutics Program, Karmanos Cancer Institute, 110 E Warren Ave, Detroit, MI 48201; e-mail: gey@karmanos.org.
References
Author notes
*H.E. and C.X. contributed equally to this study.